Abstract
The Q gene encodes an AP2-like transcription factor that played an important role in domestication of polyploid wheat. The chromosome 5A Q alleles (5AQ and 5Aq) have been well studied, but much less is known about the q alleles on wheat homoeologous chromosomes 5B (5Bq) and 5D (5Dq). We investigated the organization, evolution, and function of the Q/q homoeoalleles in hexaploid wheat (Triticum aestivum L.). Q/q gene sequences are highly conserved within and among the A, B, and D genomes of hexaploid wheat, the A and B genomes of tetraploid wheat, and the A, S, and D genomes of the diploid progenitors, but the intergenic regions of the Q/q locus are highly divergent among homoeologous genomes. Duplication of the q gene 5.8 Mya was likely followed by selective loss of one of the copies from the A genome progenitor and the other copy from the B, D, and S genomes. A recent V329-to-I mutation in the A lineage is correlated with the Q phenotype. The 5Bq homoeoalleles became a pseudogene after allotetraploidization. Expression analysis indicated that the homoeoalleles are coregulated in a complex manner. Combined phenotypic and expression analysis indicated that, whereas 5AQ plays a major role in conferring domestication-related traits, 5Dq contributes directly and 5Bq indirectly to suppression of the speltoid phenotype. The evolution of the Q/q loci in polyploid wheat resulted in the hyperfunctionalization of 5AQ, pseudogenization of 5Bq, and subfunctionalization of 5Dq, all contributing to the domestication traits.
Keywords: fate of duplicated genes, hyperfunctionilization, subfunctionilization
Polyploidy is an important evolutionary feature widespread in the plant kingdom leading to whole-genome duplication (1–3), after which individual genes follow one of many possible evolutionary fates, including nonfunctionalization (deletion or pseudogenization), neofunctionalization, or subfunctionalization (4, 5), often modified by genetic and epigenetic interactions between homoeoalleles (1, 6). Various mechanisms affect regulation of gene expression and the fate of homoeologous genes, including altered or incompatible regulatory interactions, epigenetic modifications, gene dosage changes, partitioning, and compensation (6–11).
Wheat was among the first plant species to be domesticated and was instrumental in spawning the agricultural revolution and the establishment of human civilization. The widespread cultivation of wheat resulted from founder evolutionary events including the acquisition of domestication traits such as a nonbrittle rachis to prevent spikelet shattering and soft glumes and hull-less seed for ease of threshing, as well as allopolyploidization events (12) that resulted in species with better agronomic performance and wide adaptability.
The widely cultivated allohexaploid Triticum aestivum L. (2n = 6x = 42, AABBDD), known as common or bread wheat, originated as the result of two separate amphiploidization events (Fig. S1) (13–16). The tetraploid wheat Triticum turgidum L., (2n = 4x = 28, AABB), or pasta wheat, arose <0.5 million years ago as a result of hybridization between Triticum urartu Tumanian ex Gandylian (2n = 2x = 14, AA) and an unidentified diploid Aegilops species of the section Sitopsis, thought to be Aegilops speltoides Tausch (2n = 2x = 14, SS) or a close relative thereof, as the donor of the B genome (12, 13, 17–22). A spontaneous hybridization that occurred between a primitive domesticated tetraploid T. turgidum subspecies and the diploid goatgrass Aegilops tauschii Coss (2n = 2x = 14, DD) about 10,000 years ago gave rise to T. turgidum (14, 15).
The wheat Q gene, which arose through a spontaneous mutation in allopolyploid wheat, is arguably the most important domestication gene in cultivated wheat because it confers the free-threshing character (Fig. 1) and influences a number of other domestication-related traits such as rachis fragility, glume toughness, spike architecture, flowering time, plant height, and others (23, 24). The q allele in wild wheats confers a speltoid spike phenotype characterized by a lax head with elongated rachis and non–free-threshing seed (Fig. 1). The mutation that gave rise to Q had a profound effect on agriculture because it allowed early farmers to efficiently harvest their grain on a grand scale, one of the features that led to rapid spread of polyploid wheat cultivation around the world.
Fig. 1.
Comparison of effects of 5AQ on morphological and domestication-related traits of hexaploid wheat cv. CS spikes. (A) Presence of 5AQ confers a square-shaped spike (Left), and the absence of 5AQ (in the deletion mutant CS del143) (Right) results in a speltoid spike characterized by a much longer and spear-shaped spike compared with the spike of CS primarily due to the elongation of rachis segments. (B) Seed of CS with 5AQ (Left) is free threshing because the seed is loosely held and abscission occurs at the base of the glumes, leaving the rachis intact. CS lacking the 5AQ gene (Right) is not free threshing because the glumes hold the seed more tightly and abscission occurs within the rachis, causing disarticulation of the entire spikelet with a portion of the rachis left attached to the base of the spikelet. (C) Detailed illustration of a single spikelet of CS when 5AQ is present (Left) and absent (Right). Both spikelets have three fertile florets (first, second, and third) and each spikelet has a pair of glumes (1). Each floret has a lemma (2), seed (3), and palea (4). Note the looseness of the seeds due to the difference in the position of the glumes in 5AQ compared with the mutant lacking 5AQ. The abscission zones are indicated by the blue arrows (the lower spikelet in the 5AQ mutant was removed so that the abscission zone could be observed).
The Q gene, encoding a member of the AP2 family of transcription factors (23), resides on the long arm of wheat chromosome 5A (5AQ) (24, 25). Homoeologous q loci reside on chromosome arms 5BL (5Bq) and 5DL (5Dq). The Q allele mutation resulted in a single-amino-acid substitution changing properties of the transcription factor (23), probably affecting expression of multiple genes, thus explaining its pleiotropic nature. The free-threshing phenotype, however, is only expressed in polyploid wheat (26), indicating that genome composition, cross-talk between homoeoalleles, or other changes caused by polyploidization, are essential as well.
Here, we evaluate the structure, evolution, and function of the homoeologous Q/q loci to establish a molecular basis for their contribution to the domestication phenotype in the genetic background of diploid and allopolyploid wheat.
Results
Organization of the Q/q Gene Locus in Different Wheat Genomes and Species.
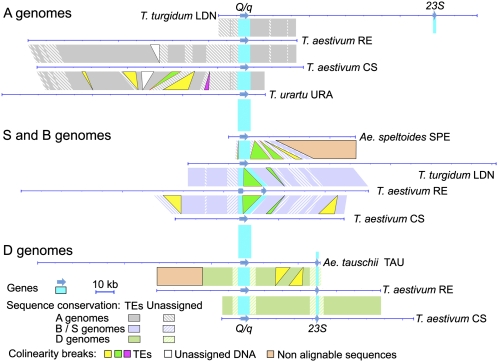
The sequences of 11 homologous and homoeologous genomic regions spanning the Q/q loci from five wheat species (A, B, S, and D genomes and three ploidy levels) were compared: the A-genome locus from the diploid T. urartu (URA) accession TMU138, the tetraploid T. turgidum cv. Langdon (LDN) and the hexaploid T. turgidum cvs. Renan (RE) and Chinese Spring (CS); the B-genome locus from LDN, RE, and CS; the S-genome locus from the diploid Ae. speltoides (SPE) accession SPELT1; and the D-genome locus from the diploid Ae. tauschii (TAU) accession AL8/78, as well as from RE and CS (Fig. 2 and Fig. S2).
Fig. 2.
Schematic presentation of the 11 sequenced homologous and homoeologous genomic regions, spanning the Q/q loci from five wheat species. Line diagrams illustrate sequenced DNA segments. Color-coded block diagrams illustrate results of pairwise sequence comparisons. Additional information is provided in Fig. S2.
Q/q was the only gene identified in the sequenced fragments of the B- and S-genome loci. A 23S ribosomal protein-like gene was identified 40 kb downstream of 5Dq in the D genomes and 100 kb downstream of 5Aq in the A genome of LDN. This gene is located 55 kb from the 5Aq gene in Triticum monococcum and 9 kb from the q ortholog in rice chromosome 3 (27).
Eighty percent of the sequenced genomic regions consist of nested transposable elements (TEs) interspersed with stretches of unassigned sequences (Fig. 2 and Fig. S2). TEs are different when comparing homoeologous loci, i.e., between A, B/S, and D loci, highlighting their dynamic nature. Most TEs are conserved among A, B, and D homologous loci where 19 large genome-specific indels and breaks in genome colinearity due to recent events such as transposon insertions were revealed (Fig. 2 and Fig. S2). TE annotation and insertion dates estimated for 52 retrotransposons with complete long-terminal repeats (LTRs) are presented in Fig. S2 and Table S1.
All Q/q homoeoalleles Are Transcriptionally Active, but 5Bq Is a Pseudogene That Does Not Encode a Full-Length q Protein.
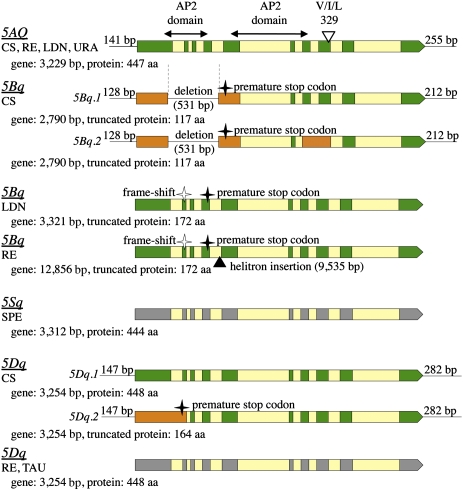
The intron/exon structure of the 5AQ gene (Fig. 3) was previously established (23). Sequences of the 5AQ coding regions of LDN and CS were identical and differed from the RE sequence by only two substitutions in introns. The coding sequence of URA 5Aq differed from 5AQ coding sequence of CS, RE, and LDN by five substitutions including two nonsynonymous substitutions, one being the V329I mutation (numbering as in 5AQ) proposed to be responsible for the phenotypic differences between 5AQ and 5Aq alleles (23).
Fig. 3.
An overview of the intron/exon structure of the Q/q homoeoalleles determined by RT-PCR cloning and sequencing. Exons conforming to the splicing pattern found for the 5AQ gene are shown in green (deduced from cDNA sequences) and gray (predicted), alternative exons are shown in orange, and introns are shown in yellow. 5′- and 3′-UTR lengths (L) by RACE are shown for CS mRNAs. Gene sizes shown are from the translation start to the stop codon. Exons encoding the AP2 domains and the position of the codon for amino acid residue 329 in 5AQ (I), and the corresponding codons in 5Aq (V) and 5Dq (L) are also shown.
For 5Bq, two transcript variants, 5Bq.1 and 5Bq.2, were identified for CS (Fig. 3). 5Bq.1 contains only six exons, corresponding to exons 1 and 5–10 of 5AQ. The first exon ends 45 bp upstream and then continues from 79 bp upstream of the corresponding 3′- and 5′-splice sites of exons 1 and 5 of 5AQ, respectively, due to a 531-bp deletion including sequences corresponding to exons 2–4 as well as parts of exon 1 and intron 4 of 5AQ. 5Bq.2 has the same structure as 5Bq.1, except that the sequence corresponding to intron 7 of 5AQ is not spliced out. 5Bq.1 and 5Bq.2 encode truncated 354-bp ORFs with a stop codon located at a position equivalent to nucleotides 63–61 upstream of the corresponding 5′-splice site of 5AQ exon 5.
The 531-bp deletion was not found in 5Bq of LDN and RE, but a frameshift caused by a 2-bp deletion in predicted exon 2 leads to a stop codon 69 bp from the beginning of predicted exon 4 and a 519-bp truncated ORF (Fig. 3). The 2-bp deletion is not found in CS 5Bq because of the larger 531-bp deletion. The 5Bq 2-bp deletion is present in hexaploid wheats and in most domesticated tetraploids, but it is much less prevalent among wild tetraploids (T. turgidum ssp. dicoccoides), suggesting that it occurred after tetraploidization and was perhaps selected upon domestication. The 531-bp deletion was only found in some hexaploid wheat lines, suggesting that it occurred after allohexaploidization.
For LDN, one 5Bq transcript type with the 5AQ splicing pattern was confirmed (Fig. 3). For RE, five different transcript types were detected by RT-PCR cloning and sequencing. These transcripts include two to four additional exons spliced out of a 9,535-bp helitron inserted two bp upstream of the 5′-splice site of the predicted exon 5 of the 5Bq gene (Fig. 3). The entire helitron sequence was presumably transcribed. Some 5Bq transcripts from RE lacked the canonical exon 5 or used an alternative 3′ splicing site for intron 4. One transcript lacked the canonical exon 9. The RE sequence differed from CS by one synonymous substitution and they both differed from LDN by two more synonymous and one nonsynonymous substitutions, if the 5AQ splicing pattern was assumed and the frameshift-causing deletions were ignored.
5Bq of LDN, CS, and RE are transcriptionally active, but do not encode full-length functional q proteins. Therefore, 5Bq is a pseudogene in all three species.
The 9,536-bp helitron transposon in RE is the first complete helitron identified in wheat (more information is provided in Fig. S3). The helitron insertion was traced in the RE pedigree to Mironovskaya 808, a T. turgidum winter wheat cultivar of Russian origin. This insertion has not been found in any other hexaploid nor in any wild or domesticated tetraploid wheat line (Table S2), suggesting that it occurred only very recently.
Two transcript variants were identified for the CS 5Dq gene. 5Dq.1 results from splicing of 10 exons matching the splicing pattern of 5AQ, but for transcript 5Dq.2, retention of the first intron leads to a premature stop codon and a 495-bp truncated ORF (Fig. 3). The coding parts of 5Dq of CS and REN are identical and differ from the TAU sequence by only one synonymous substitution and three intron substitutions. 5Sq in SPE encodes a full-length q protein, on the basis of the splicing pattern of 5AQ (Fig. 3).
A q Gene Duplication in the Wheat Ancestral Genome Was Probably Followed by Differential Loss of One Copy.
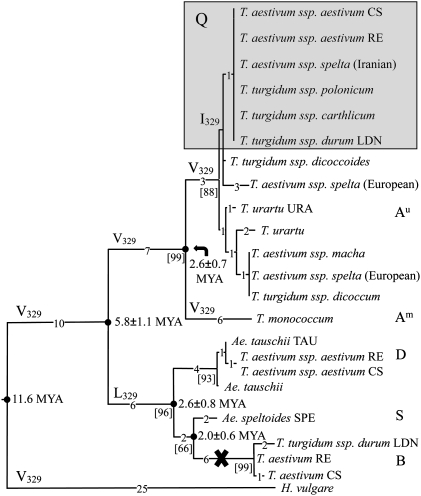
Full-length Triticeae Q/q coding sequences were used in the phylogenetic analysis (Fig. 4). For the 5Bq pseudogenes, the frameshifts caused by the deletions were adjusted to generate coding sequences corresponding to full-length ORFs found in other Q/q genes, even though such mRNAs and corresponding proteins are not made. The null hypothesis of equal evolutionary rate (third codon positions) throughout the tree was not rejected under any of several models tested. The molecular clock was calibrated using 11.6 million years ago (Mya) for the divergence time between barley and wheat (12).
Fig. 4.
Phylogenetic relationships and divergence times of the Q/q genes. The phylogenetic tree for synonymous substitutions was constructed by the neighbor-joining method. Branch lengths are shown by the number of substitutions. Bootstrap values (%) are shown in square brackets. The molecular clock was calibrated using 11.6 Mya for the time of divergence between barley and wheat (12). Species with the Q allele carrying I329 are shown on a gray background. Genomes are indicated by letters. The cross indicates that 5Bq is a pseudogene. Numbering of amino acid residues is as in 5AQ.
It has been shown previously that 5AQ and 5Aq alleles and corresponding phenotypes are correlated with a V329I mutation (23). This mutation occurred very recently and is present in the A genome of only some wheats (Fig. 4). V is the ancestral state at residue 329 and it mutated to L in the B/D/S q lineage.
The 2.6 ± 0.8 Mya divergence time between the D and B/S genomes (S genome appears to be more closely related to B than to D genome) (Fig. 4) is consistent with our previous estimates (12, 13, 28). The divergence time of 5Aq of T. monococcum from 5AQ/q of T. urartu and polyploid wheats was estimated at 2.6 ± 0.7 Mya, an earlier date than similar estimates for the Acc-1 gene (13). In the absence of any evidence of an accelerated substitution rate, an ancestral q gene duplication some 5.8 Mya (Fig. 4) followed by a selective loss of one of the copies in the A-genome progenitor and the other one in the progenitor of the B, D, and S genomes is the most plausible explanation of the early divergence of the A gene. Thus, the duplication occurred much earlier than the divergence of the homoeologous genomes (12, 13, 28).
In addition to the exon and intron sequences, a 2-kb segment at the 5′ ends and a 1-kb segment at the 3′ ends of the genes, including 5′- and 3′-UTRs, respectively, consist of blocks of sequence conserved in the A, B, D, and S genomes, defining the boundaries of the gene (Fig. 2). Sequence conservation between the A and D genomes extends by 2 kb upstream of Q and between the B, S, and D genomes by 2 kb downstream of the gene. Consistent with the phylogenetic relationships inferred from the coding sequences, divergence of 5Aq from 5Bq/5Dq/5Sq is higher than the divergence between B, D, and S homoeologs for all these sequences.
Pairwise comparisons of intron sequences concatenated with 12 kb of the sequence upstream of the start codon and 11 kb of the sequence downstream of the stop codon (Fig. S4) reveal haplotype variation with cross-over points within or near the Q/q gene, as indicated by the patterns of single-nucleotide substitutions for pairs of closely related A and B sequences (Fig. S4 A and B) and by different nucleotide substitution rates in gene-flanking regions for A and D sequences from different species (Fig. S4 A, Bottom, and C). The nucleotide substitution rate in the intergenic region is up to six times higher than either the intron substitution rate or the synonymous substitution rate due to the haplotype variation, consistent with observations for the wheat ACC loci (12).
Expression Interaction of Q/q Genes in Wild Type and Mutant Hexaploid Wheat Lines.
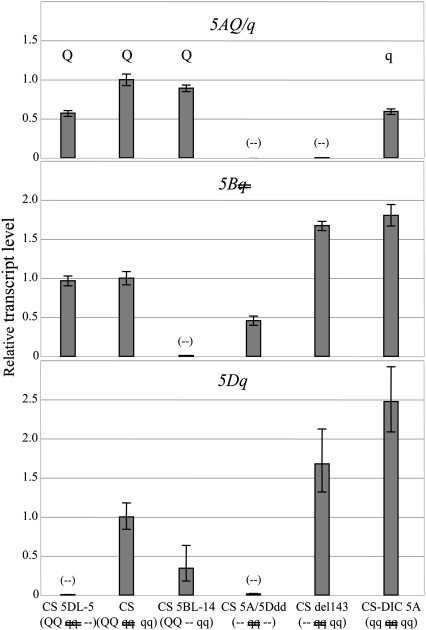
Expression of the Q/q homoeologs in hexaploid wheat was assessed in a series of lines altered at one or two Q/q loci as shown in Fig. S5. Lack of significant RQ-PCR signal for CS-del143, CS 5BL-14, and CS 5DL-5 with 5AQ-, 5Bq-, and 5Dq primers, respectively, confirmed specificity of the mRNA measurements (Fig. 5). Deletion of 5AQ (CS-del143) or its replacement with 5Aq (CS-DIC 5A) resulted in increased transcription of 5Bq and 5Dq relative to CS (Fig. 5), suggesting a repressing activity of the 5AQ allele. Deletion of 5Dq (CS 5DL-5) had no effect on the 5Bq transcript level, but lowered the level of the 5AQ transcript. Deletion of the 5Bq pseudogene (CS 5BL-14) resulted in a lower level of 5Dq transcript and a small reduction of the 5AQ transcript level (Fig. 5). Finally, in the 5AQ and 5Dq double-deletion line (CS-5A/5Ddd), in which no functional Q or q protein is made, the 5Bq transcript level was lower than in CS as well as in CS Fndel-143. In both cases this is a different outcome than in the corresponding single-deletion lines.
Fig. 5.
Steady-state level of 5AQ/q, 5Bq, and 5Dq transcripts in mutant lines relative to the wild-type wheat CS. The Q/q genotype of wheat lines is shown in parentheses (AABBDD), where  indicates the q pseudogene and – indicates a chromosomal deletion encompassing the Q/q locus.
indicates the q pseudogene and – indicates a chromosomal deletion encompassing the Q/q locus.
Role of Q/q in Controlling Morphology and Domestication Traits of Hexaploid Wheat.
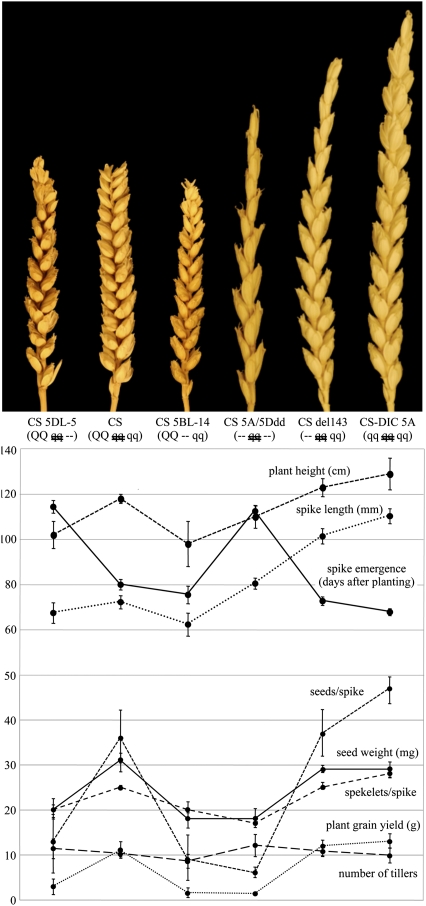
Correlation between Q/q loci and various traits revealed pleiotropic effects of 5AQ influencing glume toughness, threshability, rachis fragility, spike length and shape, spike emergence time, and plant height (Fig. 6 and Table S3). Deletion of 5AQ (CS-del143) or replacement with 5Aq (CS-DIC 5A) had similar effects on all traits compared with CS. CS has soft, papery glumes and free-threshing seed, whereas CS-DIC 5A and CS-del143 have tougher, thicker, and more heavily keeled glumes tightly enveloping the seed that was not free threshing. CS-DIC 5A and CS-del143 plants are taller and have longer, lax, and tapering (speltoid) spikes compared with CS (Fig. 6 and Table S3). Spike emergence is 7 d earlier in CS-del143 and 12 d earlier in CS-DIC 5A than CS, which is consistent with the previously identified role of 5AQ in delaying heading time (23). CS-DIC 5A has more spikelets and seeds per spike than CS-del143 and CS, but grain yield per plant is similar for all three due to more tillers in CS and CS-del143.
Fig. 6.
Spike morphology and domestication traits of genetic stocks with various combinations of Q/q homoeoalleles and chromosomal deletions encompassing the Q/q locus. Q/q genotype of wheat lines is shown in parentheses (AABBDD), where  indicates q pseudogene and – indicates a chromosomal deletion encompassing the Q/q locus.
indicates q pseudogene and – indicates a chromosomal deletion encompassing the Q/q locus.
The spikes of CS 5BL-14 (5Bq deletion) and CS 5DL-5 (5Dq deletion) showed slightly more speltoid phenotypes compared with CS, primarily due to reduced spikelet density in the lower portions of the spikes (Fig. 6). The shape of the upper portion of the spikes of CS 5BL-14 and CS 5DL-5 was intermediate between the speltoid spike shape of CS-del143 and the square-headed spike shape of CS. Glumes of CS 5BL-14 and CS 5DL-5 were tougher and more keeled compared with those of CS, but they were not as tough as the CS-del143 glumes. Disarticulation in CS 5BL-14 and CS 5DL-5 occurred at the base of the glumes, as it does in CS. Compared with CS, the number of spikelets per spike and seed weight were reduced in CS 5BL-14 and CS 5DL-5 (Fig. 6 and Table S3). The number of seeds per spike was reduced by 67–75% due to low fertility. All these changes led to substantially reduced grain yield. These observations suggest that 5Dq and 5Bq contribute to threshability and the suppression of the speltoid characters, but to a lesser degree than 5AQ.
Compared with the parental lines CS-del143 and CS 5DL-5, the spikes of the double deletion line CS-5A/5Ddd, which produces no functional Q or q protein, were much more speltoid in appearance (Fig. 6) with extremely tough glumes suggesting that the genetic effects of 5AQ and 5Dq are additive. The double deletion line had fewer spikelets per spike and lower fertility than its parental lines (Table S3), which resulted in the lowest grain yield among all of the stocks tested. Spikes of CS 5DL-5 and CS-5A/5Ddd emerged more than 30 d later than spikes of CS, most likely caused by the absence of gene(s) other than q in the terminal deletion of chromosome 5D.
Discussion
It has long been understood that the Q allele on wheat chromosome 5A has had a profound influence on wheat domestication. Here, we characterize the structure, evolution, and function of the Q/q loci on chromosomes 5A, 5B (and 5S), and 5D, to understand their contribution to domestication traits.
First, the intergenic sequences flanking the Q/q loci, composed primarily of TEs, have profoundly diverged by nested insertion and deletion among the A, B, D, and S genomes. This is a common feature of the intergenic regions of the wheat genome (12, 28–31). This important comparative study showed thus no putative regulating conserved sequence, except the ∼2 and ∼1 kb at 5′- and 3′ ends of the Q/q gene, respectively (Fig. 2).
Second, 5AQ/q is a cryptoparalog of the q gene on B, S, and D genomes, a result of gene duplication in the progenitor of the diploid wheats followed by selective loss of one copy of the gene. Functional interactions of paralogs may be substantially different from interactions of less divergent homoeologs.
Third, 5AQ/q, 5Dq, and 5Sq are expected to produce functional Q/q proteins, but 5Bq is a pseudogene encoding truncated proteins without the AP2 DNA-binding domains. It is unknown whether these truncated proteins serve a function. Both 5Bq and 5Dq produce multiple transcript variants. One of the 5Dq transcripts encodes a full-length q protein, whereas the other transcript encodes a truncated protein without the AP2 domains. About 20% of plant genes produce multiple transcripts, but their functions often remain unknown (32).
Fourth, the 5Bq pseudogene remains transcriptionally active. Plant pseudogenes derived from recently duplicated genes often remain transcriptionally active, presumably due to insufficient time for promoter degradation (33). We showed however that the 5Bq pseudogene participates in regulation of expression of the 5AQ and 5Dq genes at the transcript level, suggesting an active interaction between the homoeoalleles (see below).
Fifth, the 5Bq pseudogenization occurred after allotetraploidization, possibly upon selection imposed by domestication. This suggests that 5Bq plays a role in regulating expression of the domestication traits in tetraploid and hexaploid wheat.
Given the current findings, we propose that expression of the free-threshing phenotype and other domestication traits associated with the Q allele requires not only the V329I amino acid substitution in the Q protein, but also a combination of the divergent Q/q loci, with features we have identified, brought together in polyploid wheat. This also provides an explanation for the specificity of the free-threshing character to polyploid wheat and its absence among the diploids (26).
To investigate expression regulation and interactions between different Q/q homoeoalleles and their roles in domestication of hexaploid wheat, we analyzed different wheat lines altered for the Q/q homoeoalleles. Whereas the deletion of 5AQ includes a small genomic segment of ∼2 Mb (34), deletions of 5Bq and 5Dq encompass genomic segments representing 25% and 24% of chromosomes 5B and 5D, respectively (35), suggesting that deletion of other genes could also influence Q/q expression or the morphological traits that we evaluated. However, loci affecting these traits have not been reported on the deleted segments of chromosomes 5B or 5D. Good evidence of the appropriateness of the genetic material to study interactions between the different Q/q homoeoalleles was provided by the observation that the transcript levels and phenotypes for lines lacking 5Bq and 5Dq, which both down-regulate expression of 5AQ, were nearly the same.
Our transcriptional expression analysis indicated complex interactions among the Q/q homoeoalleles. Interpretation was complicated by the fact that 5Bq is a transcriptionally active pseudogene (no q protein is made) and 5AQ/q is a cryptoparalog of 5Bq and 5Dq. Clearly, deletion of 5Dq or 5Bq leading to reduced expression of 5AQ suggests that they indirectly up-regulate expression of 5AQ, whereas the latter plays a repressor role for both 5Bq and 5Dq expression. Our results, consistent with previous research (23), demonstrate that 5AQ is transcribed at higher levels than 5Aq. The V329I substitution was shown to be responsible for enhanced homodimer formation in 5AQ, possibly leading to self up-regulation of 5AQ transcription (23). Our results show that the expression of 5Bq and 5Dq increases when 5AQ is deleted or replaced by 5Aq, suggesting that the V329I mutation in 5Aq/Q is also responsible for the down-regulation of 5Bq and 5Dq (comparison of expression in lines CS, CS-del143, and CS-DIC 5A). This result could be explained by dosage balance or complementary effects in regulation among the Q/q homoeoalleles (36), but more work is needed to decipher the mechanism of this transcriptional regulation.
It has been recently proposed that interactions between mRNAs, transcribed pseudogenes, and long noncoding RNAs form a large-scale regulatory network across the transcriptome using microRNA response elements (11). A putative miRNA172 binding site in exon 10 of the Q/q gene (23) points to a possible role of miRNA in regulation of Q/q gene expression. The miRNA172 binding site is also present in the 5Bq pseudogene transcripts.
Interesting correlations were observed between expression variation of Q/q homoeoalleles and variations of domestication and agronomic traits. Our study confirms previous findings that the 5AQ/q genes influence numerous domestication traits (23, 25, 34, 37, 38), and it shows that along with the 5AQ/q alleles, the 5Dq and 5Bq alleles contribute to speltoid suppression, but their function depends on the allelic state of 5AQ/q. The contribution of 5Bq to these traits is indirect because it is a pseudogene, but we demonstrated that its deletion significantly reduced transcription of 5Dq. This apparent coregulation of the q genes largely explains the similar phenotype observed when 5Dq (CS 5DL-5) or 5Bq (CS 5BL-14) are deleted. The phenotypic differences correlated with expression variations observed between CS-del143 and CS-5A/5Ddd suggest that 5Dq plays a more important role in controlling spike shape, spikelet number, and glume toughness when 5AQ is absent.
Subfunctionalization (evolution of partitioned ancestral functions among alleles or homoeoalleles) and neofunctionalization (evolution of novel functions among alleles or homoeoalleles) were suggested as mechanisms leading to the preservation of the duplicated genes after polyploidization (5). Here, we provide a rare example of the fate of a set of homoeologous genes, associated with plant morphology and domestication in hexaploid wheat. A recent mutation in 5Aq led to “hyperfunctionalization” of 5AQ; 5Bq underwent pseudogenization, but remains transcriptionally active and influences regulation of expression of the homoeologs; and 5Dq has been subfunctionalized because it contributes to many of the domestication traits.
Materials and Methods
A full description of the materials and methods used in this work is provided in SI Materials and Methods. This information includes BAC library screening, sequencing, and annotation; cDNA amplification and sequencing; transcript and expression analysis; phylogenetic analysis; phenotypic analysis; sequences of primer (Table S4); and descriptions of the wheat genotypes used in this research.
Supplementary Material
Acknowledgments
We thank Joseph Jahier (Institut National de la Recherche Agronomique) and Moshe Feldman (Weizmann Institute of Science) for valuable discussions and for providing some of the wheat samples. This project was supported by US Department of Agriculture (USDA), Agricultural Research Service, Current Research Information Systems Project 5442-22000-033-00D and the National Center for Sequencing (Centre National de Séquençage-Génoscope)/APCNS2003 Project: Triticum Species Comparative Genome Sequencing in Wheat. PCR-based tracing of retrotransposon insertions was funded by the Agence Nationale pour la Recherche (ANR) Biodiversiteé Project (ANR-05-BDIV-015) and the ANR-05-Blanc Project impact of transposable elements on gene regulation.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JF01612–JF01622.)
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1110552108/-/DCSupplemental.
References
- 1.Adams KL, Wendel JF. Novel patterns of gene expression in polyploid plants. Trends Genet. 2005;21:539–543. doi: 10.1016/j.tig.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 2.Soltis PS, Soltis DE. The role of hybridization in plant speciation. Annu Rev Plant Biol. 2009;60:561–588. doi: 10.1146/annurev.arplant.043008.092039. [DOI] [PubMed] [Google Scholar]
- 3.Van de Peer Y, Fawcett JA, Proost S, Sterck L, Vandepoele K. The flowering world: A tale of duplications. Trends Plant Sci. 2009;14:680–688. doi: 10.1016/j.tplants.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Prince VE, Pickett FB. Splitting pairs: The diverging fates of duplicated genes. Nat Rev Genet. 2002;3:827–837. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- 5.Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Comai L. The advantages and disadvantages of being polyploid. Nat Rev Genet. 2005;6:836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- 7.Chen ZJ, Ni Z. Mechanisms of genomic rearrangements and gene expression changes in plant polyploids. Bioessays. 2006;28:240–252. doi: 10.1002/bies.20374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birchler JA, Auger DL, Riddle NC. In search of the molecular basis of heterosis. Plant Cell. 2003;15:2236–2239. doi: 10.1105/tpc.151030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaudhary B, et al. Reciprocal silencing, transcriptional bias and functional divergence of homoeologs in polyploid cotton (gossypium) Genetics. 2009;182:503–517. doi: 10.1534/genetics.109.102608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hovav R, et al. Partitioned expression of duplicated genes during development and evolution of a single cell in a polyploid plant. Proc Natl Acad Sci USA. 2008;105:6191–6195. doi: 10.1073/pnas.0711569105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chalupska D, et al. Acc homoeoloci and the evolution of wheat genomes. Proc Natl Acad Sci USA. 2008;105:9691–9696. doi: 10.1073/pnas.0803981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang S, et al. Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc Natl Acad Sci USA. 2002;99:8133–8138. doi: 10.1073/pnas.072223799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kihara H. Discovery of the DD-analyser, one of the ancestors of vulgare wheats. Ag. Hort. (Tokyo) 1944;19:889–890. [Google Scholar]
- 15.McFadden ES, Sears ER. The origin of Triticum spelta and its free-threshing hexaploid relatives. J Hered. 1946;37:81–89, 107. doi: 10.1093/oxfordjournals.jhered.a105590. [DOI] [PubMed] [Google Scholar]
- 16.Nesbitt M, Samuel D. From the staple crop to extinction? The archaeology and history of hulled wheats. In: Padulosi S, Hammer K, Heller J, editors. Hulled Wheats. Proceedings of the First International Workshop on Hulled Wheats. Rome: International Plant Genetic Resources Institute; 1996. pp. 41–100. [Google Scholar]
- 17.Blake NK, Lehfeldt BR, Lavin M, Talbert LE. Phylogenetic reconstruction based on low copy DNA sequence data in an allopolyploid: The B genome of wheat. Genome. 1999;42:351–360. [PubMed] [Google Scholar]
- 18.Dvorák J, Terlizzi P, Zhang H-B, Resta P. The evolution of polyploid wheats: Identification of the A genome donor species. Genome. 1993;36:21–31. doi: 10.1139/g93-004. [DOI] [PubMed] [Google Scholar]
- 19.Dvorák J, Zhang HB. Variation in repeated nucleotide sequences sheds light on the phylogeny of the wheat B and G genomes. Proc Natl Acad Sci USA. 1990;87:9640–9644. doi: 10.1073/pnas.87.24.9640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riley R, Unrau J, Chapman V. Evidence on the origin of the B genome of wheat. J Hered. 1958;49:91–98. [Google Scholar]
- 21.Takumi S, Nasuda S, Liu YG, Tsunewaki K. Wheat phylogeny determined by RFLP analysis of nuclear DNA. I. Einkorn wheat. Jpn J Genet. 1993;68:73–79. [Google Scholar]
- 22.Talbert LE, Blake NK, Storlie EW, Lavin M. Variability in wheat based on low-copy DNA sequence comparisons. Genome. 1995;38:951–957. doi: 10.1139/g95-125. [DOI] [PubMed] [Google Scholar]
- 23.Simons KJ, et al. Molecular characterization of the major wheat domestication gene Q. Genetics. 2006;172:547–555. doi: 10.1534/genetics.105.044727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faris JD, Simons KJ, Zhang Z, Gill BS. The wheat super domestication gene Q. Frontiers of wheat Bioscience. Wheat Information Service. 2005;100(Memorial Issue):129–148. [Google Scholar]
- 25.Faris JD, Gill BS. Genomic targeting and high-resolution mapping of the domestication gene Q in wheat. Genome. 2002;45:706–718. doi: 10.1139/g02-036. [DOI] [PubMed] [Google Scholar]
- 26.Sharma HC, Waines JG. Attempted gene transfer from tetraploids to diploids in Triticum. Can J Genet Cytol. 1982;23:639–645. [Google Scholar]
- 27.Faris JD, Zhang Z, Fellers JP, Gill BS. Micro-colinearity between rice, Brachypodium, and Triticum monococcum at the wheat domestication locus Q. Funct Integr Genomics. 2008;8:149–164. doi: 10.1007/s10142-008-0073-z. [DOI] [PubMed] [Google Scholar]
- 28.Salse J, et al. New insights into the origin of the B genome of hexaploid wheat: Evolutionary relationships at the SPA genomic region with the S genome of the diploid relative Aegilops speltoides. BMC Genomics. 2008;9:555. doi: 10.1186/1471-2164-9-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chantret N, et al. Molecular basis of evolutionary events that shaped the hardness locus in diploid and polyploid wheat species (Triticum and Aegilops) Plant Cell. 2005;17:1033–1045. doi: 10.1105/tpc.104.029181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dvorak J, Akhunov ED, Akhunov AR, Deal KR, Luo MC. Molecular characterization of a diagnostic DNA marker for domesticated tetraploid wheat provides evidence for gene flow from wild tetraploid wheat to hexaploid wheat. Mol Biol Evol. 2006;23:1386–1396. doi: 10.1093/molbev/msl004. [DOI] [PubMed] [Google Scholar]
- 31.Isidore E, Scherrer B, Chalhoub B, Feuillet C, Keller B. Ancient haplotypes resulting from extensive molecular rearrangements in the wheat A genome have been maintained in species of three different ploidy levels. Genome Res. 2005;15:526–536. doi: 10.1101/gr.3131005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barbazuk WB, Fu Y, McGinnis KM. Genome-wide analyses of alternative splicing in plants: opportunities and challenges. Genome Res. 2008;18:1381–1392. doi: 10.1101/gr.053678.106. [DOI] [PubMed] [Google Scholar]
- 33.Zou C, et al. Evolutionary and expression signatures of pseudogenes in Arabidopsis and rice. Plant Physiol. 2009;151:3–15. doi: 10.1104/pp.109.140632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faris JD, Fellers JP, Brooks SA, Gill BS. A bacterial artificial chromosome contig spanning the major domestication locus Q in wheat and identification of a candidate gene. Genetics. 2003;164:311–321. doi: 10.1093/genetics/164.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Endo TR, Gill BS. The deletion stocks of common wheat. J Hered. 1996;87:295–307. [Google Scholar]
- 36.Birchler JA, Riddle NC, Auger DL, Veitia RA. Dosage balance in gene regulation: Biological implications. Trends Genet. 2005;21:219–226. doi: 10.1016/j.tig.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 37.Kato K, Miura H, Sawada S. QTL mapping of genes conrolling ear emergence time and plant height on chromosome 5A of wheat. Theor Appl Genet. 1999;98:472–477. [Google Scholar]
- 38.Muramatsu M. Dosage effect of the spelta gene Q of hexaploid wheat. Genetics. 1963;48:469–482. doi: 10.1093/genetics/48.4.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.