Abstract
Protein degradation by the 26S proteasome is a fundamental process involved in a broad range of cellular activities, yet how proteasome activity is regulated remains poorly understood. We report here that ubiquitin-like domain-containing C-terminal domain phosphatase 1 (UBLCP1) is a 26S proteasome phosphatase that regulates nuclear proteasome activity. UBLCP1 directly interacts with the proteasome via its UBL domain and is exclusively localized in the nucleus. UBLCP1 dephosphorylates the 26S proteasome and inhibits proteasome activity in vitro. Knockdown of UBLCP1 in cells promotes 26S proteasome assembly and selectively enhances nuclear proteasome activity. Our results describe the first identified proteasome-specific phosphatase and uncover a unique mechanism for phosphoregulation of the proteasome.
The ubiquitin–proteasome system is responsible for degrading the majority of proteins in eukaryotic cells, and deregulation of this system is widely associated with human diseases (1–4). The 26S proteasome consists of a 20S core particle (CP) and one or two 19S regulatory particles (RP) (5, 6). The cylindrical 20S CP houses peptidase activities and is formed by two sets of α-subunits (α1-7) and two sets of β-subunits (β1-7). The 19S RP consists of at least 19 subunits. Six of them (Rpt1-6) are AAA+-type ATPases, which directly bind the CP and form the “base” of RP together with the non-ATPase subunits Rpn1, 2, and 13. The base is connected to the “lid” composed of the remaining Rpn proteins (Rpn3, 5–12, and 15). The 19S RP caps and activates the 20S CP by capturing ubiquitinated substrates and driving their translocation into the catalytic chamber.
With its enormous size and complexity, the proteasome provides a sophisticated platform for dynamic interactions with a great number of proteasome-interacting proteins (PIPs) (7, 8). Among the PIPs are a group of ubiquitin-like (UBL) domain-containing proteins, including the substrate shuttle proteins Rad23A/B and Dsk2/PLIC, the E3 ubiquitin ligase Parkin, and the deubiquitinating enzyme Ubp6/USP14 (9, 10). Despite low similarity in amino acid sequence, UBL domains and ubiquitin (Ub) share a similar core structure known as the β-grasp fold and are recognized by a variety of ubiquitin/UBL-binding proteins, including Rpn1, 10, and 13 of the 19S RP (11–16).
The abundance, localization, composition, and activity of the proteasome are dynamically regulated in response to various stimuli. Numerous reports have documented proteasome phosphorylation in vivo (7, 17–30). Most phosphorylation sites on the proteasome have been identified in proteomic screens, which generally afford little information about their biological meanings. Some phosphorylation sites have been found from independent studies and are evolutionarily conserved, suggesting that they are likely of biological importance. In contrast to numerous publications of kinases that phosphorylate the proteasome, few reports exist on proteasome regulation by phosphatases (31, 32).
Work from our laboratory and others has defined a family of haloacid dehalogenase (HAD)-like serine/threonine phosphatases (refs. 33 and 34 and Fig. S1A). The hallmark of these phosphatases is a DXDXT catalytic motif, in which both aspartic acid residues are critical for enzyme activity. There are at least seven family members in humans: SCP1, SCP2, SCP3, FCP1, Dullard, UBLCP1, and HSPC129. The founding members—SCP1 and FCP1—dephosphorylate the C-terminal domain (CTD) of the largest subunit of RNA polymerase II (Pol II) (35, 36) and play critical roles during Pol II-mediated gene transcription (37, 38). Some family members contain domains that are important for substrate binding and/or intracellular localization. For example, FCP1 has a C-terminal BRCT (breast cancer gene 1 C-terminal domain) domain that is involved in CTD recognition (39). We have recently shown that Dullard has an N-terminal transmembrane region that targets the protein to the nuclear envelope, where it controls membrane biosynthesis (34). UBLCP1 (ubiquitin-like domain-containing CTD phosphatase 1, previously called MGC10067) contains a putative N-terminal UBL domain (40), although the biological function of this protein has not been characterized.
Here we show that UBLCP1 is a proteasome-specific phosphatase known to exist in mammalian cells. It directly binds the 19S RP via the UBL domain and dephosphorylates the proteasome. UBLCP1 selectively represses nuclear proteasome activity in a phosphatase-dependent manner, suggesting a unique mechanism for nuclear proteasome regulation.
Results
UBLCP1 Interacts with the Proteasome.
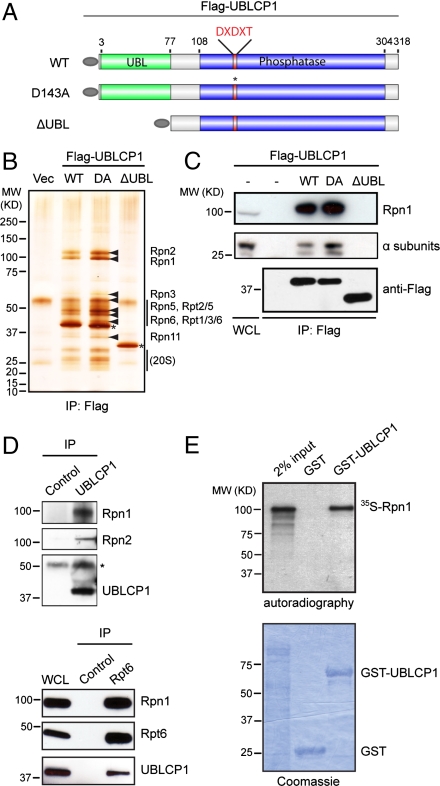
To understand the cellular function of UBLCP1, we tried to identify its interacting proteins by coimmunoprecipitation experiments followed by mass spectrometry analysis (IP/MS). Flag-tagged WT human UBLCP1 and mutants were stably expressed in HEK293T cells (Fig. 1A). Mutation of the first aspartic acid in the DXDXT motif to an alanine (D143A, or DA) renders UBLCP1 inactive, whereas deletion of the putative UBL domain (∆UBL) was predicted to interfere with protein–protein interactions. After anti-FLAG immunoprecipitation, a number of proteins were visualized by silver staining that coprecipitated with full-length UBLCP1 (WT and DA). These proteins were absent in the control and the ∆UBL immunoprecipitates. Intriguingly, all of these interacting proteins were identified as regulatory subunits of the 26S proteasome (Fig. 1B, Table S1, and Dataset S1). Western blot analysis confirmed that both RP and CP subunits coprecipitated with FLAG-tagged full-length UBLCP1 but not the ∆UBL mutant (Fig. 1C). Binding also occurs between endogenous UBLCP1 and the proteasome as evidenced by reciprocal immunoprecipitation experiments using an affinity-purified UBLCP1 antibody (Fig. 1D). In a separate study, we also found that UBLCP1 copurified with the 26S proteasome using Rpn11-HTBH as bait (8). Together, these results demonstrate that UBLCP1 is a bona fide proteasome-interacting protein, and this interaction is dependent on the UBL domain of UBLCP1.
Fig. 1.
UBLCP1 binds the 26S proteasome. (A) A schematic representation of UBLCP1 used for generating 293T stable lines used in B. All constructs contain a N-terminal Flag tag (gray oval). (B) UBLCP1-interacting proteins visualized by silver staining. Proteasome subunits identified by MS are indicated by arrowheads. 20S subunits were verified by Western blot. Asterisks indicate the bait Flag-UBLCP1 proteins. (C) Coimmunoprecipitation of Flag-UBLCP1 with RP (Rpn1) and CP (α subunits). (D) Binding between endogenous UBLCP1 and the proteasome. Preimmune rabbit serum or normal mouse IgG was used as controls for UBLCP1 and Rpt6 IP, respectively. Asterisk indicates the heavy chain of rabbit IgG. (E) In vitro binding between UBLCP1 and Rpn1.
Ubiquitin and UBL domain proteins are captured by the 19S RP of the proteasome. Among the 19 RP subunits, only Rpn1 (also known as PSMD2) showed specific interaction with UBLCP1 in an in vitro binding assay, consistent with Rpn1’s role as a UBL receptor on the proteasome (Fig. 1E and Fig. S2). In addition, an interaction between Rpn1/PSMD2 and UBLCP1 was observed in a genome-wide yeast two-hybrid screen (41). Therefore, our results suggest that UBLCP1 is recruited to the proteasome through direct interaction between its UBL domain and Rpn1.
Lysine44 in the UBL Domain of hUBLCP1 Is Essential for Proteasome Binding.
We further analyzed the UBL domain of UBLCP1 (designated here as UBLCP1_N) by comparing it with ubiquitin and other known UBL domains. Sequence alignment and phylogenetic analysis suggest that UBLCP1_N belongs to the UBL domain family (Fig. S1B). It is most similar to the UBL domain of Ubp6/USP14 (Ubp_N), a deubiquitinating enzyme that also binds Rpn1 (42–44). In addition, we solved the crystal structure of Drosophila UBLCP1 at 1.9-Å resolution (Fig. 2A and Fig. S3A, PDB ID code 3SHQ). Fly and human UBLCP1 share a high degree of sequence homology (74% similar, 60% identical). The full-length protein is organized into an N-terminal UBL domain and a C-terminal phosphatase domain that are connected by a flexible linker region (Fig. S3A). Like ubiquitin and other UBL domains, DmUBLCP1_N adopts a typical β-grasp fold in the crystal structure and contains conserved residues that may form the hydrophobic patch involved in Ub/UBL recognition, including Leu48 (Leu46 of human UBLCP1) that corresponds to Ile44 of ubiquitin (45, Fig. 2A). In addition, Lys44 of human UBLCP1 (Lys46 of fly UBLCP1) is invariant in all species and a basic residue is common at the corresponding position in ubiquitin and other UBLs, suggesting functional relevance (Fig. S1C).
Fig. 2.
Functional importance of the UBL domain. (A) Structural comparison of ubiquitin (PDB ID code 1UBQ) and the UBL domains of Drosophila UBLCP1 (PDB ID code 3SHQ), yeast Dsk2 (PDB ID code 2BWF), and human Rad23A (PDB ID code 2WYQ). The side chains of the KLL motif (Lys46/Leu47/Leu48) in DmUBLCP1 and its counterparts in the other structures are shown. (B) 293T cells were transfected with Flag-UBLCP1 constructs (G10E, L46A, K44E, and K44A/L45A/L46A) and were subjected to coimmunoprecipitation assays. (C) Immunostaining of endogenous UBLCP1 and Rpt6 in HaCaT cells (human keratinocytes). Scale bar = 10 μm. (D) Localization of GFP-tagged UBLCP1 and mutants in HeLa cells. Nuclear contours are depicted with white dotted lines based on DAPI images. Scale bar = 10 μm.
In contrast to the ubiquitin I44A mutation that prevents ubiquitin recognition (45), replacement of the corresponding Leu46 of human UBLCP1 with alanine (L46A) had no effect on proteasome binding. Similarly, Gly10 of hUBLCP1, which corresponds to Leu8 of ubiquitin, was mutated to Glu and the resulting G10E mutant still interacted with the proteasome, albeit to a lesser extent (Fig. 2B). However, mutation of Lys44 alone (K44E) or in combination with adjacent residues (K44A/L45A/L46A, “KLL”) completely abolished the association between hUBLCP1 and the proteasome (Fig. 2B). Therefore, UBLCP1 binding to the proteasome requires a functional UBL domain, and Lys44 is critical for this interaction.
UBLCP1 Is a Nuclear Protein and Colocalizes with Nuclear Proteasomes.
We next determined the intracellular localization of UBLCP1 by fluorescence microscopy. Endogenous UBLCP1 exhibits strong nuclear staining, which can be completely eliminated by UBLCP1 RNAi or antibody preblocking (Fig. 2C and Fig. S4). The proteasome is also enriched in the nucleus (Fig. 2C, 46) and colocalizes with endogenous UBLCP1, further supporting the physical interaction between this phosphatase and the proteasome.
Because UBLCP1 is a relatively small protein (37 KD) and does not have a canonical nuclear localization signal (NLS), we wondered whether its intracellular distribution could be a result of proteasome binding. Using GFP fusion proteins, we first confirmed that both wild-type UBLCP1 and the D143A mutant are exclusively localized to the nucleus. This nuclear pattern was also seen with the UBL domain alone but lost with the ΔUBL mutant, suggesting that the UBL domain is both necessary and sufficient for nuclear localization (Fig. 2D). Moreover, those UBL domain mutants that retain the ability to bind the proteasome (G10E and L46A) also remained nuclear, whereas the proteasome binding-deficient Lys44 mutants displayed a diffuse, pancellular localization and were even excluded from the nucleus in a subset of cells (Fig. 2D). These data strongly suggest that UBLCP1 is a nuclear protein whose localization is coupled to its interaction with nuclear proteasomes.
UBLCP1 Dephosphorylates the Proteasome in Vitro.
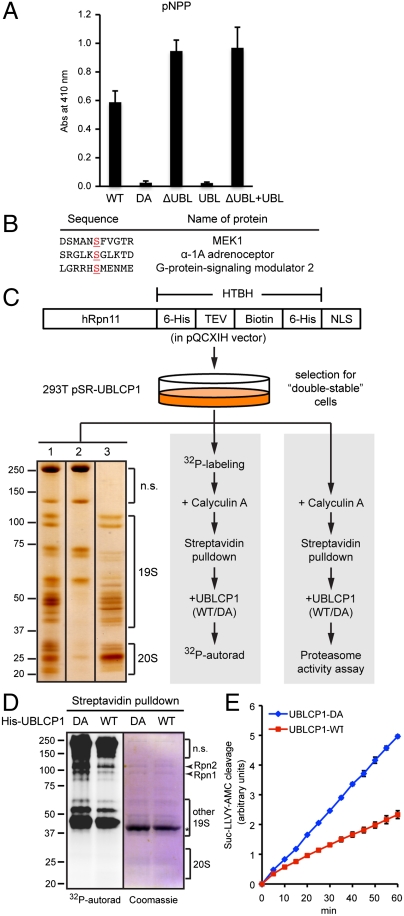
Purified recombinant UBLCP1 could dephosphorylate the phosphatase substrate p-nitrophenyl phosphate (pNPP), and the activity was moderately enhanced upon removal of the UBL domain (Fig. 3A). Because we have found no evidence for intra- or intermolecular interactions between the UBL and phosphatase domains, it is possible that UBLCP1 phosphatase activity may be allosterically regulated when its UBL domain docks onto the proteasome. UBLCP1 was also able to dephosphorylate several peptides containing phosphoserine (Fig. 3B), consistent with the notion that this family of proteins functions as pSer/Thr phosphatases.
Fig. 3.
UBLCP1 dephosphorylates the proteasome in vitro. (A) pNPP assay with the indicated UBLCP1 proteins. Data are shown as average ± SD. (B) Phospho-peptides significantly dephosphorylated by UBLCP1 in vitro, with the phospho-serine residues underlined. This phospho-peptide array (Millipore) does not include any proteasome peptides. (C) 26S proteasome isolation and analysis. 293T double-stable line was used for high-purity proteasome preparation (Left), 32P-labeling and in vitro phosphatase assay (Middle), or in vitro proteasome activity assay (Right). (Left) A representative silver-staining image of streptavidin-immobilized proteasomes (lane 1), purified proteasomes after tobacco etch virus (TEV) protease treatment (lane 3), and nonspecific (n.s.) proteins remaining on the beads (lane 2). See Materials and Methods for details. (D) Metabolically radio-labeled, affinity-purified nuclear proteasomes were treated in vitro with purified UBLCP1 (shown by an asterisk). Proteasome subunits were visualized by Coomassie blue staining and 32P-autoradiography. (E) Peptidase activity assay of the 26S proteasome purified and treated as in D. AMC fluorescence is represented as the average ± SD.
Given all these observations, we hypothesized that the proteasome could be a direct substrate of UBLCP1. Nuclear proteasomes can be enriched and affinity-purified from 293T cells using Rpn11-HTBH-NLS as bait (7) (Fig. 3C), which allows us to examine proteasome dephosphorylation in vitro. We radiolabeled cells with 32P-orthophsophate and isolated nuclear proteasomes, which contained a number of in vivo phosphorylated proteins corresponding to 19S RP subunits based on their migration during SDS-PAGE (Fig. 3D). Several, but not all, of these 32P-labeled proteasomal proteins were markedly dephosphorylated by purified wild-type UBLCP1 but not the catalytically inactive D143A mutant (Fig. 3D). Importantly, under the same condition, UBLCP1-mediated dephosphorylation led to a significant reduction (30–50%) in proteasome activity as determined using a fluorogenic peptide substrate (Fig. 3E). As a control, alkaline phosphatase treatment reduced the activity of purified 26S proteasome by approximately 70% (Fig. S5). These results suggest that UBLCP1 can directly dephosphorylate a subset of proteasome subunits and negatively regulate proteasomal activity in vitro.
UBLCP1 Knockdown Enhances Nuclear Proteasome Activity.
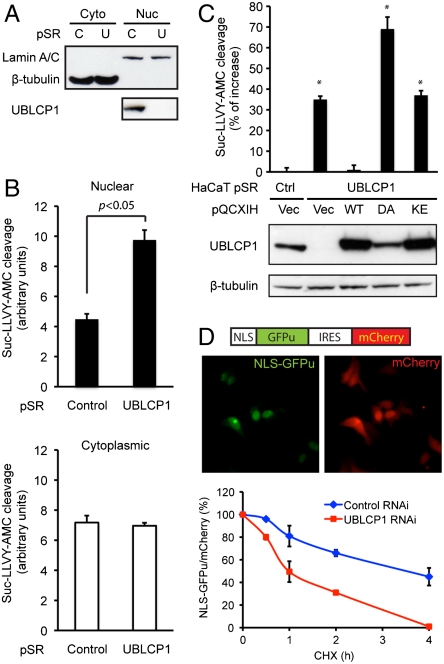
Considering the nuclear localization of UBLCP1 and its ability to dephosphorylate the proteasome, we reasoned that UBLCP1 would selectively regulate nuclear proteasome activity in cells. Overexpression of wild-type UBLCP1 in cells did not significantly alter proteasome activity, probably due to the high level of endogenous protein. Therefore, we relied on a loss-of-function strategy to explore the physiological function of UBLCP1. Several types of cells were engineered to stably express a control shRNA or a UBLCP1-specific shRNA that reduced the protein level by more than 90% (Fig. 4A). Control and UBLCP1 knockdown cells were fractionated, and nuclear and cytoplasmic proteasome activity was measured using fluorogenic peptide substrates (Fig. 4 A and B). Nuclear proteasome activity was significantly higher in UBLCP1 knockdown cells than in control cells (Fig. 4B), which could be reduced by in vitro treatment of UBLCP1 (Fig. S6A). These are consistent with the earlier result that UBLCP1 is a negative regulator of the proteasome (Fig. 3E). As expected, cytoplasmic proteasome activity was unchanged by UBLCP1 RNAi (Fig. 4B). We also developed an “in-well” assay for efficient measurement of nuclear proteasome activity from cells grown in 96-well plates (see SI Materials and Methods). The activating effect of UBLCP1 knockdown on nuclear proteasome activity was reproducible in this assay system, and again cytoplasmic proteasome activity was unchanged (Fig. S6B). Similar experiments were performed using different peptide substrates, and the results showed that all three types of peptidase activity of the proteasome (caspase-like, trypsin-like, and chymotrypsin-like) were enhanced upon UBLCP1 depletion (Fig. S6C).
Fig. 4.
UBLCP1 knockdown enhances nuclear proteasome activity. (A) Cytoplasmic (Cyto) and nuclear (Nuc) fractionation of ZR751 breast cancer cells stably expressing either control (C) or UBLCP1-specific (U) shRNA built in the pSuperRetro vector (pSR). Lamin A/C and β-tubulin were used as loading controls for nuclear and cytoplasmic fractions, respectively. (B) Proteasome activity assay using the same extracts as in A. Normalized AMC fluorescence is represented as the average ± SD. (C) UBLCP1 rescue experiments with HaCaT double-stable lines. UBLCP1 expression levels are determined by Western blotting. Nuclear proteasome activity was measured by in-well assays. Data are presented as the percent increase in activity versus control. *, p < 0.05. (D, Top) A schematic of the NLS-GFPu reporter in an IRES-mCherry backbone (26) and localization of the fluorescent proteins in 293T cells. (Bottom) GFPu/mCherry ratio during cycloheximide (CHX) treatment time course, with the starting value at time 0 being 100%. Data were analyzed from three independent experiments.
Next, we performed rescue experiments by expressing different versions of UBLCP1 in the knockdown cells. Reexpression of the wild-type protein fully reversed the increase in nuclear proteasome activity (Fig. 4C), confirming the specificity of UBLCP1 RNAi. Neither the D143A mutant nor the K44E mutant was able to rescue the knockdown phenotype (Fig. 4C). In fact, UBLCP1 (D143A) appeared to have a dominant-negative effect and further activated the proteasome. Based on these results, we conclude that both the phosphatase activity and direct proteasome binding are required for UBLCP1 to negatively regulate nuclear proteasome activity.
Furthermore, we monitored nuclear proteasome activity in live cells using GFP-based reporters. Fusion of the CL1 degron (a constitutive degradation signal) to the C terminus of GFP (GFPu) leads to its rapid ubiquitination and proteasomal degradation (47), and a N-terminal NLS successfully targets the GFPu reporter to the nucleus (Fig. 4D). UBLCP1 depletion significantly shortened the half-life of NLS-GFPu, indicative of accelerated protein degradation and elevated nuclear proteasome activity (Fig. 4D). The same effect was seen with a nuclear-targeted UbG76V-GFP reporter (48, Fig. S6D). Therefore, loss of UBLCP1 enhances nuclear proteasome activity against folded proteins in cells.
UBLCP1 Regulates RP-CP Association.
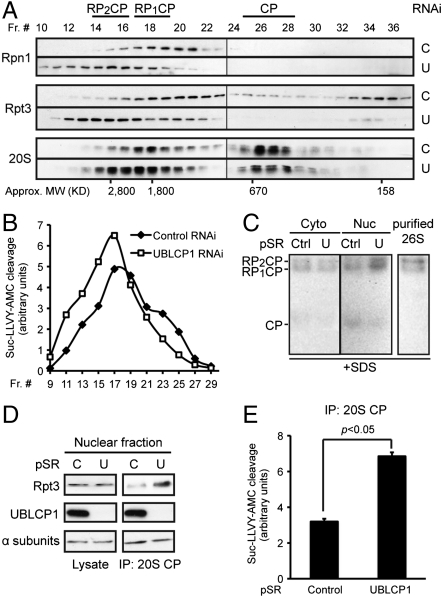
Finally, we investigated the underlying mechanism by which UBLCP1 regulates the proteasome. Overexpression or knockdown of UBLCP1 did not affect the abundance or subcellular localization of the proteasome (Figs. S3 and S4). Previous studies have suggested that phosphorylation of certain subunits promotes the assembly of RP and CP into the mature 26S proteasome, thereby increasing the efficiency of substrate degradation (21, 23). To test the possibility of UBLCP1 regulating 26S proteasome formation, we analyzed endogenous proteasome complexes from control and UBLCP1 knockdown cells using size-exclusion chromatography. The results showed a considerable increase in the size of proteasomal complexes upon UBLCP1 knockdown, with more doubly capped proteasomes (RP2CP) formed at the expense of free 20S CP (Fig. 5A). This shift in proteasome size distribution was paralleled by a similar change in the proteasome activity profile across the fractions (Fig. 5B). The increased activity of higher-order proteasomes in UBLCP1 RNAi cells was further illustrated by gel overlay assays, and the results again indicate that UBLCP1 selectively regulates the assembly/activity of nuclear proteasomes (Fig. 5C). Finally, we directly assessed the association between RP and CP in the nucleus by coimmunoprecipitation experiments. Although the protein level of RP and CP subunits was comparable between control and UBLCP1 knockdown cells, a stronger RP-CP interaction was observed with UBLCP1 RNAi (Fig. 5D). Correspondingly, immunoprecipitated nuclear proteasomes from UBLCP1 knockdown cells exhibited significantly higher activity than those from the control cells (Fig. 5E), recapitulating the results described earlier (Fig. 4). Consistently, treatment of purified 26S proteasome with UBLCP1 in vitro led to partial dissociation of RP and CP (Fig. S6E). Taken together, these data demonstrate that UBLCP1 negatively regulates the RP-CP assembly in the nucleus thereby restricting the overall nuclear proteasome activity.
Fig. 5.
UBLCP1 regulates RP-CP association. (A) Size-exclusion chromatography analysis of proteasome complexes from 293T cells stably expressing control or UBLCP1 shRNA. Proteasome subunits within each fraction were monitored by Western blot. The different forms of proteasome are shown (Top) based on their subunit composition and their expected sizes (Bottom). (B) Proteasome activity in gel-filtration fractions from A after normalization against the protein content within each fraction. (C) Gel overlay assay. Equal amounts of cytosolic and nuclear extracts from HaCaT pSuperRetro stable lines were separated on a 4.5% native gel. U, UBLCP1 shRNA. Purified 26S proteasome (1 μg; see Fig. 3C) was included as a positive control. The gel was then overlaid with proteasome assay buffer containing Suc-LLVY-AMC and 0.02% SDS (to detect 20S activity). After incubation in the dark at 37 °C for 30 min, the gel was imaged under UV light. (D and E) Coimmunoprecipitation of nuclear CP and RP from HaCaT pSuperRetro stable lines. Anti-CP immunoprecipitates were divided and used for Western blot (D) and proteasome activity assay (E). AMC fluorescence was normalized against the amount of immunoprecipitated CP.
Discussion
The pivotal role of the proteasome is well established in eukaryotic cells, and yet the regulation of proteasome activity is only beginning to be understood. Here we uncover a previously undescribed mechanism for controlling proteasome activity by the unique phosphatase, UBLCP1, which specifically associates with and dephosphorylates the proteasome. It is highly conserved from fly to human and is the only phosphatase in the human genome that contains a UBL domain. Among the family members, orthologs of SCP1, FCP1, and Dullard are present in all eukaryotic species, whereas UBLCP1 appeared much later in evolution and is not found in yeast or Caenorhabditis elegans. On the other hand, proteasome composition and function are largely conserved from yeast to human. It will be interesting to determine whether yeast and worm proteasomes are subjected to similar regulation by a phosphatase equivalent to UBLCP1.
It has been reported that UBLCP1 dephosphorylates recombinant Pol II CTD in vitro (40). However, we found that in cells there is no significant change in phosphorylation of recombinant or endogenous CTD upon UBLCP1 overexpression or depletion (Fig. S3 B and C). Nor was UBLCP1 found to interact with Pol II. Therefore, UBLCP1 does not appear to use Pol II CTD as a substrate in vivo. Our crystallography study offers a clue for understanding such distinction between UBLCP1 and SCP1/FCP1. Despite the similar fold of their phosphatase domains with almost identical catalytic site structure (including the DXDXT motif and the coordinated Mg2+ ion), the UBLCP1 phosphatase domain lacks a β-hairpin-like “insertion domain” found in both SCP1 and FCP1 (Fig. S3A), which is thought to be important for substrate binding (49, 50). This structural difference suggests that UBLCP1 may have a unique substrate specificity.
Phosphorylation of the proteasome often increases its activity, whereas in vitro dephosphorylation by nonspecific phosphatases inhibits proteasome function (19, 22, 24, 26, 28, 51, Fig. S5). Our work supports this notion by presenting evidence that a physiological proteasome phosphatase negatively regulates nuclear proteasome activity both in vitro and in vivo. Through a series of structural and biochemical studies, we propose that UBLCP1 is anchored to Rpn1 via its UBL domain and dephosphorylates Rpn1 and/or other proteasome subunits within vicinity, most likely those of the base of the 19S RP. Because the base constitutes the interface between the RP and the CP, changing the phosphorylation status of base subunits may affect the RP-CP interaction and 26S proteasome assembly, which is what we observed in UBLCP1 knockdown cells (Fig. 5). It is likely that UBLCP1-mediated dephosphorylation is required to maintain the equilibrium between different pools of proteasome subcomplexes, thereby keeping the nuclear proteasome activity at a proper level. This, though, might be one of several possible mechanisms by which UBLCP1 regulates the proteasome, considering that more than one in vivo phosphorylated proteasome subunit/PIP could be dephosphorylated by UBLCP1 in vitro (Fig. 3D). Lacking knowledge on the relevant kinases and the low stoichiometry of proteasome phosphorylation in general has imposed technical challenges on our attempts to identify the specific sites dephosphorylated by UBLCP1 in vivo. In any event, further efforts will be taken to identify UBLCP1 substrates and to elucidate the link between proteasome phosphorylation and proteasome activity.
The nuclear localization of UBLCP1 explains, at least in part, selective regulation of nuclear proteasome activity by this phosphatase. Our data indicate that UBLCP1 localization is tightly coupled with its binding to the nuclear proteasome, both depending on Lys44 within the UBL domain. This result is intriguing but also somewhat puzzling, because the proteasome is present in both the cytoplasm and the nucleus. Cytoplasmic and nuclear proteasomes have distinct binding partners and undergo different modifications (52). The exact reason why UBLCP1 preferentially binds nuclear proteasomes remains to be determined. Furthermore, the distribution of proteasomes varies between cell types, changes during cell cycle, and is influenced by growth/stress conditions (52). Whether UBLCP1 regulates the proteasome in the same manner under different circumstances is an interesting issue that merits further investigation.
Our work suggests that pharmacological inhibition of UBLCP1 will lead to an activation of nuclear proteasomes. The idea of proteasome regulation by inhibiting its associated proteins has proven to be promising as a possible therapeutic approach. For example, an inhibitor of USP14 enhanced proteasome activity and reduced toxicity of misfolded/damaged proteins in cells (53). The crystal structure of UBLCP1 should be valuable for structure-guided drug discovery, and we are in the process of screening for UBLCP1 inhibitors. Recently, a HAD-like phosphatase inhibitor, rabeprazole, was identified to selectively block SCP1 activity in vitro (50). Interestingly, rabeprazole binding depends on the insertion domain of SCP1, which is absent in UBLCP1. This distinction could be used to design/search for UBLCP1-specific inhibitors that would preferentially target cells with deregulated proteasome phosphorylation.
Materials and Methods
32P-Labeling, 26S Proteasome Purification, and in Vitro Phosphatase Assays.
For in vitro proteasome dephosphorylation, one 6-cm plate of 293T “double-stable” cells expressing hRpn11-HTBH-NLS and UBLCP1 shRNA were labeled with 1.0 mCi/mL 32P-orthophosphate (PerkinElmer) for 4 h at 37 °C and treated with 50 nM calyculin A for 30 min before harvesting. Cells were lysed in NETN buffer (150 mM NaCl, 0.5 mM EDTA, 50 mM Tris, pH 7.5, 0.5% NP-40) with 1 mM DTT, 5 mM ATP, and inhibitors of proteases and phosphatases. Cell lysates were incubated with high-capacity streptavidin agarose beads (Thermo) at 4 °C for 30 min. Immobilized 26S proteasomes were washed three times with NETN buffer and once with 1× phosphatase buffer (100 mM NaOAc, 50 mM bis-Tris, 50 mM Tris-HCl, pH 6.0, 1 mM DTT, 10 mM MgCl2, 10 mM NaF, 50 nM calyculin A). The beads were divided into two tubes, and 5 μg of His-UBLCP1 (WT or D143A) was added to each tube. The reactions were carried out at 30 °C for 1 h and terminated by boiling in Laemmli loading buffer. Samples were resolved by SDS-PAGE, stained with Coomassie blue and visualized by 32P autoradiography.
Measurement of Proteasome Activity.
Equal amounts of cytoplasmic or nuclear extracts (5–10 μg) were aliquoted in triplicate to a black 96-well plate, and endogenous 26S proteasome activity was measured with 100-μM fluorogenic peptide substrates according to a standard protocol (54). Nonproteasome protease activities were determined by fluorescence release in the presence of proteasome inhibitors MG-132 (100 μM) or epoxomicin (20 μM) and have been subtracted for data presentation. Statistic significance was determined by Student’s t test.
To monitor proteasome-dependent protein degradation in vivo, 293T cells were transfected with the GFP-based reporter construct. After 24 h, cycloheximide (50 μg/mL) was added at the indicated time points, and cell lysates were collected and transferred to a black 96-well plate for GFP and mCherry measurement. Background signals detected from untransfected cells were subtracted. More information can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We appreciate Dr. Shigeo Murata (University of Tokyo, Tokyo, Japan) for generously providing the expression constructs of the 19S subunits and David King and Sharleen Zhou for protein identification. We thank Dr. Zhiping Wang for help with image processing and Dr. Carolyn Worby for critical reading of the manuscript. This work was supported by National Institutes of Health (NIH) Grant DK018849-36 (to J.E.D.), NIH Grant GM-74830 (to L.H.), NIH/National Cancer Institute Training Grant T32 CA009523 (to V.S.T.), and Susan G. Komen postdoctoral fellowship KG111280 (to X.G.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3SHQ).
See Commentary on page 18573.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113170108/-/DCSupplemental.
References
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Bossy-Wetzel E, Schwarzenbacher R, Lipton SA. Molecular pathways to neurodegeneration. Nat Med. 2004;10(Suppl):S2–S9. doi: 10.1038/nm1067. [DOI] [PubMed] [Google Scholar]
- 3.Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458:438–444. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]
- 4.Bingol B, Sheng M. Deconstruction for reconstruction: The role of proteolysis in neural plasticity and disease. Neuron. 2011;69:22–32. doi: 10.1016/j.neuron.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murata S, Yashiroda H, Tanaka K. Molecular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol. 2009;10:104–115. doi: 10.1038/nrm2630. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, et al. Mass spectrometric characterization of the affinity-purified human 26S proteasome complex. Biochemistry. 2007;46:3553–3565. doi: 10.1021/bi061994u. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, Huang L. Identifying dynamic interactors of protein complexes by quantitative mass spectrometry. Mol Cell Proteomics. 2008;7:46–57. doi: 10.1074/mcp.M700261-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Hartmann-Petersen R, Gordon C. Integral UBL domain proteins: A family of proteasome interacting proteins. Semin Cell Dev Biol. 2004;15:247–259. doi: 10.1016/j.semcdb.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 10.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elsasser S, et al. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- 12.Elsasser S, Chandler-Militello D, Muller B, Hanna J, Finley D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J Biol Chem. 2004;279:26817–26822. doi: 10.1074/jbc.M404020200. [DOI] [PubMed] [Google Scholar]
- 13.Husnjak K, et al. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schreiner P, et al. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature. 2008;453:548–552. doi: 10.1038/nature06924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains—From structures to functions. Nat Rev Mol Cell Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winget JM, Mayor T. The diversity of ubiquitin recognition: Hot spots and varied specificity. Mol Cell. 2010;38:627–635. doi: 10.1016/j.molcel.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Ludemann R, Lerea KM, Etlinger JD. Copurification of casein kinase II with 20 S proteasomes and phosphorylation of a 30-kDa proteasome subunit. J Biol Chem. 1993;268:17413–17417. [PubMed] [Google Scholar]
- 18.Castano JG, Mahillo E, Arizti P, Arribas J. Phosphorylation of C8 and C9 subunits of the multicatalytic proteinase by casein kinase II and identification of the C8 phosphorylation sites by direct mutagenesis. Biochemistry. 1996;35:3782–3789. doi: 10.1021/bi952540s. [DOI] [PubMed] [Google Scholar]
- 19.Mason GG, Hendil KB, Rivett AJ. Phosphorylation of proteasomes in mammalian cells. Identification of two phosphorylated subunits and the effect of phosphorylation on activity. Eur J Biochem. 1996;238:453–462. doi: 10.1111/j.1432-1033.1996.0453z.x. [DOI] [PubMed] [Google Scholar]
- 20.Mason GG, Murray RZ, Pappin D, Rivett AJ. Phosphorylation of ATPase subunits of the 26S proteasome. FEBS Lett. 1998;430:269–274. doi: 10.1016/s0014-5793(98)00676-0. [DOI] [PubMed] [Google Scholar]
- 21.Satoh K, Sasajima H, Nyoumura K-i, Yokosawa H, Sawada H. Assembly of the 26S proteasome is regulated by phosphorylation of the p45/Rpt6 ATPase subunit. Biochemistry. 2001;40:314–319. doi: 10.1021/bi001815n. [DOI] [PubMed] [Google Scholar]
- 22.Feng Y, Longo DL, Ferris DK. Polo-like kinase interacts with proteasomes and regulates their activity. Cell Growth Differ. 2001;12:29–37. [PubMed] [Google Scholar]
- 23.Bose S, Stratford FL, Broadfoot KI, Mason GG, Rivett AJ. Phosphorylation of 20S proteasome alpha subunit C8 (alpha7) stabilizes the 26S proteasome and plays a role in the regulation of proteasome complexes by gamma-interferon. Biochem J. 2004;378(1):177–184. doi: 10.1042/BJ20031122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang F, et al. Proteasome function is regulated by cyclic AMP-dependent protein kinase through phosphorylation of Rpt6. J Biol Chem. 2007;282:22460–22471. doi: 10.1074/jbc.M702439200. [DOI] [PubMed] [Google Scholar]
- 25.Dephoure N, et al. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Djakovic SN, Schwarz LA, Barylko B, DeMartino GN, Patrick GN. Regulation of the proteasome by neuronal activity and calcium/calmodulin-dependent protein kinase II. J Biol Chem. 2009;284:26655–26665. doi: 10.1074/jbc.M109.021956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huttlin EL, et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kikuchi J, et al. Co- and post-translational modifications of the 26S proteasome in yeast. Proteomics. 2010;10:2769–2779. doi: 10.1002/pmic.200900283. [DOI] [PubMed] [Google Scholar]
- 29.Lee SH, Park Y, Yoon SK, Yoon JB. Osmotic stress inhibits proteasome by p38 MAPK-dependent phosphorylation. J Biol Chem. 2010;285:41280–41289. doi: 10.1074/jbc.M110.182188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Um JW, et al. ASK1 negatively regulates the 26 S proteasome. J Biol Chem. 2010;285:36434–36446. doi: 10.1074/jbc.M110.133777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zong C, et al. Regulation of murine cardiac 20S proteasomes: Role of associating partners. Circ Res. 2006;99:372–380. doi: 10.1161/01.RES.0000237389.40000.02. [DOI] [PubMed] [Google Scholar]
- 32.Li N, Zhang Z, Zhang W, Wei Q. Calcineurin B subunit interacts with proteasome subunit alpha type 7 and represses hypoxia-inducible factor-1alpha activity via the proteasome pathway. Biochem Biophys Res Commun. 2011;405:468–472. doi: 10.1016/j.bbrc.2011.01.055. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, et al. Determinants for dephosphorylation of the RNA polymerase II C-terminal domain by Scp1. Mol Cell. 2006;24:759–770. doi: 10.1016/j.molcel.2006.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim Y, et al. A conserved phosphatase cascade that regulates nuclear membrane biogenesis. Proc Natl Acad Sci USA. 2007;104:6596–6601. doi: 10.1073/pnas.0702099104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Archambault J, et al. An essential component of a C-terminal domain phosphatase that interacts with transcription factor IIF in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:14300–14305. doi: 10.1073/pnas.94.26.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeo M, Lin PS, Dahmus ME, Gill GN. A novel RNA polymerase II C-terminal domain phosphatase that preferentially dephosphorylates serine 5. J Biol Chem. 2003;278:26078–26085. doi: 10.1074/jbc.M301791200. [DOI] [PubMed] [Google Scholar]
- 37.Kobor MS, et al. An unusual eukaryotic protein phosphatase required for transcription by RNA polymerase II and CTD dephosphorylation in S. cerevisiae. Molecular Cell. 1999;4:55–62. doi: 10.1016/s1097-2765(00)80187-2. [DOI] [PubMed] [Google Scholar]
- 38.Yeo M, et al. Small CTD phosphatases function in silencing neuronal gene expression. Science. 2005;307:596–600. doi: 10.1126/science.1100801. [DOI] [PubMed] [Google Scholar]
- 39.Yu X, Chini CCS, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 40.Zheng H, et al. Cloning and characterization of a novel RNA polymerase II C-terminal domain phosphatase. Biochem Biophys Res Commun. 2005;331:1401–1407. doi: 10.1016/j.bbrc.2005.04.065. [DOI] [PubMed] [Google Scholar]
- 41.Rual JF, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 42.Borodovsky A, et al. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 2001;20:5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Komander D, Clague MJ, Urbe S. Breaking the chains: Structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 44.Sakata E, et al. The catalytic activity of Ubp6 enhances maturation of the proteasomal regulatory particle. Mol Cell. 2011;42:637–649. doi: 10.1016/j.molcel.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 45.Beal R, Deveraux Q, Xia G, Rechsteiner M, Pickart C. Surface hydrophobic residues of multiubiquitin chains essential for proteolytic targeting. Proc Natl Acad Sci USA. 1996;93:861–866. doi: 10.1073/pnas.93.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newton K, et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–678. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 47.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 48.Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat Biotechnol. 2000;18:538–543. doi: 10.1038/75406. [DOI] [PubMed] [Google Scholar]
- 49.Shi Y. Serine/threonine phosphatases: Mechanism through structure. Cell. 2009;139:468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 50.Zhang M, Cho EJ, Burstein G, Siegel D, Zhang Y. Selective inactivation of a human neuronal silencing phosphatase by a small molecule inhibitor. ACS Chem Biol. 2011;6:511–519. doi: 10.1021/cb100357t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bingol B, et al. Autophosphorylated CaMKII[alpha] acts as a scaffold to recruit proteasomes to dendritic spines. Cell. 2010;140:567–578. doi: 10.1016/j.cell.2010.01.024. [DOI] [PubMed] [Google Scholar]
- 52.Wojcik C, DeMartino GN. Intracellular localization of proteasomes. Int J Biochem Cell Biol. 2003;35:579–589. doi: 10.1016/s1357-2725(02)00380-1. [DOI] [PubMed] [Google Scholar]
- 53.Lee BH, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kisselev AF, Goldberg AL. Monitoring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Methods Enzymol. 2005;398:364–378. doi: 10.1016/S0076-6879(05)98030-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.