In this Review article, Zaret and Carroll discuss how pioneer transcription factors can influence chromatin structure and activation of gene expression during cellular reprogramming.
Keywords: pioneer factors, transcription, activation, competence, development, steroid hormone receptors, cancer
Abstract
Transcription factors are adaptor molecules that detect regulatory sequences in the DNA and target the assembly of protein complexes that control gene expression. Yet much of the DNA in the eukaryotic cell is in nucleosomes and thereby occluded by histones, and can be further occluded by higher-order chromatin structures and repressor complexes. Indeed, genome-wide location analyses have revealed that, for all transcription factors tested, the vast majority of potential DNA-binding sites are unoccupied, demonstrating the inaccessibility of most of the nuclear DNA. This raises the question of how target sites at silent genes become bound de novo by transcription factors, thereby initiating regulatory events in chromatin. Binding cooperativity can be sufficient for many kinds of factors to simultaneously engage a target site in chromatin and activate gene expression. However, in cases in which the binding of a series of factors is sequential in time and thus not initially cooperative, special “pioneer transcription factors” can be the first to engage target sites in chromatin. Such initial binding can passively enhance transcription by reducing the number of additional factors that are needed to bind the DNA, culminating in activation. In addition, pioneer factor binding can actively open up the local chromatin and directly make it competent for other factors to bind. Passive and active roles for the pioneer factor FoxA occur in embryonic development, steroid hormone induction, and human cancers. Herein we review the field and describe how pioneer factors may enable cellular reprogramming.
Genes in eukaryotic cells are packaged into chromatin, allowing the condensation of 2 m of DNA into a nucleus of microns in diameter. The primary means of condensation is by the wrapping of DNA around an octamer of the four core histones to create the nucleosome core particle (Kornberg 1977). Arrays of nucleosome cores can be condensed by binding of linker histone near the nucleosome dyad axis (Zhou et al. 1998), binding of corepressor complexes (Fan et al. 2004; Francis et al. 2004; Sekiya and Zaret 2007), and folding of nucleosomes into higher-order structures (Schwarz and Hansen 1994). In the process, substantial surface area of DNA faces the histone octamer (Luger et al. 1997), and DNA access can be further occluded by juxtaposition to nearby nucleosomes in chromatin fibers (Schalch et al. 2005). Within this context, transcription factors and other DNA-transacting proteins must locate their target sequences and open the path for subsequent functions, such as transcription, replication, and repair.
How do DNA-binding factors find their targets amidst the tangle of nuclear chromatin? The ability to genetically reprogram cells in the absence of DNA replication (Chiu and Blau 1984) means that the problem is not generally solved by simply having increased accessibility during chromatin assembly in S phase. What is the role of DNA sequence in destabilizing local nucleosome structure to allow factor binding? Do different transcription factors have different properties with regard to target site access? How do histone modification states and chromatin remodelers alter target accessibility? Herein we review these questions with a focus on “pioneer” factors, a special class of proteins found to be able to access their DNA target sites in chromatin when other factors cannot and to continue to access the DNA prior to the time of transcriptional activation. We consider the role of pioneer factors in endowing the competence for genes to be expressed. Finally, we discuss the role of transcriptional competence in the context of development, stem cells, and hormonal regulation, where pioneer factors play crucial roles in cell programming and conferring the ability of cells to respond to environmental cues.
Parameters affecting transcription factor access to target sites in chromatin
Genome-wide location analyses have shown that all known transcription factors physically occupy less than a few percent of their consensus target sites in the genome (Carr and Biggin 1999; Iyer et al. 2001; Yang et al. 2006; Joseph et al. 2010; Kaplan et al. 2011). This simple observation indicates that chromatin features beyond DNA sequence dictate where and when transcription factors bind their target sites. It also raises the question of what chromatin modifications may enhance transcription factor binding. Coupling consensus sequence information with the genomic location of other transcription factors in the same cell (Filion et al. 2010) can greatly improve the ability to predict whether a factor will be found to bind a given target site. The importance of cooperative binding is reflected by the observation that many transcription factors individually cannot occupy their target sites on nucleosomal DNA in vitro, but cooperative interactions among a group of such factors can allow nucleosome binding (Adams and Workman 1995; Steger and Workman 1997).
The location of histone-modifying enzymes, such as the lysine acetyltransferase p300 at enhancers (Visel et al. 2009), and histone modifications, such as histone H3K4 methylation at active enhancers and promoters (Heintzman et al. 2007; Mikkelsen et al. 2007; He et al. 2010), can also help predict whether a transcription factor target sequence will be occupied. Recent studies indicate that specific complexes of factors directly bind to specific combinations of modified histones (Bartke et al. 2010) apparently sensitive to specific histone codes (Turner 1993; Strahl and Allis 2000; Li et al. 2006). The presence of certain histone modifications in chromatin can affect both transcription factor access (e.g., Shogren-Knaak et al. 2006; Martino et al. 2009) and transcriptional initiation (Vignali et al. 2000; Li et al. 2010). Clusters of transcription factor-binding sites (e.g., at promoters and enhancers) are often DNase I-hypersensitive and thereby open for factors to bind (Boyle et al. 2011; John et al. 2011; Kaplan et al. 2011; XY Li et al. 2011; Pique-Regi et al. 2011). While current studies indicate that long, noncoding RNAs can be associated with specific genomic regions and associated regulatory factors (KC Wang et al. 2011), it remains to be determined whether such RNAs play an initiating role in targeting themselves or transcription factors to DNA.
However, all of the above observations do not address how general coregulators, chromatin modifiers, histone modifications, and hypersensitivity become localized to genomic sites in the first place and thus fail to resolve the issue of how regulatory events are initiated at particular genomic locations.
Nucleosome positioning or depletion can also affect transcription factor binding. Mechanisms for destabilizing nucleosomes in vivo, and thus increasing transcription factor access, include the presence of poly(dA-dT) sequences (Sekinger et al. 2005) and ATP-dependent nucleosome remodeling complexes (Boeger et al. 2003; Reinke and Horz 2003). The pervasiveness of apparent nucleosome-free or nucleosome-destabilized regions at promoters throughout the genome (Yuan et al. 2005; Mavrich et al. 2008) led to the hypothesis that such regions possess sequences that intrinsically destabilize the nucleosome (Segal et al. 2006). However, reconstitution of genomes into nucleosome arrays in vitro indicates that nucleosome positioning and destabilization are only partially due to intrinsic DNA sequence (Zhang et al. 2009; Valouev et al. 2011). Indeed, addition of a soluble nuclear extract to the yeast genome reconstituted into nucleosomes in vitro can recapitulate much of the nucleosomal pattern seen in vivo (Zhang et al. 2011). Histone variants, such as H3.3 and H2A.Z, at promoters can also destabilize nucleosomes (Zhang et al. 2005; Jin and Felsenfeld 2007; Jin et al. 2009). To summarize, many or most regulatory regions do not coincide with DNA sequence that assembles into intrinsically unstable nucleosomes. Thus, other mechanisms are needed to expose the DNA. These considerations once again raise the issue of how general chromatin remodeling factors and histone variants that modulate nucleosome position or stability are initially targeted to specific regions of chromatin.
Initiating events in chromatin: pioneer factors bind first
Chromatin immunoprecipitation (ChIP) and in vivo footprinting have been the primary methods for identifying transcription factors that bind silent chromatin prior to target gene activation. While ChIP-based approaches can provide genome-wide views and are currently the most frequently used methods for such analysis, they rely on prior knowledge of factors to test for binding, and each factor must be queried separately. In contrast, in vivo footprinting only scans a several-hundred-base-pair region and, based on the sequence being occupied, provides clues to binding factor identities. However, in vivo footprinting can comprehensively reveal DNA occupancy states and, importantly, can show when a region is not occupied by factors. This latter point is crucial for discovering which factors bind DNA first and thus may be pioneer factors.
For example, in vivo footprinting analysis of the liver-specific enhancer of the Alb1 gene in adult mouse liver revealed a continuous array of five factor-binding sites occupied over a 120-base-pair (bp) region that is important for enhancer activity (Liu et al. 1991; McPherson et al. 1993). Six binding sites were occupied at the same sequence in nascent liver buds in mouse embryos (Gualdi et al. 1996). However, in undifferentiated gut endoderm, in which the Alb1 gene is silent but competent for activation, only two of the six factor-binding sites were occupied (Gualdi et al. 1996). The binding sites were for FoxA and GATA factors, which are expressed in the endoderm and nascent liver (Ang et al. 1993; Arceci et al. 1993; Monaghan et al. 1993; Laverriere et al. 1994) and whose binding sites are crucial for Alb1 enhancer function (Liu et al. 1991; Bossard and Zaret 1998). FoxA1, FoxA2, and FoxA3 are highly related members of the Forkhead box (Fox) family of transcription factors (Hannenhalli and Kaestner 2009), and FoxA1 and FoxA2 are redundantly expressed in the foregut endoderm and are necessary for liver induction (Lee et al. 2005). GATA-4 and GATA-6 are redundantly expressed in the foregut endoderm and are necessary for early liver development (Holtzinger and Evans 2005; Zhao et al. 2005; Watt et al. 2007). Thus, the FoxA and GATA factors were the first detectable factors to engage the enhancer in development, and their expression is required for the induction of the liver program. While the specific dimethyl sulfate protection technique used for in vivo footprinting could have failed to detect other factors, FoxA and GATA site occupancy clearly preceded occupancy of the other enhancer-binding factors in nascent liver and occurred in progenitor cells prior to gene activation.
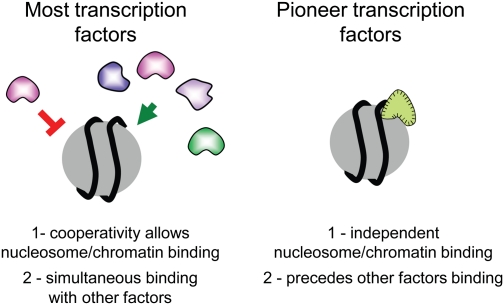
By these criteria, FoxA and GATA are initial and important chromatin-binding factors, and as such were termed pioneer factors. A distinction about the designation of pioneer factors is that they not only perform a genetic function early in the activation of transcription, but they have been physically shown to bind to the genome for a period prior to activation and prior to other factors binding and, as such, impart competence for activity (Fig. 1). Furthermore, as described below, pioneer factors have the special property of being able to bind their target sites in condensed chromatin.
Figure 1.
Properties that distinguish pioneer factors from other transcription factors. Most transcription factors cannot access their target sequences on nucleosomes or in compacted chromatin, yet can do so when binding in a highly cooperative fashion with other transcription factors. Pioneer factors can access their target sequences on nucleosomes and certain forms of compacted chromatin, although not all forms. Pioneer factor binding occurs for a stable period and precedes the binding of other transcription factors.
In the time since the discovery of FoxA and GATA factors in this context, it has become clear that homologs of both factors play crucial roles within a network for gut development that is evolutionarily conserved in all metazoans (Davidson and Erwin 2006). Notably, the activation of genes involved in pharyngeal development by PHA-4, the Caenorhabditis elegans homolog of FoxA, is, in the first approximation, in the order of PHA-4's apparent affinity to its target binding sites (Gaudet and Mango 2002; Fakhouri et al. 2010). That is, the genes with sites containing the highest similarity to the binding consensus sequence for PHA-4 are usually expressed early in development, when PHA-4 is first expressed, and genes that have sites with weaker similarity to the consensus sequence for PHA-4 are induced later in development, at the time of full PHA-4 expression. This suggests that for many PHA-4 target sites, although not all, simple affinity for DNA is a primary determinant of PHA-4 binding, rather than other features of chromatin structure.
On the other hand, careful analysis of genome-wide FoxA binding in the adult mouse liver revealed that FoxA target sites with a medium or weak consensus binding sequence were at genes that are much more liver-specific than genes with a strong consensus sequence (Tuteja et al. 2008). Simple target site affinity was not a predictor of FoxA occupancy, at least in terminally differentiated cells. While the nature of this is discussed in detail below (and see also Hoffman et al. 2010), Tuteja et al. (2008) did find that medium- and lower-affinity, liver-specific targets were highly enriched for binding of other hepatic nuclear factors, suggesting a role for cooperative binding during the maintenance of differentiation. Furthermore, in adult liver tissue, FoxA1 and FoxA2 binding was not required to maintain local nucleosome organization (Z Li et al. 2011). While the two factors together are also not necessary for late fetal hepatocyte differentiation (Li et al. 2009), they are absolutely essential for the embryonic initiation of hepatic differentiation (Lee et al. 2005). Also, from the fetal stages onward, a third homolog, FoxA3, is expressed in the liver and apparently provides compensation (Z Li et al. 2011). Indeed, FoxA3 along with GATA4 are among three factors that together can potently reprogram fibroblasts to hepatocyte-like cells (Huang et al. 2011). When the technology evolves to enable the interrogation of genomic occupancy at the single-cell level, it will be interesting to determine the impact of FoxA and GATA factors on chromatin structure in the embryonic endoderm and during the initial specification of the hepatic lineage.
In the meantime, many other examples of early chromatin binding have emerged. In B-cell development, in vivo footprinting and ChIP show that an intronic enhancer of the Pax5 gene is bound and ultimately regulated by PU.1, IRF4, IRF8, and NF-KB in multipotent hematopoietic progenitors prior to the time of Pax5 activation (Decker et al. 2009). PU.1 itself can expand the linker region between nucleosomes and promote local histone modifications, likely contributing to its ability to enhance binding of other factors (Ghisletti et al. 2010; Heinz et al. 2010). PU.1 and RUNX1 prime transcription of the c-fms/csf1R gene, with RUNX1 being needed only transiently (Krysinska et al. 2007; Hoogenkamp et al. 2009). There are other examples where initial factor binding to enhancers is only transiently required for transcriptional activation (Thomassin et al. 2001; Chong et al. 2010). Taken together with the above discussion of FoxA, it suggests that pioneer factors may function more for initiating a developmental lineage than maintaining it.
Recently, it has become appreciated that various tissue-specific genes that are activated late in differentiation are occupied by transcription factors in embryonic stem (ES) cells, when the genes are silent (Xu et al. 2007, 2009; Liber et al. 2010). These studies revealed that distinct transcription factors, including FoxD3, Sox2, and Sp1, can occupy late differentiation genes in ES cells (Ram and Meshorer 2009; Smale 2010). Notably, FoxD3 was shown to block methylation of an underlying CpG sequence at the silent Alb1 gene in ES cells, demonstrating that pioneer factor occupancy can modulate the local epigenetic state of chromatin (Xu et al. 2009). Furthermore, FoxD3 may serve as a placeholder until the later appearance of FoxA1 during gastrulation. While it is not straightforward to determine the functionality of a factor binding in ES cells for a gene such as Alb1, which is induced many developmental steps later, the emerging picture is that silent genes in progenitor cells may be generally marked for potential activity by pioneer factors. The pervasiveness of this phenomenon in ES cells underscores the importance of the hierarchical nature of factor binding and provides new insights into the basis for cell programming.
Active and passive means by which pioneer factors are useful in development
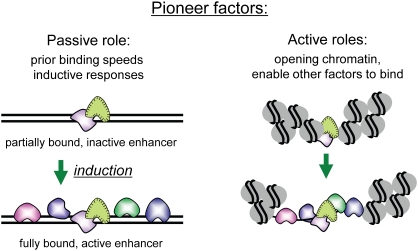
In principle, pioneer factors in embryonic progenitor and stem cells can enable development by active or passive means. Pioneer factor binding could have no mechanistic consequence, in terms of altering chromatin structure and/or allowing other factors to bind, but the factors' mere presence at a regulatory sequence could reduce the number of subsequent factor-binding events that are needed for transcriptional activation at a later time in development. This passive mode is supported by the observation that the vast majority of promoters and enhancers, particularly those involved in tissue-specific expression, require binding by a combination of transcription factors for activity (Boyer et al. 2005; MacArthur et al. 2009; Levine 2010). By having one or a few factors bound in undifferentiated progenitor cells that are not sufficient to induce transcription, a target gene can be more rapidly activated when the final rate-limiting factors appear; e.g., in response to developmental inductive signals (Fig. 2). This will allow tissue specification and embryogenesis to proceed quickly. An example of this is at the aforementioned Alb1 enhancer, where FoxA and GATA factor sites are bound in the endoderm, allowing a rapid and synchronous transcriptional response in the cell population receiving liver-inductive signals. As discussed below, after differentiation, this passive role of pioneer factors—i.e., simply “being there” prior to gene expression—can help confer rapid responses to hormonal induction.
Figure 2.
Passive and active roles for pioneer factors in endowing transcriptional competence. In the passive role, stable, prior binding of pioneer factors to a complex regulatory sequence, such as an enhancer element, reduces the number of additional factors that are needed to bind at a later time in order to create an active enhancer. Such “priming” can increase the rapidity of a transcriptional response and is seen during development and in hormonal regulation (Gualdi et al. 1996; Carroll et al. 2005). In the active role, pioneer factors can directly facilitate other factors binding to nucleosomal DNA (Cirillo and Zaret 1999) or open up the local chromatin and thereby indirectly allow other factors to bind (Cirillo et al. 2002).
Pioneer factors can also function actively by helping to open or organize the local chromatin, in turn allowing the binding of other transcription factors, chromatin modifiers, and nucleosome remodelers. For example, direct assessments of FoxA1 and GATA-4, in comparison with other Alb1 enhancer-binding proteins, showed that these pioneer factors were uniquely capable of binding to their target sites in highly compacted chromatin in vitro and generating a local region of DNase or restriction enzyme sensitivity (Cirillo et al. 2002). The FoxA factor was more active than the GATA factor in this regard, and studies of FoxE and FoxO have shown that they can similarly open compacted chromatin (Cuesta et al. 2007; Hatta and Cirillo 2007). Notably, these studies found that the opening activity is intrinsic to the transcription factor binding its target in chromatin (and see below) and does not require ATP or an ATP-dependent nucleosome remodeler. The ability of FoxA factors to open chromatin in vivo has been demonstrated by elegant studies of PHA-4, the C. elegans homolog, whose binding to reiterated sites in compacted chromatin in vivo leads to extensive decompaction of the chromosomal domain (Fakhouri et al. 2010). In early development, PHA-4 leads to the recruitment of the histone variant H2A.Z to promoters, which in turn can contribute to local chromatin opening (Updike and Mango 2006). With regard to FoxA1, a C-terminal domain of the protein, independent of the central, DNA-binding domain, binds to core histones and is necessary for chromatin opening in vitro (Cirillo et al. 2002). A simple explanation for the chromatin opening activity is that by binding si multaneously to the DNA and the core histones, FoxA1 disrupts local internucleosomal interactions that are known to stabilize chromatin higher-order structure (Schalch et al. 2005). Thus, the FoxA1 pioneer factor has evolved a special mechanism for keeping open a local domain of chromatin and allowing the entry of other factors. Chromatin opening properties have also been seen for the basic helix–loop–helix (bHLH) factor TFE3, which, in vitro, induces hypersensitivity in chromatin and mediates local nucleosome positioning (Ishii et al. 2004).
The heart of pioneer factor activity: accessing target sequences on nucleosomes
Both of the active and passive functions of pioneer factors involve an initial binding step to DNA target sites embedded in chromatin. As noted above, it is clear that cooperativity among binding partners can be sufficient for many kinds of transcription factors to simultaneously engage their target sites on nucleosomes or in compacted chromatin and activate gene expression. However, in cases in which the binding of various factors to a regulatory sequence is sequential in time, and thus not initially cooperative, intrinsic capacities to bind nucleosomal DNA and compacted chromatin can be significant.
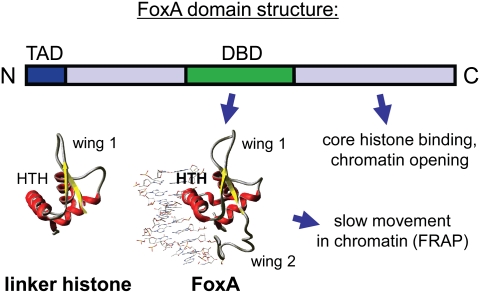
A notable feature of FoxA pioneer factors is that the crystal structure of their DNA-binding domain highly resembles that of linker histone, an avid nucleosome-binding protein (Fig. 3; Clark et al. 1993; Ramakrishnan et al. 1993). Both FoxA and linker histone use the “winged helix” motif to bind along one “face” of DNA along the long axis, which allows other proteins, such as core histones, to bind on the other side. Indeed, FoxA proteins can bind their target sites on a nucleosome core particle in vitro and in vivo, and the DNA-binding domain alone is sufficient for such binding (Cirillo et al. 1998; Chaya et al. 2001). While the FoxA factors have base-specific contacts with DNA that are necessary for site-specific binding (Clark et al. 1993; Sekiya et al. 2009), linker histone lacks such contacts and does not bind DNA sites specifically. In contrast, the globular domain of linker histone has a second DNA-binding interface that allows the protein to bind the DNA crossover point at or near the dyad axis of the nucleosome (Goytisolo et al. 1996). While FoxA factors lack the requisite basic residues on the “back” side of the globular domain to enable such binding, conversion of analogously positioned residues to basic residues in FoxA enables the factor to bind nucleosomes more tightly, similar to linker histone (Cirillo et al. 1998).
Figure 3.
FoxA factors possess features of linker histones and conventional transcription factors. The crystal structures of the DNA-binding domains of linker histone (Ramakrishnan et al. 1993) and FoxA3 (Clark et al. 1993) are shown side by side. The “winged helix” motif is evident in each, consisting of a helix–turn–helix motif (HTH) flanked by wings of polypeptide chain (wing 1 and wing 2) that make minor groove contacts along the long axis of the DNA. Such binding allows other proteins (e.g., core histones) to reside on the other side of the DNA. Like linker histone, the FoxA proteins move much more slowly in chromatin than most other transcription factors, as assessed by fluorescence recovery after photobleaching (FRAP) (Sekiya et al. 2009). Unlike linker histone, FoxA proteins make specific base contacts that direct target site binding and possess an N-terminal trans-activation domain (TAD) (Pani et al. 1992). Also, FoxA proteins possess a C-terminal domain that binds directly to core histone proteins and is necessary for the factor to open chromatin (Cirillo et al. 2002).
Notably, FoxA proteins move in nuclear chromatin much more slowly than other factors tested, implicating unusually tight chromatin binding in vivo (Sekiya et al. 2009). In certain contexts, FoxA binding can displace linker histone to allow other factors to bind chromatin (Taube et al. 2010). Also, like linker histone, FoxA and other Fox family members are retained on mitotic chromosomes and thus could serve as “bookmarking” proteins in mitosis (Yan et al. 2006; Zaret et al. 2011). In conclusion, the structure of the FoxA DNA-binding domain appears intermediate between that of linker histone and other transcription factors, and this feature contributes to pioneer activity.
There are several ways by which FoxA1 can enable the binding of other transcription factors to DNA in chromatin. The rate of association of FoxA for nucleosomes is low, apparently due to the need for proper rotational phasing of the target site on the nucleosome surface (Sekiya et al. 2009), but the rate of dissociation is much lower than that for free DNA (Cirillo and Zaret 1999). Thus, binding of FoxA1 can help stabilize a nucleosome position (Shim et al. 1998), which in turn can phase other local binding sites on and off of nucleosomes to modulate factor binding. Also, binding of FoxA to an already positioned nucleosome can enable the binding of other transcription factors that cannot bind on their own (Cirillo and Zaret 1999), apparently due to the cooperative effects described above (Adams and Workman 1995; Steger and Workman 1997). Finally, FoxA proteins have an N-terminal, trans-activation domain (Fig. 3; Pani et al. 1992) that presumably recruits coregulators, which in turn can facilitate other factors to enter the chromatin.
A distinct subset of other transcription factors has been found to bind their target sites on nucleosomes, although not necessarily via a linker histone-like fold. The glucocorticoid receptor (GR), which binds DNA via a zinc finger motif, was the first transcription factor for RNA polymerase II (Pol II) shown to bind its target site on nucleosomal DNA (Perlmann and Wrange 1988; Perlmann 1992; Li and Wrange 1993). GR binding to transcriptionally silent chromatin can initiate the formation of a nuclease-hypersensitive site (Zaret and Yamamoto 1984; Richard-Foy and Hager 1987). Although such target sequences may be a minority for GR at the genomic level (John et al. 2011), the sites where GR induces hypersensitivity are where other factors can subsequently enter the chromatin and therefore represent regions where GR serves as a pioneer factor (Voss et al. 2011).
The yeast Gal4 protein's DNA-binding domain can bind nucleosomal DNA in vitro and in vivo, the latter without replication, and stabilize distinct nucleosome positions (Taylor et al. 1991; Pazin et al. 1998; Balasubramanian and Morse 1999). In contrast, yeast heat-shock factor binds nucleosomal targets after the nucleosomes have been through early S phase (Venturi et al. 2000). The yeast Rap1 protein can bind target sites in chromatin and open the domain for other factors (Yu and Morse 1999; Koerber et al. 2009). In early Xenopus embryos, prior to the mid-blastula transition, β-catenin occupies silent genes and establishes poised chromatin states that are necessary for later transcription (Blythe et al. 2010).
In summary, pioneer transcription factors have nucleosome-binding properties that distinguish the proteins from other DNA-binding factors. Pioneer factors can thus actively help initiate the assembly of regulatory factors on the DNA by either opening the chromatin locally, positioning nucleosomes, enabling intrinsic cooperative binding effects among other DNA-binding factors, or directly recruiting other chromatin modifiers and coregulators. In addition, pioneer factors have the passive role of simply being engaged at a complex regulatory sequence, requiring fewer subsequent factors to bind for regulatory activity. Both the active and passive roles are used extensively in developmental gene induction, and in sections below, we see how they have evolved to promote timely physiologic responses to hormones and are significant in human cancers.
Pioneer factors recruiting corepressor complexes
While the emphasis of the field has been on the positive regulation of gene expression, evidence is emerging that pioneer factors are also used to establish stably silenced domains of the genome. Groucho/TLE corepressor proteins can interact specifically with FoxA (Fu et al. 2000). The interaction was found to allow FoxA to recruit TLE3/Groucho-related protein 3 (Grg3) to specific genomic target sites, causing the local chromatin to close rather than open, and thereby elicit transcriptional repression (Sekiya and Zaret 2007). Interestingly, while Grg3 tetramers by themselves avidly bind chromatin, they do not elicit closing, herein defined as conferring resistance to nuclease access (Sekiya and Zaret 2007). However, upon being recruited to a specific target site by FoxA1, Grg3 now closes three to four nucleosomes in the vicinity. Similar effects were seen by recruitment of Grg3 to DNA by HES1, the latter functioning downstream from Notch signaling.
Undifferentiated embryonic endoderm cells express Grg1 and Grg3 when the albumin gene is silent, and both Grgs are markedly down-regulated during hepatic specification when the albumin gene is activated (Santisteban et al. 2010). Forced maintenance of Grg3 expression in cultured endoderm impairs albumin induction. Taken together, it appears that FoxA may recruit a corepressor to maintain the silent, competent state, and loss of the corepressor contributes to activation that is enabled by FoxA and additional factors that bind the enhancer during liver induction (Gualdi et al. 1996). We speculate that the open state provided by FoxA binding alone could make cells more prone to inappropriate gene activation in response to random fluctuations in inductive signals, and thus recruiting a corepressor prior to activation provides insurance against premature gene expression. It will be interesting to determine the generality of the model of pioneer factors recruiting corepressors in the silent but competent state. Indeed, a recent study of FoxA-binding events in the adult liver at genes that are transcriptionally silent revealed consistent patterns of repressor proteins bound nearby (Watts et al. 2011). This suggests a network of repressors interacting with FoxA at silent genes in cells where the factor otherwise functions as an activator of transcription.
Pioneer factors enabling hormone induction in differentiated cells
Defining the pioneer factors that govern the priming of gene expression in differentiated cells, enabling other transcription factors to bind to regulatory regions, has largely revolved around steroid hormone receptors in solid human cancers. Estrogen receptor (ER) is an essential transcription factor for the development and function of female reproductive organs, such as the mammary gland (Couse and Korach 1999). In adults, ER contributes to tumor formation in almost three-quarters of breast cancer cases (Ali and Coombes 2002; Green and Carroll 2007). ER is a member of the nuclear hormone receptor family of transcription factors, a subset of which, including the androgen receptor (AR) and the GR, is not stably bound to chromatin in unstimulated cells (Glass and Rosenfeld 2000; Hager et al. 2004). Each of these nuclear receptors binds its specific steroid ligand, whereupon it binds to chromatin and activates specific target genes.
Despite intensive investigation into ER-associated proteins and the target genes regulated by ER under different hormonal conditions, our understanding of the chromatin competence that enables ER binding to chromatin was rudimentary until ∼5 years ago. An appreciation for chromatin priming and the role for pioneer factors in hormone-responsive differentiated cells evolved following the coupling of ChIP with microarrays (ChIP-on-chip) or with high-throughput sequencing (ChIP-seq) for global transcription factor mapping. Thus, ER was one of the first transcription factors mapped across both promoter and nonpromoter regions. Unbiased ER genome location mapping experiments in breast cancer cell lines revealed that the majority of ER-binding events occur at significant distances from promoters (Carroll et al. 2005, 2006; Lin et al. 2007) and that transcription involves long-distance interactions between cis-regulatory elements where ER binds and the promoters of target genes in chromatin (Fullwood et al. 2009). Computation analysis of the collection of ER-binding regions revealed various DNA-binding motifs, including those for estrogen-responsive elements (EREs) and for Fox factors (Carroll et al. 2005).
In ER-expressing (ER+) MCF-7 breast cancer cells, FoxA1 is the forkhead factor that is constitutively bound to chromatin regions that are also bound by ER following estrogen treatment (Carroll et al. 2005; Eeckhoute et al. 2006). About half of all ER-binding regions overlap with a FoxA1-binding region, suggesting significant genomic co-occupancy between the two factors (Carroll et al. 2005; Lupien et al. 2008; Hurtado et al. 2011). Knockdown of FoxA1 by RNAi results in decreased ER binding (Carroll et al. 2005; Hurtado et al. 2011), decreased cofactor recruitment (Eeckhoute et al. 2006), and decreased estrogen-stimulated transcription (Carroll et al. 2005; Laganiere et al. 2005; Eeckhoute et al. 2006). In addition, loss of FoxA1 results in cell cycle arrest of ER-positive breast cancer cells (Laganiere et al. 2005; Hurtado et al. 2011). Importantly, in ER-positive breast cancer cells, FoxA1 binding is not dependent on ER binding (Laganiere et al. 2005; Hurtado et al. 2011). The dependency of significant ER binding on FoxA1 illustrates that the latter functions as a pioneer factor for ER in breast cancer cells. FoxA1 binding is required for genes that are either induced or repressed by ER, as well as for ER binding in the presence of both an agonist (estrogen) and an antagonist (tamoxifen) (Malik et al. 2010; Hurtado et al. 2011).
In gene expression data sets from primary breast cancers, FoxA1 is one of the minimal genes that defines an ER+ luminal breast cancer (Perou et al. 2000; Sorlie et al. 2003), and its expression negatively correlates with tumor grade and is associated with better survival (Laganiere et al. 2005; Thorat et al. 2008; Albergaria et al. 2009). Recent work has suggested that the pioneering function of FoxA1 in mediating ER transcriptional activity also occurs during mammary gland development (Bernardo et al. 2010). Additionally, ER can positively regulate FoxA1 expression (and that of GATA3); thus, estrogen action could be amplified in a positive, feed-forward loop that includes ER, FoxA1, and GATA3 (Laganiere et al. 2005). By this means, the pioneer function of FoxA1 could be co-opted by breast cancer cells to promote their aggressive growth phenotype in response to estrogen signaling.
Genomic analysis of FoxA1 and chromatin modifications at target sites
The number of ER-binding events in the breast cancer genome ranges from ∼8000 to ∼15,000 (Welboren et al. 2009; Joseph et al. 2010; Ross-Innes et al. 2010; Hurtado et al. 2011), although this number increases as the sensitivity of the technology evolves. Given the fact that ∼1 million predicted EREs exist in the human genome (Vega et al. 2006), of which >220,000 are optimal binding sequences (Joseph et al. 2010), additional levels of regulation must govern ER-binding potential. The requirement for a Forkhead motif and FoxA1 binding contributes to the selectivity in ER binding. In the absence of FoxA1, ER binding can be mediated by other factors, resulting in distinct ER-binding profiles. This is observed in U20S osteosarcoma cells stably expressing ER (Monroe et al. 2003), where ER is recruited to genomic loci that are different from the ER-binding profile observed in breast cancer cells (Krum et al. 2008). Expression of FoxA1 in U20S-ER cells can induce ER binding to regions previously only observed in the breast cancer cells, and this alters the genes that estrogen–ER can transcribe (Hurtado et al. 2011). Hence, FoxA1 binding, in combination with the presence of an ERE, is a primary determinant of ER binding to chromatin.
The number of FoxA1-binding events in the MCF-7 genome ranges from ∼13,000 to ∼80,000 (Lupien et al. 2008; Hurtado et al. 2011). The wide range in numbers is again due to the increasing sensitivity of the evolving ChIP-seq technology. Interestingly, similar to ER, FoxA1 binding only occurs at a fraction of the genomic regions containing a Forkhead motif (Lupien et al. 2008), suggesting that additional levels of specificity dictate genuine FoxA1-binding events. Thus, despite being a pioneer factor, FoxA1 binding to chromatin can be enabled or restricted by other parameters. DNA methylation may contribute to impaired chromatin access by FoxA1. The Forkhead motif lacks the classic methylated GC sequence (Nakshatri and Badve 2009), and both ER- and FoxA1-binding events tend to be at hypomethylated regions (Serandour et al. 2011). As such, heavily methylated regions may negatively impact ER- and/or FoxA1-binding potential. In support of this, forced expression of FoxA1 in a FoxA1- and ER-negative breast cancer cell line results in opportunistic FoxA1 binding at hypomethylated regions of the genome (Serandour et al. 2011). In addition, specific H3K4 histone modifications correlate positively with FoxA1-binding potential. Histone 3 Lys 4 trimethylation (H3K4me3) tends to occur at promoter regions, and H3K4me1 and H3K4me2 are biased toward enhancer regions (Santos-Rosa et al. 2002; Heintzman et al. 2007). Lupien et al. (2008) confirmed that H3K4me1 and H3K4me2 marks are enriched at cis-regulatory domains that are bound by ER and FoxA1. Expression of KDM1 (LSD1), a demethylase that specifically removes methyl groups from H3K4 (Shi et al. 2004), negatively affects FoxA1 binding (Lupien et al. 2008), but specific silencing of FoxA1 does not affect the presence of H3K4me1 or H3K4me2 marks, implying that the presence of the H3K4me1 and H3K4me2 marks is important for FoxA1 binding (Lupien et al. 2008) and, therefore, ER binding (Carroll et al. 2005; Laganiere et al. 2005; Hurtado et al. 2011). Despite these considerations, expression of FoxA1 in ER-negative cells results in opportunistic binding of FoxA1 to numerous regions lacking H3K4me1 or H3K4me2 marks, followed by subsequent methylation of these histone residues (Serandour et al. 2011).
By combining FAIRE (formaldehyde-assisted isolation of regulatory elements) (Giresi et al. 2007) or DHS (DNase-hypersensitive site) analysis with high-throughput sequencing, a global understanding of chromatin structure is evolving (Eeckhoute et al. 2009; Joseph et al. 2010; Hurtado et al. 2011; John et al. 2011; Serandour et al. 2011). FoxA1-binding events in MCF-7 cells are enriched at regions with high FAIRE signal, representing nucleosome-depleted domains (Eeckhoute et al. 2009), and FoxA1 is required for maintaining active chromatin (Eeckhoute et al. 2009; Hurtado et al. 2011), possibly by inhibiting linker histone association and subsequent chromatin condensation (Cirillo et al. 2002; Taube et al. 2010). Specific silencing of FoxA1 decreases the hypersensitivity of specific ER- and FoxA1-binding regions without affecting H3K4me1 or H3K4me2 levels (Lupien et al. 2008), highlighting the fact that FoxA1 can decouple histone modifications from global chromatin accessibility.
In vitro, FoxA1 can bind efficiently to nucleosomes and chromatin made with recombinant histones that lack modifications (Sekiya and Zaret 2007; Sekiya et al. 2009). Thus, it is not yet clear whether the modulation of FoxA1 binding by the histone modification state in vivo is due to a direct effect of the modification or indirect effects of other factors recruited to the chromatin domain.
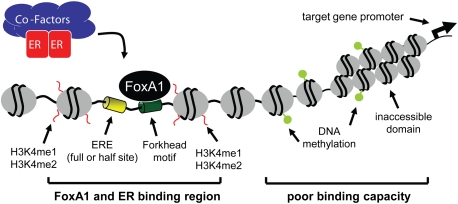
Based on all of the above, we conclude that there are defined features of chromatin that can facilitate FoxA1 binding in breast cancer cells (Fig. 4). These include the presence of the FoxA consensus motif, the absence of DNA methylation (Serandour et al. 2011), nucleosome depletion, and the presence of H3K4me1 and H3K4me2 modification (Joseph et al. 2010). Upon FoxA1 binding, chromatin condensation is relieved further, thereby allowing ER to bind to local EREs (Carroll et al. 2005). This establishes a stable platform for recruitment of ER-associated cofactors and mediating chromatin loops with promoters of target genes.
Figure 4.
Optimal chromatin features for FoxA1 binding to chromatin in breast cancer cells. The figure shows features that enhance or inhibit FoxA1 binding to chromatin in the breast cancer cell line, where parameters have been investigated in detail. FoxA1 binding in turn enables binding by ER and its cofactors, leading to the stimulation of transcription of target genes under estrogen hormonal control.
Interaction between FoxA1 and diverse nuclear receptors
A role for FoxA1 as a nuclear receptor regulatory protein and pioneer factor is not limited to ER and appears to be a more general phenomenon observed for other hormone-responsive nuclear receptors. It has been well established that both FoxA1 and FoxA2 interact with the AR, the driving transcription factor in prostate cancer cells. Mechanistically, AR-binding regions in the androgen-sensitive prostate cancer cell line LnCaP are enriched for Forkhead motifs (Wang et al. 2007; Jia et al. 2008); more than half of the AR-binding events in LnCaP cells overlap with a FoxA1-binding event (Lupien et al. 2008). Silencing of FoxA1 decreases AR binding (Wang et al. 2009) but simultaneously induces numerous new AR-binding events (D Wang et al. 2011), suggesting that FoxA1 both positively and negatively regulates AR-binding potential.
AR binding in other contexts can also require the pioneering function of FoxA1. Molecular apocrine breast cancers are ER− AR+ but express a gene expression pattern resembling an ER+ breast cancer (Farmer et al. 2005; Doane et al. 2006). In molecular apocrine breast cancer cells, AR substitutes for ER and uses FoxA1 to bind to the regulatory regions normally occupied by ER (Ni et al. 2011; Robinson et al. 2011).
In addition to ER and AR, FoxA1 may influence GR function. Much work delineating GR function has evolved from analysis of the MMTV promoter region, a GR-regulated chromatin domain (Ostrowski et al. 1983; Zaret and Yamamoto 1984; Richard-Foy and Hager 1987). FoxA1 binding alters chromatin structure regardless of whether GR is bound to the MMTV promoter or not (Holmqvist et al. 2005). GR binding to the MMTV promoter is enhanced by expression of FoxA1 (Belikov et al. 2009), supporting a model in which FoxA1 mediates optimal chromatin structure and the association of both GR and GR-associated transcription factors. Following FoxA1 and GR recruitment to the MMTV promoter, associated transcription factors, including NF1 and Oct1, can get recruited and increase transcriptional activity, but this can be dependent on the presence of FoxA1 (Belikov et al. 2009). As noted above, many GR-binding events occur at regions that are already open and accessible (John et al. 2011). However, GR binds its target site on nucleosomal DNA (Perlmann and Wrange 1988), and there are numerous genomic sites where such “pioneer” binding creates local hypersensitivity and promotes the recruitment of other transcription factors (Voss et al. 2011). Currently, it is unclear how GR can serve as a pioneer factor, but the related ER and AR steroid receptors appear to require distinct factors with “pioneer” activity.
GATA and other pioneer factors for nuclear receptors
The initial work on pioneer factors for nuclear receptors revolved around FoxA1, but there is much interest in identifying and characterizing other proteins that help nuclear receptors engage silent, condensed chromatin. Analysis of additional enriched motifs within ER-binding domains beyond those for FoxA1 (Carroll et al. 2005) revealed the overrepresentation of motifs for GATA transcription factors (Lin et al. 2007). Similar to FoxA1, GATA3 is another gene that characterizes an ER+ luminal breast cancer (Perou et al. 2000; Sorlie et al. 2003). GATA3 and FoxA1 correlate in breast cancer cells (Yamaguchi et al. 2008) and have long been implicated in a network with ER (Lacroix and Leclercq 2004). GATA3 is required for mammary gland development (Asselin-Labat et al. 2007) and maintains cellular differentiation in the adult (Kouros-Mehr et al. 2006). GATA3 is recruited to ER cis-regulatory elements and is essential for ER-mediated transcription of target genes and for growth of MCF-7 breast cancer cells (Eeckhoute et al. 2007). GATA motifs are also enriched in AR-binding events in LnCaP prostate cancer cells, and GATA2 is shown to be recruited to several AR-binding regions, where it is required for effective AR–chromatin interactions (Wang et al. 2007). Therefore, in prostate cancer cells, GATA2 plays a role as a pioneer factor for AR. In U20S-ER osteosarcoma cells, where FoxA1 and GATA3 are absent, ER binding is redirected to different regions in the genome (Krum et al. 2008), and this is mediated by GATA4 (Miranda-Carboni et al. 2011). Since GATA proteins can bind to condensed chromatin and can induce local nuclease hypersensitivity, albeit more weakly than that by FoxA1 (Cirillo et al. 2002), GATA factors appear to function as pioneer factors for ER and AR in hormone-dependent cancers.
AP-2 motifs are also enriched in ER-binding regions (Tan et al. 2011), and AP-2γ binds to a significant fraction of ER-binding events, where it is essential for estrogen-mediated transcription and ER binding (Tan et al. 2011). The ER-binding regions that are cobound by AP-2γ also tend to be occupied by FoxA1, and, interestingly, AP-2γ and FoxA1 require each other for effective binding capacity (Tan et al. 2011). As such, the emerging picture involves multiple pioneer factors that contribute to ER–chromatin interactions, with an interplay and dependency existing between the factors.
Pioneer factors as drug targets in cancer
The growth of many hormone-driven solid tumors (e.g., of breast and prostate) are mediated by nuclear receptors, and, as such, it is possible that their accompanying pioneer factors may constitute therapeutic targets (Nakshatri and Badve 2007; Fu et al. 2011; Hurtado et al. 2011). In both breast and prostate cancer, drug resistance is a frequent problem. Approximately 80% of metastases that come from an ER+ luminal breast cancer maintain ER expression (Harrell et al. 2006), and resistance to one ER antagonist does not always render a tumor resistant to additional ER antagonists (Ali and Coombes 2002). This implies that ER transcriptional activity drives cell proliferation even in a drug-resistant context, and thus it is possible that FoxA1 is still mediating ER–chromatin interactions as a pioneer factor.
Growth factor pathways contribute to endocrine resistance (Knowlden et al. 2003, 2005), and induction of AKT or EGF pathways results in acquisition of unique ER-binding events (Bhat-Nakshatri et al. 2008; Lupien et al. 2010). In the case of EGF stimulation, the growth factor-mediated ER-binding regions are enriched with Fox motifs (Lupien et al. 2010). A similar situation is seen in tamoxifen-resistant MCF-7 breast cancer derivatives (Knowlden et al. 2003), where ER-binding regions are acquired during resistance, and these correlate with a gain in FoxA1 binding (Hurtado et al. 2011). Specific silencing of FoxA1 inhibits the proliferation of tamoxifen-resistant MCF-7 cells (Hurtado et al. 2011) and of tamoxifen-resistant ER+ breast cancer cells possessing the HER2 amplification (Yamaguchi et al. 2008). Thus, an inhibitor that blocks FoxA1 or other essential pioneer factors may have therapeutic potential in these nuclear receptor-driven cancer types, where traditional therapies have failed.
Summary and perspectives for cellular reprogramming
It is now appreciated that in undifferentiated stem cells and embryonic progenitors as well as in fully differentiated cells, genes can exist in stable states where they are transcriptionally silent, yet competent for expression. Such silent yet competent states enable a rapid transcriptional response to inductive signals (Fig. 2). Many studies have revealed chromatin marks (Bernstein et al. 2006) and poised RNA polymerase (Nechaev and Adelman 2011) at silent genes that are competent for expression. Indeed, release from transcriptional pausing has emerged as a major mechanism of regulation. Yet general factors such as RNA Pol II and chromatin modifiers must be recruited by proteins that recognize specific DNA sequences and target them to particular regions of the genome. Pioneer factors, including FoxA proteins, can initiate such regulatory events, at least in part due to their resemblance to linker histone, and, hence, special nucleosome- and chromatin-binding properties. While FoxA binding to target sequences on nucleosomes is efficient in vitro with unmodified histones, in vivo the factor exhibits preferences for particular modified domains. Despite such modulation, FoxA factors still bind chromatin sites when other factors cannot, and enable other factors to bind. Further work is required to assess the roles that chromatin modifications and other regulatory complexes play in enhancing or restricting pioneer factors binding to chromatin.
Developmental and genetic studies have identified many transcription factors that are necessary for the specification of different cell types and that are likely to function as pioneer factors. Currently, there is interest in using this information to reprogram cells of one fate to another or to pluripotency by transducing transcription factor genes. The approach began with the transcription factor MyoD, which, when introduced into fibroblasts, was found to initiate the myogenic program (Davis et al. 1987). Other examples include GATA1, whose ectopic expression in monocytes induces the erythroid and megakaryocyte programs (Visvader et al. 1992; Kulessa et al. 1995); C/EBPα and C/EBPβ, whose expression in B cells activates the macrophage program (Xie et al. 2004); and, in perhaps the most dramatic example, Pax6, whose expression can cause the appearance of additional eyes growing out of a fly (Halder et al. 1995). These cases of trans-differentiation are paradigms for “direct reprogramming” approaches to create cells of biomedical interest, which include creating liver hepatocytes and pancreatic β cells (Shen et al. 2000; Sapir et al. 2005; Jarikji et al. 2007; Zhou et al. 2008; Yechoor et al. 2009). In many of the gene transduction studies, multiple factors are required for the cellular conversion, and in cases in which a single ectopic factor has been used, it converts cells from related germ layer lineages. For example, binding of MyoD to the silent myogenin promoter to induce myogenesis occurs by interaction with a prebound Pbx factor, which is widely expressed in mesodermal and other cell types (Berkes et al. 2004). These points underscore the role of cooperativity among factors in the starting population of cells.
An additional dramatic example of cellular reprogramming is the ability of ectopic Oct4, Sox2, Klf4, and c-Myc factors to convert somatic cells to pluripotency (Takahashi and Yamanaka 2006; Hochedlinger and Plath 2009). Only a small subset of the targeted cells becomes converted, suggesting that either a subset of the cells are competent or stochastic parameters are involved (Yamanaka 2009). The latter could include epigenetic factors and varying expression of other regulatory factors in the population of cells. A role for an epigenetic component is supported by the ability of inhibitors of histone-modifying enzymes to enhance reprogramming (Huangfu et al. 2008; Shi et al. 2008). Increasing the number of cell divisions during reprogramming increases the time for stochastic events to occur, and thus enhances reprogramming efficiency (Hanna et al. 2009). Still, this is in contrast to the relatively high efficiency with which somatic cell nuclear transfer to eggs and oocytes can elicit reprogramming (Gurdon et al. 1958). In all cases of reprogramming cells to pluripotency, the relevant transcription factors must engage many silent target sites and open the chromatin for transcription. However, it remains to be determined whether Oct4, Sox2, Klf4, c-Myc, and other pluripotency factors individually possess pioneer factor activities, or whether their engagement of silent chromatin sites is predominantly via simultaneous, cooperative binding, as discussed above (see Fig. 1).
As noted previously, the pioneer factors FoxA3 and GATA4 are among a group of gene regulatory proteins that enable reprogramming of fibroblasts to hepatocytes (Huang et al. 2011). The ability of the male genome to be reprogrammed efficiently upon fertilization and the high efficiency of reprogramming by somatic cell nuclear transfer illustrate that there must exist specific mechanisms by which early developmental transcription factors can efficiently gain access to chromatin and reprogram the genome (Jullien et al. 2011). We expect that detailed studies of the diverse factors being used for cellular reprogramming will reveal subsets of factors with dominant chromatin-binding functions and pioneer activity. Defining how pioneer factors act in these and other contexts to enable subsequent regulatory events in chromatin will provide insight into methods for increasing the efficiency of cellular reprogramming, an understanding of hormonal control, and a better sense of how perturbations of pioneer factor expression or activity dysregulates the genome in cancer.
Acknowledgments
We thank Eileen Pytko for help in preparing the manuscript. Work in our laboratories on pioneer factors has been supported by NIH grants R37GM36477 and R01DK082623 to K.S.Z. and an ERC starting grant to J.S.C.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.176826.111.
References
- Adams CC, Workman JL 1995. Binding of disparate transcriptional activators to nucleosomal DNA is inherently cooperative. Mol Cell Biol 15: 1405–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albergaria A, Paredes J, Sousa B, Milanezi F, Carneiro V, Bastos J, Costa S, Vieira D, Lopes N, Lam EW, et al. 2009. Expression of FOXA1 and GATA-3 in breast cancer: the prognostic significance in hormone receptor-negative tumours. Breast Cancer Res 11: R40 doi: 10.1186/bcr2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S, Coombes RC 2002. Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer 2: 101–112 [DOI] [PubMed] [Google Scholar]
- Ang S-L, Wierda A, Wong D, Stevens KA, Cascio S, Rossant J, Zaret KS 1993. The formation and maintenance of the definitive endoderm lineage in the mouse: involvement of HNF3/forkhead proteins. Development 119: 1301–1315 [DOI] [PubMed] [Google Scholar]
- Arceci R, King AAJ, Simon MC, Orkin SH, Wilson DB 1993. Mouse GATA-4: a retinoic acid-inducible GATA-binding transcription factor expressed in endodermally derived tissues and heart. Mol Cell Biol 13: 2235–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, Hartley L, Robb L, Grosveld FG, van der Wees J, et al. 2007. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol 9: 201–209 [DOI] [PubMed] [Google Scholar]
- Balasubramanian B, Morse RH 1999. Binding of Gal4p and bicoid to nucleosomal sites in yeast in the absence of replication. Mol Cell Biol 19: 2977–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T 2010. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 143: 470–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belikov S, Astrand C, Wrange O 2009. FoxA1 binding directs chromatin structure and the functional response of a glucocorticoid receptor-regulated promoter. Mol Cell Biol 29: 5413–5425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkes CA, Bergstrom DA, Penn BH, Seaver KJ, Knoepfler PS, Tapscott SJ 2004. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol Cell 14: 465–477 [DOI] [PubMed] [Google Scholar]
- Bernardo GM, Lozada KL, Miedler JD, Harburg G, Hewitt SC, Mosley JD, Godwin AK, Korach KS, Visvader JE, Kaestner KH, et al. 2010. FOXA1 is an essential determinant of ERα expression and mammary ductal morphogenesis. Development 137: 2045–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Bhat-Nakshatri P, Wang G, Appaiah H, Luktuke N, Carroll JS, Geistlinger TR, Brown M, Badve S, Liu Y, Nakshatri H 2008. AKT alters genome-wide estrogen receptor α binding and impacts estrogen signaling in breast cancer. Mol Cell Biol 28: 7487–7503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe SA, Cha SW, Tadjuidje E, Heasman J, Klein PS 2010. β-Catenin primes organizer gene expression by recruiting a histone H3 arginine 8 methyltransferase, Prmt2. Dev Cell 19: 220–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeger H, Griesenbeck J, Strattan JS, Kornberg RD 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell 11: 1587–1598 [DOI] [PubMed] [Google Scholar]
- Bossard P, Zaret KS 1998. GATA transcription factors as potentiators of gut endoderm differentiation. Development 125: 4909–4917 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Song L, Lee BK, London D, Keefe D, Birney E, Iyer VR, Crawford GE, Furey TS 2011. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res 21: 456–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr A, Biggin MD 1999. A comparison of in vivo and in vitro DNA-binding specificities suggests a new model for homeoprotein DNA binding in Drosophila embryos. EMBO J 18: 1598–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122: 33–43 [DOI] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat Genet 38: 1289–1297 [DOI] [PubMed] [Google Scholar]
- Chaya D, Hayamizu T, Bustin M, Zaret KS 2001. Transcription factor FoxA (HNF3) on a nucleosome at an enhancer complex in liver chromatin. J Biol Chem 276: 44385–44389 [DOI] [PubMed] [Google Scholar]
- Chiu CP, Blau HM 1984. Reprogramming cell differentiation in the absence of DNA synthesis. Cell 37: 879–887 [DOI] [PubMed] [Google Scholar]
- Chong MM, Simpson N, Ciofani M, Chen G, Collins A, Littman DR 2010. Epigenetic propagation of CD4 expression is established by the Cd4 proximal enhancer in helper T cells. Genes Dev 24: 659–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo LA, Zaret KS 1999. An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol Cell 4: 961–969 [DOI] [PubMed] [Google Scholar]
- Cirillo LA, McPherson CE, Bossard P, Stevens K, Cherian S, Shim E-Y, Clark EA, Burley SK, Zaret KS 1998. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J 17: 244–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo L, Lin FR, Cuesta I, Jarnik M, Friedman D, Zaret K 2002. Opening of compacted chromatin by early developmental transcription factors HNF3 (FOXA) and GATA-4. Mol Cell 9: 279–289 [DOI] [PubMed] [Google Scholar]
- Clark KL, Halay ED, Lai E, Burley SK 1993. Co-crystal structure of the HNF3/fork head DNA recognition motif resembles histone H5. Nature 364: 412–420 [DOI] [PubMed] [Google Scholar]
- Couse JF, Korach KS 1999. Estrogen receptor null mice: what have we learned and where will they lead us? Endocr Rev 20: 358–417 [DOI] [PubMed] [Google Scholar]
- Cuesta I, Zaret KS, Santisteban P 2007. The forkhead factor FoxE1 binds to the thyroperoxidase promoter during thyroid cell differentiation and modifies compacted chromatin structure. Mol Cell Biol 27: 7302–7314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, Erwin DH 2006. Gene regulatory networks and the evolution of animal body plans. Science 311: 796–800 [DOI] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB 1987. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51: 987–1000 [DOI] [PubMed] [Google Scholar]
- Decker T, Pasca di Magliano M, McManus S, Sun Q, Bonifer C, Tagoh H, Busslinger M 2009. Stepwise activation of enhancer and promoter regions of the B cell commitment gene Pax5 in early lymphopoiesis. Immunity 30: 508–520 [DOI] [PubMed] [Google Scholar]
- Doane AS, Danso M, Lal P, Donaton M, Zhang L, Hudis C, Gerald WL 2006. An estrogen receptor-negative breast cancer subset characterized by a hormonally regulated transcriptional program and response to androgen. Oncogene 25: 3994–4008 [DOI] [PubMed] [Google Scholar]
- Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M 2006. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev 20: 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeckhoute J, Krasnickas Keeton E, Lupien M, Krum SA, Carroll JS, Brown M 2007. Positive cross-regulatory loop ties GATA-3 to estrogen receptor α expression in breast cancer. Cancer Res 67: 6477–6483 [DOI] [PubMed] [Google Scholar]
- Eeckhoute J, Lupien M, Meyer CA, Verzi MP, Shivdasani RA, Liu XS, Brown M 2009. Cell-type selective chromatin remodeling defines the active subset of FOXA1-bound enhancers. Genome Res 19: 372–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhouri TH, Stevenson J, Chisholm AD, Mango SE 2010. Dynamic chromatin organization during foregut development mediated by the organ selector gene PHA-4/FoxA. PLoS Genet 6: e1001060 doi: 10.1371/journal.pgen.1001060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan JY, Rangasamy D, Luger K, Tremethick DJ 2004. H2A.Z alters the nucleosome surface to promote HP1α-mediated chromatin fiber folding. Mol Cell 16: 655–661 [DOI] [PubMed] [Google Scholar]
- Farmer P, Bonnefoi H, Becette V, Tubiana-Hulin M, Fumoleau P, Larsimont D, Macgrogan G, Bergh J, Cameron D, Goldstein D, et al. 2005. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene 24: 4660–4671 [DOI] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. 2010. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143: 212–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL 2004. Chromatin compaction by a polycomb group protein complex. Science 306: 1574–1577 [DOI] [PubMed] [Google Scholar]
- Fu M, Wang C, Reutens AT, Wang J, Angeletti RH, Siconolfi-Baez L, Ogryzko V, Avantaggiati ML, Pestell RG 2000. p300 and p300/cAMP-response element-binding protein-associated factor acetylate the androgen receptor at sites governing hormone-dependent transactivation. J Biol Chem 275: 20853–20860 [DOI] [PubMed] [Google Scholar]
- Fu X, Huang C, Schiff R 2011. More on FOX News: FOXA1 on the horizon of estrogen receptor function and endocrine response. Breast Cancer Res 13: 307 doi: 10.1186/bcr2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, et al. 2009. An oestrogen-receptor-α-bound human chromatin interactome. Nature 462: 58–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet J, Mango SE 2002. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science 295: 821–825 [DOI] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. 2010. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity 32: 317–328 [DOI] [PubMed] [Google Scholar]
- Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD 2007. FAIRE (formaldehyde-assisted isolation of regulatory elements) isolates active regulatory elements from human chromatin. Genome Res 17: 877–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev 14: 121–141 [PubMed] [Google Scholar]
- Goytisolo FA, Gerchman SE, Yu X, Rees C, Graziano V, Ramakrishnan V, Thomas JO 1996. Identification of two DNA-binding sites on the globular domain of histone H5. EMBO J 15: 3421–3429 [PMC free article] [PubMed] [Google Scholar]
- Green KA, Carroll JS 2007. Oestrogen-receptor-mediated transcription and the influence of co-factors and chromatin state. Nat Rev Cancer 7: 713–722 [DOI] [PubMed] [Google Scholar]
- Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman JR, Zaret KS 1996. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev 10: 1670–1682 [DOI] [PubMed] [Google Scholar]
- Gurdon JB, Elsdale TR, Fischberg M 1958. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature 182: 64–65 [DOI] [PubMed] [Google Scholar]
- Hager GL, Nagaich AK, Johnson TA, Walker DA, John S 2004. Dynamics of nuclear receptor movement and transcription. Biochim Biophys Acta 1677: 46–51 [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Gehring WJ 1995. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science 267: 1788–1792 [DOI] [PubMed] [Google Scholar]
- Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R 2009. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 462: 595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannenhalli S, Kaestner KH 2009. The evolution of Fox genes and their role in development and disease. Nat Rev Genet 10: 233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrell JC, Dye WW, Allred DC, Jedlicka P, Spoelstra NS, Sartorius CA, Horwitz KB 2006. Estrogen receptor positive breast cancer metastasis: altered hormonal sensitivity and tumor aggressiveness in lymphatic vessels and lymph nodes. Cancer Res 66: 9308–9315 [DOI] [PubMed] [Google Scholar]
- Hatta M, Cirillo LA 2007. Chromatin opening and stable perturbation of core histone:DNA contacts by FoxO1. J Biol Chem 282: 35583–35593 [DOI] [PubMed] [Google Scholar]
- He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, et al. 2010. Nucleosome dynamics define transcriptional enhancers. Nat Genet 42: 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K, Plath K 2009. Epigenetic reprogramming and induced pluripotency. Development 136: 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman BG, Robertson G, Zavaglia B, Beach M, Cullum R, Lee S, Soukhatcheva G, Li L, Wederell ED, Thiessen N, et al. 2010. Locus co-occupancy, nucleosome positioning, and H3K4me1 regulate the functionality of FOXA2-, HNF4A-, and PDX1-bound loci in islets and liver. Genome Res 20: 1037–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmqvist P-H, Belikov S, Zaret KS, Wrange Ö 2005. FoxA1 binding to the MMTV LTR modulates chromatin structure and transcription. Exp Cell Res 304: 593–603 [DOI] [PubMed] [Google Scholar]
- Holtzinger A, Evans T 2005. Gata4 regulates the formation of multiple organs. Development 132: 4005–4014 [DOI] [PubMed] [Google Scholar]
- Hoogenkamp M, Lichtinger M, Krysinska H, Lancrin C, Clarke D, Williamson A, Mazzarella L, Ingram R, Jorgensen H, Fisher A, et al. 2009. Early chromatin unfolding by RUNX1—a molecular explanation for differential requirements during specification versus maintenance of the hematopoietic gene expression program. Blood 114: 299–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, Hu Y, Wang X, Hui L 2011. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature 475: 386–389 [DOI] [PubMed] [Google Scholar]
- Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA 2008. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol 26: 795–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS 2011. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet 43: 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii H, Sen R, Pazin MJ 2004. Combinatorial control of DNase I-hypersensitive site formation and erasure by immunoglobulin heavy chain enhancer-binding proteins. J Biol Chem 279: 7331–7338 [DOI] [PubMed] [Google Scholar]
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409: 533–538 [DOI] [PubMed] [Google Scholar]
- Jarikji ZH, Vanamala S, Beck CW, Wright CV, Leach SD, Horb ME 2007. Differential ability of Ptf1a and Ptf1a-VP16 to convert stomach, duodenum and liver to pancreas. Dev Biol 304: 786–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L, Berman BP, Jariwala U, Yan X, Cogan JP, Walters A, Chen T, Buchanan G, Frenkel B, Coetzee GA 2008. Genomic androgen receptor-occupied regions with different functions, defined by histone acetylation, coregulators and transcriptional capacity. PLoS ONE 3: e3645 doi: 10.1371/journal.pone.0003645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Felsenfeld G 2007. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev 21: 1519–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G 2009. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet 41: 941–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA 2011. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet 43: 264–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph R, Orlov YL, Huss M, Sun W, Kong SL, Ukil L, Pan YF, Li G, Lim M, Thomsen JS, et al. 2010. Integrative model of genomic factors for determining binding site selection by estrogen receptor-α. Mol Syst Biol 6: 456 doi: 10.1038/msb.2010.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien J, Pasque V, Halley-Stott RP, Miyamoto K, Gurdon JB 2011. Mechanisms of nuclear reprogramming by eggs and oocytes: a deterministic process? Nat Rev Mol Cell Biol 12: 453–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan T, Li XY, Sabo PJ, Thomas S, Stamatoyannopoulos JA, Biggin MD, Eisen MB 2011. Quantitative models of the mechanisms that control genome-wide patterns of transcription factor binding during early Drosophila development. PLoS Genet 7: e1001290 doi: 10.1371/journal.pgen.1001290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlden JM, Hutcheson IR, Jones HE, Madden T, Gee JM, Harper ME, Barrow D, Wakeling AE, Nicholson RI 2003. Elevated levels of epidermal growth factor receptor/c-erbB2 heterodimers mediate an autocrine growth regulatory pathway in tamoxifen-resistant MCF-7 cells. Endocrinology 144: 1032–1044 [DOI] [PubMed] [Google Scholar]
- Knowlden JM, Hutcheson IR, Barrow D, Gee JM, Nicholson RI 2005. Insulin-like growth factor-I receptor signaling in tamoxifen-resistant breast cancer: a supporting role to the epidermal growth factor receptor. Endocrinology 146: 4609–4618 [DOI] [PubMed] [Google Scholar]
- Koerber RT, Rhee HS, Jiang C, Pugh BF 2009. Interaction of transcriptional regulators with specific nucleosomes across the Saccharomyces genome. Mol Cell 35: 889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD 1977. Structure of chromatin. Annu Rev Biochem 46: 931–954 [DOI] [PubMed] [Google Scholar]
- Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z 2006. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell 127: 1041–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krum SA, Miranda-Carboni GA, Lupien M, Eeckhoute J, Carroll JS, Brown M 2008. Unique ERα cistromes control cell type-specific gene regulation. Mol Endocrinol 22: 2393–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysinska H, Hoogenkamp M, Ingram R, Wilson N, Tagoh H, Laslo P, Singh H, Bonifer C 2007. A two-step, PU.1-dependent mechanism for developmentally regulated chromatin remodeling and transcription of the c-fms gene. Mol Cell Biol 27: 878–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulessa H, Frampton J, Graf T 1995. GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes Dev 9: 1250–1262 [DOI] [PubMed] [Google Scholar]
- Lacroix M, Leclercq G 2004. About GATA3, HNF3A, and XBP1, three genes co-expressed with the oestrogen receptor-α gene (ESR1) in breast cancer. Mol Cell Endocrinol 219: 1–7 [DOI] [PubMed] [Google Scholar]
- Laganiere J, Deblois G, Lefebvre C, Bataille AR, Robert F, Giguere V 2005. Location analysis of estrogen receptor α target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc Natl Acad Sci 102: 11651–11656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laverriere AC, MacNeill C, Mueller C, Poelmann RE, Burch JBE, Evans T 1994. GATA-4/5/6, a subfamily of three transcription factors transcribed in developing heart and gut. J Biol Chem 269: 23177–23184 [PubMed] [Google Scholar]
- Lee CS, Friedman JR, Fulmer JT, Kaestner KH 2005. The initiation of liver development is dependent on Foxa transcription factors. Nature 435: 944–947 [DOI] [PubMed] [Google Scholar]
- Levine M 2010. Transcriptional enhancers in animal development and evolution. Curr Biol 20: R754–R763 doi: 10.1016/j.cub.2010.06.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Wrange Ö 1993. Translational positioning of a nucleosome; glucocorticoid response element modulates glucocorticoid receptor affinity. Genes Dev 7: 2471–2482 [DOI] [PubMed] [Google Scholar]
- Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ 2006. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442: 91–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, White P, Tuteja G, Rubins N, Sackett S, Kaestner KH 2009. Foxa1 and Foxa2 regulate bile duct development in mice. J Clin Invest 119: 1537–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Margueron R, Hu G, Stokes D, Wang YH, Reinberg D 2010. Highly compacted chromatin formed in vitro reflects the dynamics of transcription activation in vivo. Mol Cell 38: 41–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Thomas S, Sabo PJ, Eisen MB, Stamatoyannopoulos JA, Biggin MD 2011. The role of chromatin accessibility in directing the widespread, overlapping patterns of Drosophila transcription factor binding. Genome Biol 12: R34 doi: 10.1186/gb-2011-12-4-r34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Schug J, Tuteja G, White P, Kaestner KH 2011. The nucleosome map of the mammalian liver. Nat Struct Mol Biol 18: 742–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liber D, Domaschenz R, Holmqvist PH, Mazzarella L, Georgiou A, Leleu M, Fisher AG, Labosky PA, Dillon N 2010. Epigenetic priming of a pre-B cell-specific enhancer through binding of Sox2 and Foxd3 at the ESC stage. Cell Stem Cell 7: 114–126 [DOI] [PubMed] [Google Scholar]
- Lin CY, Vega VB, Thomsen JS, Zhang T, Kong SL, Xie M, Chiu KP, Lipovich L, Barnett DH, Stossi F, et al. 2007. Whole-genome cartography of estrogen receptor α binding sites. PLoS Genet 3: e87 doi: 10.1371/journal.pgen.0030087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JK, DiPersio CM, Zaret KS 1991. Extracellular signals that regulate liver transcription factors during hepatic differentiation in vitro. Mol Cell Biol 11: 773–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251–255 [DOI] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M 2008. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132: 958–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M, Meyer CA, Bailey ST, Eeckhoute J, Cook J, Westerling T, Zhang X, Carroll JS, Rhodes DR, Liu XS, et al. 2010. Growth factor stimulation induces a distinct ERα cistrome underlying breast cancer endocrine resistance. Genes Dev 24: 2219–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur S, Li XY, Li J, Brown JB, Chu HC, Zeng L, Grondona BP, Hechmer A, Simirenko L, Keranen SV, et al. 2009. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol 10: R80 doi: 10.1186/gb-2009-10-7-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Jiang S, Garee JP, Verdin E, Lee AV, O'Malley BW, Zhang M, Belaguli NS, Oesterreich S 2010. Histone deacetylase 7 and FoxA1 in estrogen-mediated repression of RPRM. Mol Cell Biol 30: 399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino F, Kueng S, Robinson P, Tsai-Pflugfelder M, van Leeuwen F, Ziegler M, Cubizolles F, Cockell MM, Rhodes D, Gasser SM 2009. Reconstitution of yeast silent chromatin: multiple contact sites and O-AADPR binding load SIR complexes onto nucleosomes in vitro. Mol Cell 33: 323–334 [DOI] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, et al. 2008. Nucleosome organization in the Drosophila genome. Nature 453: 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson CE, Shim E-Y, Friedman DS, Zaret KS 1993. An active tissue-specific enhancer and bound transcription factors existing in a precisely postioned nucleosomal array. Cell 75: 387–398 [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Carboni GA, Guemes M, Bailey S, Anaya E, Corselli M, Peault B, Krum SA 2011. GATA4 regulates estrogen receptor-α-mediated osteoblast transcription. Mol Endocrinol 25: 1126–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan AP, Kaestner KH, Grau E, Schütz G 1993. Postimplantation expression patterns indicate a role for the mouse forkhead/HNF-3 α, β, and γ genes in determination of the definitive endoderm, chordamesoderm and neuroectoderm. Development 119: 567–578 [DOI] [PubMed] [Google Scholar]
- Monroe DG, Getz BJ, Johnsen SA, Riggs BL, Khosla S, Spelsberg TC 2003. Estrogen receptor isoform-specific regulation of endogenous gene expression in human osteoblastic cell lines expressing either ERα or ERβ. J Cell Biochem 90: 315–326 [DOI] [PubMed] [Google Scholar]
- Nakshatri H, Badve S 2007. FOXA1 as a therapeutic target for breast cancer. Expert Opin Ther Targets 11: 507–514 [DOI] [PubMed] [Google Scholar]
- Nakshatri H, Badve S 2009. FOXA1 in breast cancer. Expert Rev Mol Med 11: e8 doi: 10.1017/S1462399409001008 [DOI] [PubMed] [Google Scholar]
- Nechaev S, Adelman K 2011. Pol II waiting in the starting gates: regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta 1809: 34–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Chen Y, Lim E, Wimberly H, Bailey ST, Imai Y, Rimm DL, Shirley Liu X, Brown M 2011. Targeting androgen receptor in estrogen receptor-negative breast cancer. Cancer Cell 20: 119–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski MC, Richard-Foy H, Wolford RG, Berard DS, Hager GL 1983. Glucocorticoid regulation of transcription at an amplified, episomal promoter. Mol Cell Biol 3: 2045–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pani L, Overdier DG, Porcella A, Qian X, Lai E, Costa RH 1992. Hepatocyte nuclear factor 3b contains two transcriptional activation domains, one of which is novel and conserved with the Drosophila fork head protein. Mol Cell Biol 12: 3723–3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazin MJ, Hermann JW, Kadonaga JT 1998. Promoter structure and transcriptional activation with chromatin templates assembled in vitro. A single Gal4-VP16 dimer binds to chromatin or to DNA with comparable affinity. J Biol Chem 273: 34653–34660 [DOI] [PubMed] [Google Scholar]
- Perlmann T 1992. Glucocorticoid receptor DNA-binding specificity is increased by the organization of DNA in nucleosomes. Proc Natl Acad Sci 89: 3884–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlmann T, Wrange O 1988. Specific glucocorticoid receptor binding to DNA reconstituted in nucleosome. EMBO J 7: 3073–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. 2000. Molecular portraits of human breast tumours. Nature 406: 747–752 [DOI] [PubMed] [Google Scholar]
- Pique-Regi R, Degner JF, Pai AA, Gaffney DJ, Gilad Y, Pritchard JK 2011. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res 21: 447–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram EV, Meshorer E 2009. Transcriptional competence in pluripotency. Genes Dev 23: 2793–2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan V, Finch JT, Graziano V, Lee PL, Sweet RM 1993. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature 362: 219–224 [DOI] [PubMed] [Google Scholar]
- Reinke H, Horz W 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol Cell 11: 1599–1607 [DOI] [PubMed] [Google Scholar]
- Richard-Foy H, Hager GL 1987. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J 6: 2321–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JL, Macarthur S, Ross-Innes CS, Tilley WD, Neal DE, Mills IG, Carroll JS 2011. Androgen receptor driven transcription in molecular apocrine breast cancer is mediated by FoxA1. EMBO J 30: 3019–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Holmes KA, Schmidt D, Spyrou C, Russell R, Massie CE, Vowler SL, Eldridge M, Carroll JS 2010. Cooperative interaction between retinoic acid receptor-α and estrogen receptor in breast cancer. Genes Dev 24: 171–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santisteban P, Recacha P, Metzger DE, Zaret KS 2010. Dynamic expression of Groucho-related genes Grg1 and Grg3 in foregut endoderm and antagonism of differentiation. Dev Dyn 239: 980–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419: 407–411 [DOI] [PubMed] [Google Scholar]
- Sapir T, Shternhall K, Meivar-Levy I, Blumenfeld T, Cohen H, Skutelsky E, Eventov-Friedman S, Barshack I, Goldberg I, Pri-Chen S, et al. 2005. Cell-replacement therapy for diabetes: generating functional insulin-producing tissue from adult human liver cells. Proc Natl Acad Sci 102: 7964–7969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalch T, Duda S, Sargent DF, Richmond TJ 2005. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature 436: 138–141 [DOI] [PubMed] [Google Scholar]
- Schwarz PM, Hansen JC 1994. Formation and stability of higher order chromatin structures. Contributions of the histone octamer. J Biol Chem 269: 16284–16289 [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J 2006. A genomic code for nucleosome positioning. Nature 442: 772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekinger EA, Moqtaderi Z, Struhl K 2005. Intrinsic histone–DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol Cell 18: 735–748 [DOI] [PubMed] [Google Scholar]
- Sekiya T, Zaret KS 2007. Repression by Groucho/TLE/Grg proteins: genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol Cell 28: 291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya T, Muthurajan UM, Luger K, Tulin AV, Zaret KS 2009. Nucleosome-binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes Dev 23: 804–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serandour AA, Avner S, Percevault F, Demay F, Bizot M, Lucchetti-Miganeh C, Barloy-Hubler F, Brown M, Lupien M, Metivier R, et al. 2011. Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res 21: 555–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen CN, Slack JM, Tosh D 2000. Molecular basis of transdifferentiation of pancreas to liver. Nat Cell Biol 2: 879–887 [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119: 941–953 [DOI] [PubMed] [Google Scholar]
- Shi Y, Do JT, Desponts C, Hahm HS, Scholer HR, Ding S 2008. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell 2: 525–528 [DOI] [PubMed] [Google Scholar]
- Shim EY, Woodcock C, Zaret KS 1998. Nucleosome positioning by the winged-helix transcription factor HNF3. Genes Dev 12: 5–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL 2006. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311: 844–847 [DOI] [PubMed] [Google Scholar]
- Smale ST 2010. Pioneer factors in embryonic stem cells and differentiation. Curr Opin Genet Dev 20: 519–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et al. 2003. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci 100: 8418–8423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger DJ, Workman JL 1997. Stable co-occupancy of transcription factors and histones at the HIV-1 enhancer. EMBO J 16: 2463–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD 2000. The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Tan SK, Lin ZH, Chang CW, Varang V, Chng KR, Pan YF, Yong EL, Sung WK, Cheung E 2011. AP-2γ regulates oestrogen receptor-mediated long-range chromatin interaction and gene transcription. EMBO J 30: 2569–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taube JH, Allton K, Duncan SA, Shen L, Barton MC 2010. Foxa1 functions as a pioneer transcription factor at transposable elements to activate Afp during differentiation of embryonic stem cells. J Biol Chem 285: 16135–16144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor ICA, Workman JL, Schuetz TJ, Kingston RE 1991. Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: differential function of DNA-binding domains. Genes Dev 5: 1285–1298 [DOI] [PubMed] [Google Scholar]
- Thomassin H, Flavin M, Espinas ML, Grange T 2001. Glucocorticoid-induced DNA demethylation and gene memory during development. EMBO J 20: 1974–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorat MA, Marchio C, Morimiya A, Savage K, Nakshatri H, Reis-Filho JS, Badve S 2008. Forkhead box A1 expression in breast cancer is associated with luminal subtype and good prognosis. J Clin Pathol 61: 327–332 [DOI] [PubMed] [Google Scholar]
- Turner BM 1993. Decoding the nucleosome. Cell 75: 5–8 [PubMed] [Google Scholar]
- Tuteja G, Jensen ST, White P, Kaestner KH 2008. Cis-regulatory modules in the mammalian liver: composition depends on strength of Foxa2 consensus site. Nucleic Acids Res 36: 4149–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike DL, Mango SE 2006. Temporal regulation of foregut development by HTZ-1/H2A.Z and PHA-4/FoxA. PLoS Genet 2: e161 doi: 10.1371/journal.pgen.0020161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A 2011. Determinants of nucleosome organization in primary human cells. Nature 474: 516–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega VB, Lin CY, Lai KS, Kong SL, Xie M, Su X, Teh HF, Thomsen JS, Yeo AL, Sung WK, et al. 2006. Multi-platform genome-wide identification and modeling of functional human estrogen receptor binding sites. Genome Biol 7: R82 doi: 10.1186/gb-2006-7-9-r82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturi CB, Erkine AM, Gross DS 2000. Cell cycle-dependent binding of yeast heat shock factor to nucleosomes. Mol Cell Biol 20: 6435–6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignali M, Steger DJ, Neely KE, Workman JL 2000. Distribution of acetylated histones resulting from Gal4-VP16 recruitment of SAGA and NuA4 complexes. EMBO J 19: 2629–2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457: 854–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE, Elefanty AG, Strasser A, Adams JM 1992. GATA-1 but not SCL induces megakaryocytic differentiation in an early myeloid line. EMBO J 11: 4557–4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss TC, Schiltz RL, Sung MH, Yen PM, Stamatoyannopoulos JA, Biddie SC, Johnson TA, Miranda TB, John S, Hager GL 2011. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell 146: 544–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Li W, Liu X-S, Carroll JS, Janne OA, Krasnickas Keeton E, Chinnaiyan AM, Pienta KJ, Brown M 2007. A hierarchical network of transcription factors governs androgen receptor dependent prostate cancer growth. Mol Cell 27: 380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, Chen Z, Beroukhim R, Wang H, Lupien M, et al. 2009. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell 138: 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. 2011. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474: 390–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. 2011. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472: 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt AJ, Zhao R, Li J, Duncan SA 2007. Development of the mammalian liver and ventral pancreas is dependent on GATA4. BMC Dev Biol 7: 37 doi: 10.1186/1471-213X-7-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JA, Zhang C, Klein-Szanto AJ, Kormish JD, Fu J, Zhang MQ, Zaret KS 2011. Study of FoxA pioneer factor at silent genes reveals Rfx-repressed enhancer at Cdx2 and a potential indicator of esophageal adenocarcinoma development. PLoS Genet 7: e1002277 doi: 10.1371/journal.pgen.1002277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welboren WJ, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FC, Span PN, Stunnenberg HG 2009. ChIP-seq of ERα and RNA polymerase II defines genes differentially responding to ligands. EMBO J 28: 1418–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Ye M, Feng R, Graf T 2004. Stepwise reprogramming of B cells into macrophages. Cell 117: 663–676 [DOI] [PubMed] [Google Scholar]
- Xu J, Pope SD, Jazirehi AR, Attema JL, Papathanasiou P, Watts JA, Zaret KS, Weissman IL, Smale ST 2007. Pioneer factor interactions and unmethylated CpG dinucleotides mark silent tissue-specific enhancers in embryonic stem cells. Proc Natl Acad Sci 104: 12377–12382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Watts JA, Pope SD, Gadue P, Kamps M, Plath K, Zaret KS, Smale ST 2009. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev 23: 2824–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Ito E, Azuma S, Honma R, Yanagisawa Y, Nishikawa A, Kawamura M, Imai J, Tatsuta K, Inoue J, et al. 2008. FoxA1 as a lineage-specific oncogene in luminal type breast cancer. Biochem Biophys Res Commun 365: 711–717 [DOI] [PubMed] [Google Scholar]
- Yamanaka S 2009. Elite and stochastic models for induced pluripotent stem cell generation. Nature 460: 49–52 [DOI] [PubMed] [Google Scholar]
- Yan J, Xu L, Crawford G, Wang Z, Burgess SM 2006. The forkhead transcription factor FoxI1 remains bound to condensed mitotic chromosomes and stably remodels chromatin structure. Mol Cell Biol 26: 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Zhu Z, Kapranov P, McKeon F, Church GM, Gingeras TR, Struhl K 2006. Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol Cell 24: 593–602 [DOI] [PubMed] [Google Scholar]
- Yechoor V, Liu V, Espiritu C, Paul A, Oka K, Kojima H, Chan L 2009. Neurogenin3 is sufficient for transdetermination of hepatic progenitor cells into neo-islets in vivo but not transdifferentiation of hepatocytes. Dev Cell 16: 358–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Morse RH 1999. Chromatin opening and transactivator potentiation by RAP1 in Saccharomyces cerevisiae. Mol Cell Biol 19: 5279–5288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309: 626–630 [DOI] [PubMed] [Google Scholar]
- Zaret KS, Yamamoto KR 1984. Reversible and persistent changes in chromatin structure accompany activation of a glucocoriticoid-dependent enhancer element. Cell 258: 1780–1784 [DOI] [PubMed] [Google Scholar]
- Zaret KS, Caravaca JM, Tulin A, Sekiya T 2011. Nuclear mobility and mitotic chromosome binding similarities between pioneer transcription factor FoxA and linker histone H1. Cold Spring Harb Symp Quant Biol 75: 219–226 [DOI] [PubMed] [Google Scholar]
- Zhang H, Roberts DN, Cairns BR 2005. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123: 219–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, Liu XS, Struhl K 2009. Intrinsic histone–DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol 16: 847–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wippo CJ, Wal M, Ward E, Korber P, Pugh BF 2011. A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science 332: 977–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Watt AJ, Li J, Luebke-Wheeler J, Morrisey EE, Duncan SA 2005. GATA6 is essential for embryonic development of the liver but dispensable for early heart formation. Mol Cell Biol 25: 2622–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y-B, Gerchman SE, Ramakrishnan V, Travers A, Muyldermans S 1998. Position and orientation of the globular domain of linker histone H5 on the nucleosome. Nature 395: 402–405 [DOI] [PubMed] [Google Scholar]
- Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA 2008. In vivo reprogramming of adult pancreatic exocrine cells to β-cells. Nature 455: 627–632 [DOI] [PMC free article] [PubMed] [Google Scholar]