Histone methylation is an essential regulator of gene expression and chromatin structure. In this Perspective highlighting an article published in this issue, the authors discuss how demethylases achieve substrate specificity (see Sengoku and Yokoyama).
Keywords: epigenetics, UTX/KDM6A, histone demethylase, jumonji domain, crystal structure, H3K27 methylation
Abstract
Histone methylations are highly regulated by site-specific histone methyltransferases and demethylases. In this issue of Genes & Development, Sengoku and Yokoyama (pp. 2266–2277) demonstrate that a novel Zn-binding domain and the Jumonji domain of UTX/KDM6A (Lys demethylase 6A) recognize histone H3 and together function as a substrate specificity determinant for H3K27 demethylation. This study demonstrates the mechanism of site-specific demethylation by UTX/KDM6A and implicates that histone demethylases use diverse methods to accomplish target specificity.
Histone methylations in chromatin regulate gene expression by either recruiting chromatin modifiers or altering chromatin structure (Chi et al. 2010). The biological consequences of histone methylations differ depending on the site of modification. Methylations of histone H3K4 and H3K36 are highly correlated with transcription activation, while methylations of histone H3K9 and H3K27 are associated with transcription repression. Therefore, it is essential to regulate histone methylation site-specifically. Several site-specific histone methyltransferases are involved in writing methyl marks. For example, the mixed-lineage leukemia (MLL) complex methylates histone H3K4, and the Polycomb-repressive complex 2 methylates histone H3K27 (Muller et al. 2002; Yokoyama et al. 2004). Although an SET domain-containing subunit in the complexes is capable of catalyzing methylation, other subunits in the complexes also contribute to the activity in vivo. In addition to the location of methylation, the level of methylation adds to the complexity of the methyl marks. An ɛ-amino group of Lys can be modified with mono-, di-, and trimethyl groups, and the different levels of methylation on the same residue lead to different biological consequences. A single methyltransferase is capable of catalyzing mono-, di-, and trimethylation of the same residue, but the level of methylation is highly regulated (Shahbazian et al. 2005). For example, histone H3K4 trimethylation (H3K4me3) is regulated by another histone modification, H2B ubiquitylation. However, the exact mechanism of regulating the level of methylation remains unknown.
Histone methylation was once thought to be a permanent mark that could not be erased enzymatically due to the strong N–C bonding. Therefore, alternative mechanisms were proposed, such as histone clipping and histone replacement. However, dynamic changes in the methylation pattern during gene expression indicated the existence of histone demethylase enzymes (Borun et al. 1972; Annunziato et al. 1995). The first histone demethylase identified was LSD1, which was discovered by Yang Shi's group (Shi et al. 2004). LSD1 was isolated as a component of the CtBP corepressor complex and belongs to the amineoxidase family. LSD1 removes mono- and dimethyl groups from methylated histone H3K4. To date, many histone demethylases have been identified and are classified into two families, the LSD1 demethylase family and the Jumonji (Jmj) domain-containing demethylase family, depending on their domain structures and reaction mechanisms (Mosammaparast and Shi 2010). Each family contains several subfamilies based on their enzymatic target residues. The two demethylase families use radically different mechanisms to remove methyl groups. The LSD1 demethylase family removes a methyl group via an amine oxidation process in the presence of a FAD cofactor. Because the amine oxidation process requires a protonated nitrogen at the ɛ-amino group of Lys, LSD1 cannot remove a trimethyl group from the methylated Lys. The Jmj domain-containing demethylase family uses α-ketoglutarate and Fe(II) as cofactors for hydroxylating a methyl group, eventually leading to demethylation. Therefore, the Jmj domain-containing demethylase family can remove the trimethyl group. The different levels of methylation at the same residue are removed by different enzymes belonging to the two different families. LSD1 acts at the mono- and dimethylated histone H3K4, whereas the Jmj domain-containing protein JARID1 is responsible for demethylating di- and trimethylated histone H3K4 (Seward et al. 2007).
Considering the different biological consequences resulting from the various locations and levels of methylation, the histone demethylase activity needs to be highly specific and should discriminate between the different locations and levels of methylation on histone Lys residues. Despite the importance of target specificity, several biochemical studies have shown that histone demethylases show promiscuous specificity for target sites. Crystal structures of Jmj domains have been solved and show that the Jmj domain recognizes histone substrates mainly by interacting with the backbone of the histone, illustrating multiple target specificity for these enzymes (Chen et al. 2007; Couture et al. 2007; Ng et al. 2007). However, considering the importance of demethylase specificity, it might be tightly regulated in vivo by other unknown mechanisms.
UTX/KDM6A (Lys demethylase 6A) belongs to the Jmj domain-containing family and has been identified to regulate HOX gene expression by demethylating di- and trimethylated H3K27 (Agger et al. 2007; Hong et al. 2007). UTX/KDM6A is highly conserved among species and contains two conserved domains: a TRP repeat and a Jmj domain. Considering that H3K27 and H3K9 residues reside in identical sequences—“A-R-Kme-S”—it is important that UTX/KDM6A specifically recognizes H3K27 over H3K9. However, the mechanism behind the target specificity of UTX/KDM6A was unknown until now. In this issue of Genes & Development, Sengoku and Yokoyama (2011) report the crystal structure of UTX/KDM6A and demonstrate that a novel Zn-binding domain in UTX/KDM6A together with the Jmj catalytic domain confers target specificity. This work illustrates the diverse mechanisms for target specificity of histone-modifying enzymes.
The catalytic domain of UTX/KDM6A binds histone H3 in a bifurcated manner
Sengoku and Yokoyama (2011) present the crystal structure of the catalytic domain of UTX, including the Jmj domain and the highly conserved C-terminal region. The structure shows that the highly conserved C-terminal region contains a helical domain and a Zn-binding domain, which had not been detected by sequence analysis. These two domains, together with the Jmj domain, comprise the UTX/KDM6A catalytic domain. The Jmj domain of UTX/KDM6A contains a jellyroll fold flanked by α helices, which is similar to other Jmj domains in the demethylase family. The Zn-binding domain is inserted in the middle of the helical domain and is tightly associated with the other two domains to stabilize the overall structure, positioning the Jmj domain in proximity to the Zn-binding domain.
Sengoku and Yokoyama (2011) also determined the complex structure of the UTX/KDM6A catalytic domain with a histone H3 substrate, illustrating the mechanism for the substrate specificity of UTX/KDM6A. In the complex structure, histone H3 bound to the UTX/KDM6A catalytic domain adopts an extended conformation, interacting with the Jmj domain and the Zn-binding domain in a bifurcated manner. The C-terminal portion of histone H3 (H3K25 to H3G33) is recognized by the Jmj domain, and the N-terminal portion of histone H3 (H3A17 to H3T22) is recognized by the Zn-binding domain. The Jmj domain interacts with histone H3 via both the backbone and side chains of histone H3. The Leu 1127 and Gln 1133 residues of the Jmj domain hold the backbone of the substrate. Consequently, the target residue H3K27me3 is sharply bent and inserted into the catalytic pocket. This target bending was also observed in the JMJD2A–H3 complex structure, indicating that bending of the target residue may be a general mechanism used to accommodate target residues in a small catalytic cavity (Couture et al. 2007). Due to the bending, histone H3A25 and H3A29 are restricted toward the small pockets in the Jmj domain. These pockets are too small to accommodate residues other than alanine, contributing to the target specificity because the target sequence flanked by two alanines, “A-R-Kme-S-A,” is unique to histone H3K27. The Jmj domain also recognizes the side chains of the substrate through its extensive interactions with the H3A26 side chain. This binding mode enables UTX/KDM6A to discriminate histone H3K27 over H3K9 because “A-R-Kme-S-A” is the unique sequence around H3K27. Sengoku and Yokoyama (2011) show that these residues are critical for UTX/KDM6A to demethylate the substrate using molecular modeling and mutagenesis studies.
The most notable finding of this study is that the novel Zn-binding domain is involved in recognizing an N-terminal portion (H3A17 to H3T22) of the histone H3 target site. The Zn-binding domain adopts a novel fold, and the four conserved cysteine residues coordinate a Zn ion to stabilize the structure. Interestingly, the Zn-binding domain undergoes a significant conformational change upon binding to the N-terminal portion of histone H3, and this change exposes a hydrophobic patch composed of His 1320, His 1329, Leu 1342, and Val 1356 by displacing Tyr 1354, which was masking this hydrophobic patch. Among the residues in the N-terminal portion of histone H3, H3A17 and H3L20 exhibit extensive interactions with the hydrophobic patch. Because H3L20 is found only in the context of the H3K27 target, the Zn-binding domain is likely to serve as a substrate determinant for the UTX/KDM6A demethylase. The notion that the UTX/KDM6A recognizes a relatively large portion of histone H3 with two domains highlights the highly specific activity of UTX/DKM6A toward H3K27.
Diverse ways to be specific
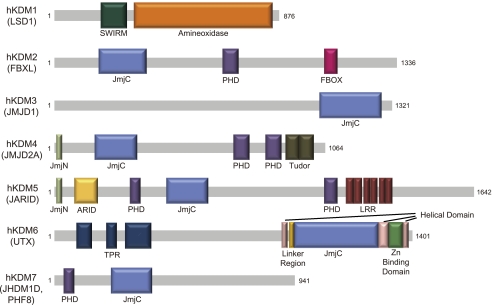
Histone methylation is a complex modification process due to the combination of locations and levels. Therefore, tight regulation of target specificity is needed. Histone demethylases are categorized into seven classes depending on their domain structures and targets (Fig. 1). Several demethylases use other domains to recognize other histone modifications, thereby increasing their specificities. KDM7 (JHDM1D and PHF8) contains a plant homeodomain (PHD) and demethylates H3K9me2 and H3K27me2. Structural and biochemical studies showed that the PHD domain binds trimethylated H3K4 and that PHF8 demethylates the substrate better when the histone substrate contains H3K4me3. However, the presence of trimethylated H3K4 decreases the activity of JHDM1D toward H3K9me2 demethylation but does not affect the H3K27me3 demethylase activity (Horton et al. 2010). Another demethylase, KDM4 (JMJD2A), contains two PHDs and a double Tudor domain, and demethylates both H3K9me2/3 and H3K36me2/3. The double Tudor domain binds to H3K4me3 and H4K20me3, implicating that these modifications affect JMJD2A activity (Huang et al. 2006). Other domains in the demethylases are likely to contribute to the demethylase activity by recognizing other histone modifications or by targeting the demethylases to the target locus. In addition to the conserved domains, there are highly conserved regions outside of the identified domains. Because most structural and biochemical studies are limited to the identified domains, the functions and structures of these highly conserved regions are unknown. The study by Sengoku and Yokoyama (2011) illustrates that other unidentified conserved regions may be involved in histone recognition.
Figure 1.
Histone demethylases. Histone demethylases are classified into seven classes depending on their reaction mechanisms and domain structures. Human KDM1 (hKDM1) contains an amineoxidase domain, and the other demethylases (hKDM2 to hKDM7) have a JmjC domain as the catalytic domain. Histone demethylases contain other conserved domains involved in various functions.
In histone methyltransferases, modifications in the residues in proximity to the target residue affect the histone methyltransferase activity, suggesting cross-talk between histone modifications. The notion that UTX/KDM6A recognizes the side chains of histone H3 and that some of these side chains can be methylated raises a possibility of cross-talk in histone demethylation. Specifically, histone H3A17 and H3A26 can be methylated by CARM1/PRMT4, and the Zn-binding domain and the Jmj domain interact with Arg 17 and Arg 26, respectively. Because UTX/KDM6A tightly holds the charged side chains of the histone H3A17 and H3A26 residues, methylation of Arg 17 and Arg 26 would affect the UTX/KDM6A demethylase activity. Sengoku and Yokoyama (2011) showed that, indeed, the methylation of Arg 17 or Arg 26 in the substrate decreased the demethylase activity of UTX/KDM6A. It would be interesting to examine whether there is cross-talk between the demethylation of histone H3K27 and the methylation of Arg 17 and Arg 26 in vivo.
In vivo, histone demethylases do not function alone; they form large complexes with other histone-modifying enzymes (Cho et al. 2007; Issaeva et al. 2007; Lee et al. 2007; Pasini et al. 2008). UTX/KDM6A is reported to form a complex with the MLL histone H3K4 methyltransferase complex, suggesting that UTX/KDM6A removes the histone H3K27 methyl mark when the MLL complex methylates histone H3K4, which are antagonistic methyl marks for gene expression (Lee et al. 2007). However, the contribution of other subunits in the demethylase complex to the demethylase activity needs to be investigated.
Conclusion
Accomplishing target specificity is a great challenge for histone demethylases due to the complex combination of target locations and levels. To overcome this challenge, histone demethylases adopt diverse strategies. In addition to identified domains, histone demethylases contain many highly conserved regions, which may contribute to the target specificity of histone demethylases in unexpected ways, as shown in the study by Sengoku and Yokoyama (2011).
Acknowledgments
We thank Sei Young Lee for the comments on the manuscript. J.S. is partially supported by the WCU program (R31-2008-000-10071-0) and a grant (A100525) from the Ministry of Health Welfare and Family Affairs.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.179473.111.
References
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K 2007. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449: 731–734 [DOI] [PubMed] [Google Scholar]
- Annunziato AT, Eason MB, Perry CA 1995. Relationship between methylation and acetylation of arginine-rich histones in cycling and arrested HeLa cells. Biochemistry 34: 2916–2924 [DOI] [PubMed] [Google Scholar]
- Borun TW, Pearson D, Paik WK 1972. Studies of histone methylation during the HeLa S-3 cell cycle. J Biol Chem 247: 4288–4298 [PubMed] [Google Scholar]
- Chen Z, Zang J, Kappler J, Hong X, Crawford F, Wang Q, Lan F, Jiang C, Whetstine J, Dai S, et al. 2007. Structural basis of the recognition of a methylated histone tail by JMJD2A. Proc Natl Acad Sci 104: 10818–10823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG 2010. Covalent histone modifications—miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer 10: 457–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, Guszczynski T, Dressler GR, Copeland TD, Kalkum M, et al. 2007. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem 282: 20395–20406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couture JF, Collazo E, Ortiz-Tello PA, Brunzelle JS, Trievel RC 2007. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat Struct Mol Biol 14: 689–695 [DOI] [PubMed] [Google Scholar]
- Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K 2007. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci 104: 18439–18444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JR, Upadhyay AK, Qi HH, Zhang X, Shi Y, Cheng X 2010. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat Struct Mol Biol 17: 38–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM 2006. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science 312: 748–751 [DOI] [PubMed] [Google Scholar]
- Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, Croce CM, Nakamura T, Mazo A, Eisenbach L, Canaani E 2007. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol 27: 1889–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di Croce L, Shiekhattar R 2007. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 318: 447–450 [DOI] [PubMed] [Google Scholar]
- Mosammaparast N, Shi Y 2010. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem 79: 155–179 [DOI] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197–208 [DOI] [PubMed] [Google Scholar]
- Ng SS, Kavanagh KL, McDonough MA, Butler D, Pilka ES, Lienard BM, Bray JE, Savitsky P, Gileadi O, von Delft F, et al. 2007. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature 448: 87–91 [DOI] [PubMed] [Google Scholar]
- Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K 2008. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes Dev 22: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengoku T, Yokoyama S 2011. Structural basis for histone H3 Lys 27 demethylation by UTX/KDM6A. Genes Dev (this issue). doi: 10.1101/gad.172296.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seward DJ, Cubberley G, Kim S, Schonewald M, Zhang L, Tripet B, Bentley DL 2007. Demethylation of trimethylated histone H3 Lys4 in vivo by JARID1 JmjC proteins. Nat Struct Mol Biol 14: 240–242 [DOI] [PubMed] [Google Scholar]
- Shahbazian MD, Zhang K, Grunstein M 2005. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol Cell 19: 271–277 [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119: 941–953 [DOI] [PubMed] [Google Scholar]
- Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, Herr W, Cleary ML 2004. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol 24: 5639–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]