The removal of H2BK123ub by the ubiquitin-specific proteases Ubp8 and Ubp10 may have nonredundant roles. The authors comprehensively compare H2BK123ub levels in wild type with those in ubp8Δ and ubp10Δ mutants and reveal that Ubp8 and Ubp10 have site-specific roles, with Ubp8 deubiquinating H2BK123ub at H3K4me3-marked regions and Ubp10 removing H2BK123ub at H3K79me3-enriched sites. In addition, Kobor et al. propose a more complete model of the H2BK123ub cross-talk.
Keywords: chromatin, histone modifications, histone cross-talk, H2B monoubiquitination, deubiquitination
Abstract
Monoubiquitination of H2BK123 (H2BK123ub), catalyzed by Rad6/Bre1, is a transient histone modification with roles in transcription and is essential for establishing H3K4 and H3K79 trimethylations (H3K4me3 and H3K79me3). Here, we investigated the chromatin network around H2BK123ub by examining its localization and co-occurrence with its dependent marks as well as the transcription elongation mark H3K36me3 across the genome of Saccharomyces cerevisiae. In yeast, H2BK123ub is removed by the deubiquitinases Ubp8 and Ubp10, but their genomic target regions remain to be determined. Genome-wide maps of H2BK123ub in the absence of Ubp8 and Ubp10 revealed their distinct target loci, which were genomic sites enriched for H3K4me3 and H3K79me3, respectively. We propose an extended model of the H2BK123ub cross-talk by integrating existing relationships with the substrate specificities of Ubp8 and Ubp10 reported here.
In eukaryotic cells, chromatin packages DNA into the nucleus and affects various aspects of genome function. As the fundamental components of chromatin, histone proteins are subject to a variety of post-translational modifications, including methylation, monoubiquitination, and acetylation (Shilatifard 2006; Smith and Shilatifard 2010). In a process referred to as histone cross-talk, some histone modifications may trigger others, resulting in distinct and coordinated localization patterns that regulate processes such as transcription, DNA replication, and repair (Lee et al. 2010). An evolutionarily conserved regulatory component of the transcription cycle is the cross-talk between H2BK123 monoubiquitination (H2BK123ub) and H3K4 and H3K79 trimethylation (H3K4me3 and H3K79me3), whereby H2BK123ub is essential to establish the trimethylation marks (Shilatifard 2006).
While previous studies revealed aspects of this cross-talk and identified major components of the associated enzymatic machinery, a comprehensive genome-wide understanding of H2BK123ub localization, including the co-occurrence with its dependent marks and the rules governing its removal, is still lacking. H2BK123ub is a transient histone mark, which is established by the Rad6/Bre1 ubiquitin ligase complex during transcription initiation and elongation (Shilatifard 2006). In Saccharomyces cerevisiae, H2BK123ub localizes to promoter and coding regions of PMA1, ADH1, and PYK1, as well as to GAL1 during gene induction (Dover et al. 2002; Henry et al. 2003; Wood et al. 2003; Kao et al. 2004; Xiao et al. 2005; Schulze et al. 2009), while in human cells it is enriched at transcribed regions of highly transcribed genes (Minsky et al. 2008). Improper monoubiquitination of H2B disturbs transcription elongation at GAL1 in S. cerevisiae (Shukla and Bhaumik 2007) and alters the localization of RNA polymerase II (RNAPII) and histones in coding regions of hem2+ and sod2+ in Schizosaccharomyces pombe (Tanny et al. 2007).
An integral part of the H2BK123ub cross-talk is the removal of H2BK123ub by the ubiquitin-specific proteases Ubp8 and Ubp10 (Henry et al. 2003; Emre et al. 2005). Ubp8 is a subunit of the Spt–Ada–Gcn5–acetyltransferase (SAGA) complex, with the integrity of SAGA being required for Ubp8 deubiquitination activity (Henry et al. 2003). Ubp8 acts mainly at early steps of transcription and regulates the transition between initiation and elongation (Henry et al. 2003; Daniel et al. 2004). In contrast, Ubp10 (also known as Dot4) acts independently of SAGA and has been associated with telomeric silencing (Kahana and Gottschling 1999; Emre et al. 2005; Gardner et al. 2005; Smith and Shilatifard 2009). Based on these functional differences and bulk protein blotting results showing increased ubiquitin levels in the ubp8Δubp10Δ double mutant compared with single-deletion strains (Emre et al. 2005; Gardner et al. 2005), we propose that Ubp8 and Ubp10 have nonredundant roles in the H2BK123ub cross-talk and act in a site-specific manner.
To test this hypothesis, we comprehensively compare H2BK123ub levels in wild type with ubp8Δ and ubp10Δ mutants. Furthermore, we extend our previous study describing the colocalization of H2BK123ub with H3K79me3 (Schulze et al. 2009) by determining the co-occurrences of H2BK123ub with both its dependent modifications and the transcription elongation mark H3K36me3. We reveal that Ubp8 and Ubp10 have site-specific roles, with Ubp8 deubiquitinating H2BK123ub at H3K4me3-marked regions and Ubp10 removing H2BK123ub at H3K79me3-enriched sites. In addition, detailed analyses identified region-specific co-occurrences of H2BK123ub and H3 methylation marks, allowing us to propose a more complete model of the H2BK123ub cross-talk.
Results and Discussion
Genome-wide distribution of H2BK123ub and H3 methylation marks with respect to gene length and transcriptional frequency
To define the chromatin network around H2BK123ub in S. cerevisiae, we mapped H2BK123ub; its dependent marks, H3K4me3 and H3K79me3; and the transcription elongation mark H3K36me3 using high-resolution tiling arrays. We also included H3K79me2 as a control for specificity. H2BK123ub and the H3 methylation marks were strongly enriched in genomic regions transcribed by RNAPII but mostly absent from other genomic features, such as telomeres, centromeres, the rRNA locus, ARSs, and tRNAs (Supplemental Table S1). Therefore, we focused our analysis on RNAPII transcripts and developed a compact yet comprehensive visualization approach (CHROMATRA) to assess the distribution of histone modifications across all transcripts at once while accounting for gene length and transcriptional frequency (T Hentrich, JM Schulze, E Emberly, and MS Kobor, in prep.).
Our work extends existing studies on single genes to encompass the entire genome and finds H2BK123ub to predominantly cover coding sequences of genes as well as some promoters (Supplemental Fig. S1). Its dependent mark, H3K4me3, peaked sharply downstream from the transcription start site (TSS); H3K79me3 covered the body of mainly longer transcripts; H3K79me2 marked mainly shorter genes; and H3K36me3 was enriched beginning at the +3 nucleosome throughout the body of transcripts (Supplemental Fig. S1).
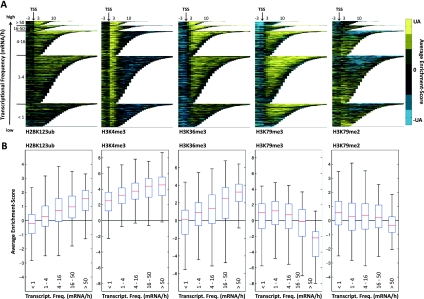
To examine the correlation between the modifications of the H2BK123ub network and transcriptional gene activity, all transcripts were grouped into five classes according to their transcriptional frequency (Holstege et al. 1998). H2BK123ub was present in all classes (Fig. 1A), and the relative occupancy was positively correlated with the transcriptional frequency of genes (Fig. 1B). H3K4me3 showed a trend similar to H2BK123ub but with higher enrichment levels (Fig. 1A,B) and, as recently described (Zhang et al. 2011), was enriched at almost all genes despite earlier proposals that it is limited to highly transcribed genes (Santos-Rosa et al. 2002; Ng et al. 2003; Liu et al. 2005). The most highly transcribed genes were instead specifically enriched for a combination of H2BK123ub, H3K4me3, and H3K36me3 (Fig. 1A). In contrast, enrichment levels of H3K79me3 and H3K79me2 strongly decreased for genes with higher transcriptional frequency (Fig. 1A), rendering the most actively transcribed genes devoid of these marks (Fig. 1B). Interestingly, the association of H3K36me3 and transcriptional activity was nearly identical to that found for H2BK123ub (Fig. 1), supporting a role of H2BK123ub in transcriptional elongation.
Figure 1.
Association of H2BK123ub and H3 methylation marks with transcriptional frequency. (A) Enrichment of H2BK123ub, H3K4me3, H3K36me3, H3K79me3, and H3K79me2 across all transcripts sorted by their length and transcriptional frequency and aligned by their TSSs. The normalized ChIP-on-chip model-based analysis of tiling arrays (MAT) scores were binned into segments of 150 base pairs (bp), and the average enrichment value for each bin was color-coded and plotted. The upper adjacent (UA) of the MAT score distribution was used for color bar limits. Transcripts were grouped into five classes according to their number of transcripts per hour (Holstege et al. 1998) (Transcriptome 2005, http://web.wi.mit.edu/young/pub/holstege.html). (B) Box plots indicating the association of histone marks with transcriptional frequencies. As in A, transcripts were grouped into five classes according to their transcriptional frequency. For all modifications and each transcript, the average enrichment score was calculated as the average MAT score of all probes between transcript start and end. For each transcription class, the average scores were plotted as standard box plots (see the Materials and Methods). For H3K4me3, which peaks downstream from the TSSs around the +2 and +3 nucleosome, average scores were calculated for 300 bp in that region.
H2BK123ub positively correlates and colocalizes with its dependent marks, H3K4me3 and H3K79me3, but also with H3K36me3
Consistent with its role as an upstream regulator, the H2BK123ub profile positively correlated with those of its dependent marks, H3K4me3 and H3K79me3, although the correlation was much stronger with H3K79me3 (r = 0.67) than with H3K4me3 (r = 0.26) (Supplemental Fig. S2A). Supporting our earlier qualitative statement, H2BK123ub correlated positively with the elongation mark H3K36me3 (r = 0.63), consistent with the role of H2BK123ub in transcriptional elongation. To examine the relationship of H2BK123ub and H3 methylation in more detail, genes were partitioned into five segments (promoter, TSS-proximal, 5′ coding sequence (CDS) mid-CDS, and 3′CDS), similar to an approach used previously (Liu et al. 2005), and the average enrichment score for each modification was calculated, hierarchically clustered, and visualized as a heat map (Supplemental Fig. S2B). Among several distinctive combinations of modification patterns, H2BK123ub strongly colocalized with H3K4me3 in the TSS-proximal segment, whereas H2BK123ub and H3K79me3, together with H3K36me3, co-occurred predominantly in the mid-CDS segment (Supplemental Fig. S2B; Supplemental Table S2).
Although nearly all regions enriched for H2BK123ub were also marked by H3K4me3 or H3K79me3, not all sites enriched for H3K4me3 and H3K79me3 also carried H2BK123ub (Supplemental Fig. S2B). This observation might be attributable to the transient nature of H2BK123ub and its mode of removal by the deubiquitinases Ubp8 and Ubp10, and likely explains the rather low rank correlation of r = 0.26 between H2BK123ub and H3K4me3.
Site-specific removal of H2BK123ub by its deubiquitinases, Ubp8 and Ubp10
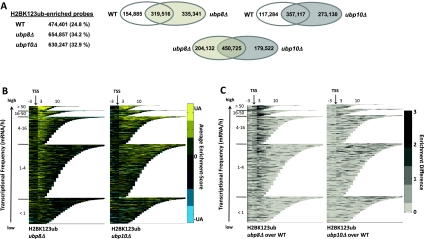
To determine the genomic regions that Ubp8 and Ubp10 act on, we mapped H2BK123ub across the genome in strains lacking either Ubp8 or Ubp10. As expected from studies based on bulk protein levels (Emre et al. 2005; Gardner et al. 2005), the number of H2BK123ub-enriched probes within transcripts strongly increased in both deletion strains from 24.8% in wild-type cells to 34.2% in the ubp8Δ strain and 32.9% in the ubp10Δ strain (Fig. 2A). Supporting our hypothesis that Ubp8 and Ubp10 act at distinct genomic loci, newly enriched probes for H2BK123ub were different between the two deletion strains, with ∼11% of all transcribed probes being uniquely deubiquitinated by Ubp8 and 9% being uniquely deubiquitinated by Ubp10 (Fig. 2A). To assess the location of ubiquitinated regions in both deletion strains, CHROMATRA was used to visualize the H2BK123ub distribution across all transcripts (Fig. 2B; Supplemental Fig. S3). In the ubp8Δ strain, H2BK123ub peaked downstream from the TSS, but was reduced in the body of transcripts compared with wild-type cells (Fig. 2B; Supplemental Fig. S3). In contrast, H2BK123ub localized to the coding sequence of mainly longer genes in the ubp10Δ strain (Fig. 2B; Supplemental Fig. S3). To better visualize the localization of newly enriched sites along the transcripts, we subtracted wild type from deletion profiles and color-coded positive-definite results (Fig. 2C). The resulting profiles clearly indicated a site-specific removal of H2BK123ub by Ubp8 in the TSS-proximal region and by Ubp10 in the gene-coding region.
Figure 2.
Site-specific removal of H2BK123 ubiquitin by Ubp8 and Ubp10. (A) Number of probes enriched for H2BK123ub within transcripts in wild-type as well as ubp8 and ubp10 deletion strains. Venn diagrams comparing the overlap of these probes between the different strains. (B) Distribution of H2BK123ub in wild-type as well as ubp8 and ubp10 deletion strains across all transcripts, sorted by their transcriptional frequencies. Calculations and plotting as in Figure 1A. (C) Differences in the enrichment of H2BK123ub in ubp8 and ubp10 deletion strains. Enrichment scores for H2BK123ub in ubp8 and ubp10 deletion strains were subtracted from wild-type enrichment scores and only positive-definite results were color-coded. Average enrichment was calculated and transcripts were sorted as in Figure 1A.
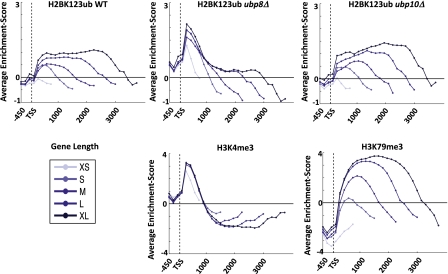
By averaging the enrichment profiles of H2BK123ub and its dependent marks in a length-dependent manner similar to a previous approach (Mayer et al. 2010), we noticed that the H2BK123ub profile was very different from H3K4me3 but comparable in its lateral distribution and overall shape with H3K79me3 (Fig. 3). Upon deletion of Ubp8, the H2BK123ub profile changed dramatically, now showing a striking similarity to H3K4me3. Consistently, the rank correlation between H2BK123ub and H3K4me3 increased from r = 0.26 in wild-type cells to r = 0.45 in the ubp8Δ strain. The ubp10 deletion profile, however, had relatively modest changes, although the degree of resemblance to H3K79me3 further increased (Fig. 3). These data suggest that Ubp8 acts primarily in the 5′CDS marked by H3K4me3, whereas Ubp10 deubiquitinates H2BK123 in the body of transcripts marked by H3K79me3.
Figure 3.
H2BK123ub profiles in ubp8 and ubp10 deletion strains resembled H3K4me3 and H3K79me3 profiles, respectively. All genes with known TSSs were divided into five length classes, and the average enrichment for H2BK123ub wild-type and ubp8 and ubp10 deletion strains as well as H3K4me3 and H3K79me3 were plotted in 150-bp increments. The H2BK123ub profiles in the ubp8Δ and ubp10Δ strains resembled the averaged profiles of H3K4me3 and H3K79me3, respectively.
Given that in wild-type cells with both deubiquitinases present the measured H2K123ub level reflects the least transient fraction captured by chromatin immunoprecipitation (ChIP), our results further indicate that H2BK123ub was more transient in the 5′CDS than in the body of transcripts. In addition, we observed a strong reduction of H2BK123ub in coding regions in the ubp8 deletion strain (Fig. 3). This might suggest that nonremoval of the ubiquitin tag at the 5′CDS caused by loss of Ubp8 hinders the transcriptional machinery to properly elongate and ubiquitinate downstream regions, thus resulting in strongly reduced ubiquitination of H2BK123 in the body of transcripts.
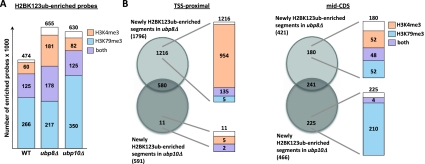
To further investigate the connection between the deubiquitinases and the H2BK123ub-dependent marks, we examined H2BK123ub-enriched probes for co-occupancy with H3K4me3 or H3K79me3 in wild type, ubp8Δ, and ubp10Δ (Fig. 4A). While the number of H2BK123ub-enriched probes co-occurring with H3K4me3 strongly increased in the ubp8Δ strain, probes co-occurring with H3K79me3 mainly increased in the ubp10Δ strain (Fig. 4A). Since the observed changes occurred mostly in the TSS-proximal and mid-CDS regions, we analyzed these gene segments further and determined the overlap of deubiquitinated segments by Ubp8 and Ubp10 with segments marked by H3K4me3 and H3K79me3 (Fig. 4B). Supporting our hypothesis, regions downstream from the TSS uniquely deubiquitinated by Ubp8 were mainly marked by H3K4me3, while very few of these regions were solely enriched for H3K79me3. In contrast, mid-CDS segments uniquely deubiquitinated by Ubp10 were mainly marked by H3K79me3, with H3K4me3 being mostly absent at these sites (Fig. 4B).
Figure 4.
Ubp8 removed H2BK123ub at sites enriched for H3K4me3, whereas Ubp10 acted on H3K79me3-marked regions. (A) All probes enriched for H2BK123ub within transcripts in wild-type as well as ubp8 and ubp10 deletion strains were compared with the number of these probes enriched for H3K4me3 and H3K79me3. (B) Venn diagrams comparing the number of TSS-proximal and mid-CDS segments newly enriched for H2BK123ub in ubp8 and ubp10 deletion strains. Circles represent the overlap of segments newly enriched for H2BK123ub in ubp8 or ubp10 deletion strains. Bars indicate segments marked by H3K4me3 (light red), H3K79me3 (light blue), or both (purple).
Despite the distinct lateral localization of H3K4me3 and H3K79me3 along genes, they overlapped at the transition of TSS-proximal and mid-CDS (Figs. 1, 4). This suggested that both Ubp8 and Ubp10 might be active at these sites despite their otherwise exclusive target regions.
Circuitry of H2BK123ub and its dependent marks, H3K4me3 and H3K79me3
To further dissect the circuitry of H2BK123 ubiquitination/deubiquitination and its dependent marks, we tested whether loss of Ubp8 or Ubp10 had any consequences on the genome-wide distribution of H3K4me3 and H3K79me3. Extending previous observations based on bulk protein blot analyses (Daniel et al. 2004; Gardner et al. 2005; Song and Ahn 2010), we found that the genome-wide distribution of H3K4me3 and H3K79me3 remained largely unchanged, and the site-specific location of these marks was unaffected upon loss of either Ubp8 or Ubp10 (Supplemental Fig. S4). These findings suggest a temporal relation in which the transient H2BK123ub mark triggers the establishment of the relatively stable H3K4me3 and H3K79me3 marks prior to its removal by Ubp8 and Ubp10.
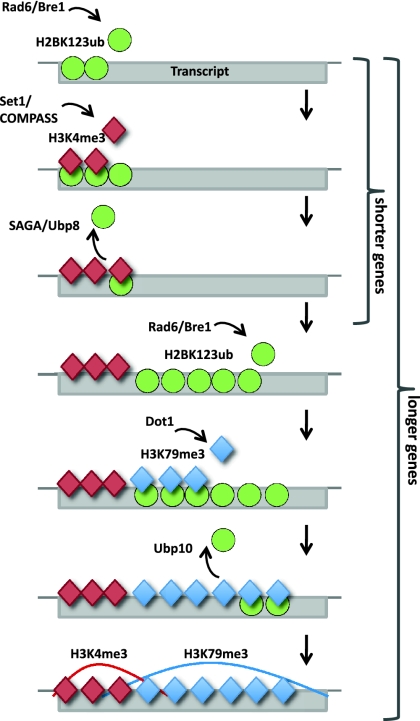
Integrating our findings with the current understanding of the H2BK123ub cross-talk, we propose the following model (Fig. 5). Initially, Rad6/Bre1 is recruited to promoters through interactions with transcriptional activators, catalyzing the monoubiquitination of H2BK123 (Smith and Shilatifard 2010). Together with the surrounding histone residues, H2BK123ub then provides a molecular “tag,” attracting Set1/COMPASS, which subsequently trimethylates H3K4 (Lee et al. 2007; Kim et al. 2009; Zheng et al. 2010). Eventually, Ubp8 removes the bulky H2BK123 monoubiquitin group, and H3K4me3 remains as a memory mark of recent transcriptional initiation (Gerber and Shilatifard 2003; Krogan et al. 2003; Ng et al. 2003; Muramoto et al. 2010). In longer genes, which require an extensive transcription elongation phase, Rad6/Bre1 stay associated with the elongating form of RNAPII and monoubiquitinate H2BK123 throughout the CDS. This provides a molecular “tag” recognized by Dot1, which binds stronger to and resides longer at these sites to specifically trimethylate H3K79 (McGinty et al. 2008; Schulze et al. 2009). In these regions, Ubp10 removes H2BK123ub after establishment of H3K79me3 by Dot1, and the stable H3K79me3 mark remains.
Figure 5.
Model depicting the circuitry of H2BK123ub and its dependent marks, H3K4me3 and H3K79me3, in short and long genes. In the 5′ end of short and long genes, H2B is monoubiquitinated by Rad6/Bre1, resulting in the recruitment of Set1/COMPASS to trimethylate H3K4me3 (Shilatifard 2006). After H3K4me3 is established, Ubp8 removes H2BK123ub. In longer genes, depending on an extensive elongation phase, H2B is monoubiquitinated in the body of transcripts, which recruits Dot1 to trimethylate H3K79me3. In these regions, H2BK123ub is removed by Ubp10.
The site specificity of Ubp8 and Ubp10 raises the question of how target regions are recognized for H2BK123ub removal. Since Sus1 is required for recruitment of Ubp8 to promoters and forms a module with Sgf11 and Sgf73 as part of the SAGA complex (Henry et al. 2003; Daniel et al. 2004; Kohler et al. 2010; Samara et al. 2010), it might help Ubp8 to be recruited to H3K4me3-marked regions specifically (Kohler et al. 2006). Ubp10 has not been identified to be part of any complex, and the mechanism of its recruitment to chromatin remains to be determined. Although Ubp10 was proposed to play a role in telomeric silencing (Emre et al. 2005; Gardner et al. 2005), we detected no H2BK123ub in telomeric regions, including the TG repeat, X and Y′ elements in wild-type and ubp8 and ubp10 deletion srains (Supplemental Fig. S5; Supplemental Table S1), and we recently demonstrated a limited role of H3K79 methylation in natural telomeric silencing (Takahashi et al. 2011). Furthermore, levels of H2BK123ub in subtelomeric regions showed minimal site-specific changes upon loss of Ubp8 or Ubp10 when compared with wild-type cells (Supplemental Fig. S5). A recent study suggests that Ubp10's role at subtelomeric regions is context-dependent and that it only exerts its function at subtelomeres upon impairment of the H2A.Z chaperone Chz1 (Wan et al. 2010). Supporting its broader role in euchromatic regions (Gardner et al. 2005), we revealed that Ubp10 specifically removes H2BK123ub from H3K79 trimethylated coding regions and provide a connection of Ubp10 (Dot4) and Dot1 across the genome.
Proper addition and removal of H2BK123ub were proposed to be essential for optimal gene expression. However, impaired removal of H2BK123ub only leads to moderate effects on transcription levels (Gardner et al. 2005; Lenstra et al. 2011), and loss of Ubp8 does not alter recruitment of RNAPII to GAL1 during gene activation (Wyce et al. 2007). Here, we confirm and extend these observations by showing that the genome-wide localization of the Rpb3 subunit of RNAPII was modestly affected by loss of either Ubp8 or Ubp10 (Supplemental Fig. S6). Furthermore, the distribution of the key elongation mark H3K36me3 was not altered upon loss of Ubp8 (Supplemental Fig. S4), despite the proposed function of H2B monoubiquitination in stimulating the rate of transcription elongation (Shukla and Bhaumik 2007; Tanny et al. 2007). Together, these findings suggest that transcription elongation still takes place and that the cell is able to sufficiently transcribe genes despite impaired removal of the ubiquitin moiety on H2B. We speculate that an indirect removal of the ubiquitin mark occurs through eviction of H2A and H2B during transcription and/or histone turnover. Also, it is possible that H2BK123ub in the body of the gene may play a role in positioning of nucleosomes in front or in the wake of the transcribing RNAPII and that the observed role of H2BK123ub in elongation could be due to its role in nucleosomal positioning. Indeed, in an accompanying study, Pugh and colleagues (Batta et al. 2011) have uncovered a role of H2BK123ub in nucleosomal organization in genic regions.
Taken together, here we comprehensively describe the chromatin network around H2BK123ub and how it relates to both dependent marks and the transcription cycle. Most importantly, our results point toward distinct roles of Ubp8 and Ubp10 in the deubiquitination machinery of eukaryotic cells and are in agreement with previous models describing deubiquitinases as major molecular regulators (D'Andrea 2010).
Materials and methods
Details of the ChIP-on-chip experiments and the genome-wide localization analyses are described in the Supplemental Material. In short, ChIPs were performed using antibodies directed against the indicated histone modifications, with a blocking peptide being added for the experiments interrogating H2BK123ub. Precipitated DNA was amplified with two rounds of T7 RNA polymerase amplification (Schulze et al. 2009) and hybridized to Affymetrix 1.0R S. cerevisiae tiling microarrays. An adapted version of the model-based analysis of tiling arrays (MAT) algorithm (Droit et al. 2010) was used to reliably detect enriched regions as described previously (Schulze et al. 2009), using mostly input DNA for normalization. In the case of H2BK123ub, data were normalized to profiles obtained under identical conditions from a H2BK123A mutant yeast strain. Custom-written scripts were developed and used for subsequent data analysis as described in the Supplemental Material.
Accession numbers
Data files may be accessed online at http://www.ebi.ac.uk/arrayexpress under the accession number E-MEXP-3217.
Acknowledgments
We thank Alice Wang, Grace Leung, Edwin Smith, and Alexander Garrett for critical reading of the manuscript. Furthermore, we thank Hunter Fraser for help with the statistical analysis, Harm van Bakel for providing the detailed ChIP-on-chip protocol as well as the TSS list, and Frank Holstege for helpful discussions. J.M.S. was supported by a fellowship from the Child and Family Research Institute, and S.N. is a Leukemia and Lymphoma Society fellow. M.S.K. is a Scholar of the Michael Smith Foundation for Health Research, the Canadian Institute for Advanced Research, and the Mowafaghian Foundation. Research for this study in the laboratory of M.S.K. was supported by an operating grant from the Canadian Institute of Health Research (MOP-79442). Studies in the Shilatifard laboratory were supported by a fund provided by the National Institutes of Health (R01GM069905) to A.S.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.177220.111.
References
- Batta K, Zhang Z, Yen K, Goffman DB, Pugh BF 2011. Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev (this issue). doi: 10.1101/gad.177238.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Andrea AD. 2010. Susceptibility pathways in Fanconi's anemia and breast cancer. N Engl J Med 362: 1909–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JA, Torok MS, Sun Z-W, Schieltz D, Allis CD, Yates JR, Grant PA 2004. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J Biol Chem 279: 1867–1871 [DOI] [PubMed] [Google Scholar]
- Dover J, Schneider J, Tawiah-Boateng M, Wood A, Dean K, Johnston M, Shilatifard A 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem 277: 28368–28371 [DOI] [PubMed] [Google Scholar]
- Droit A, Cheung C, Gottardo R 2010. rMAT—an R/Bioconductor package for analyzing ChIP–chip experiments. Bioinformatics 26: 678–679 [DOI] [PubMed] [Google Scholar]
- Emre NCT, Ingvarsdottir K, Wyce A, Wood A, Krogan NJ, Henry KW, Li K, Marmorstein R, Greenblatt JF, Shilatifard A, et al. 2005. Maintenance of low histone ubiquitylation by Ubp10 correlates with telomere-proximal Sir2 association and gene silencing. Mol Cell 17: 585–594 [DOI] [PubMed] [Google Scholar]
- Gardner RG, Nelson ZW, Gottschling DE 2005. Ubp10/Dot4p regulates the persistence of ubiquitinated histone H2B: distinct roles in telomeric silencing and general chromatin. Mol Cell Biol 25: 6123–6139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber M, Shilatifard A 2003. Transcriptional elongation by RNA polymerase II and histone methylation. J Biol Chem 278: 26303–26306 [DOI] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo W-S, Duggan LJ, Emre NCT, Kao C-F, Pillus L, Shilatifard A, Osley MA, Berger SL 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev 17: 2648–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege F, Jennings E, Wyrick J, Lee T, Hengartner C, Green M, Golub T, Lander E, Young R 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728 [DOI] [PubMed] [Google Scholar]
- Kahana A, Gottschling DE 1999. DOT4 links silencing and cell growth in Saccharomyces cerevisiae. Mol Cell Biol 19: 6608–6620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CF, Hillyer C, Tsukuda T, Henry K, Berger S, Osley MA 2004. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev 18: 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG 2009. RAD6-mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell 137: 459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Pascual-Garcia P, Llopis A, Zapater M, Posas F, Hurt E, Rodriguez-Navarro S 2006. The mRNA export factor Sus1 is involved in Spt/Ada/Gcn5 acetyltransferase-mediated H2B deubiquitinylation through its interaction with Ubp8 and Sgf11. Mol Biol Cell 17: 4228–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Zimmerman E, Schneider M, Hurt E, Zheng N 2010. Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell 141: 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell 11: 721–729 [DOI] [PubMed] [Google Scholar]
- Lee J, Shukla A, Schneider J, Swanson S, Washburn M, Florens L, Bhaumik S, Shilatifard A 2007. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 131: 1084–1096 [DOI] [PubMed] [Google Scholar]
- Lee JS, Smith E, Shilatifard A 2010. The language of histone crosstalk. Cell 142: 682–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenstra TL, Benschop JJ, Kim T, Schulze JM, Brabers NA, Margaritis T, van de Pasch LA, van Heesch SA, Brok MO, Groot Koerkamp MJ, et al. 2011. The specificity and topology of chromatin interaction pathways in yeast. Mol Cell 42: 536–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ 2005. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol 3: e328 doi: 10.1371/jounral.pbio.0030328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P 2010. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol 17: 1272–1278 [DOI] [PubMed] [Google Scholar]
- McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW 2008. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature 453: 812–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M 2008. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol 10: 483–488 [DOI] [PubMed] [Google Scholar]
- Muramoto T, Müller I, Thomas G, Melvin A, Chubb JR 2010. Methylation of H3K4 is required for inheritance of active transcriptional states. Curr Biol 20: 397–406 [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 11: 709–719 [DOI] [PubMed] [Google Scholar]
- Samara NL, Datta AB, Berndsen CE, Zhang X, Yao T, Cohen RE, Wolberger C 2010. Structural insights into the assembly and function of the SAGA deubiquitinating module. Science 328: 1025–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NCT, Schreiber SL, Mellor J, Kouzarides T 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419: 407–411 [DOI] [PubMed] [Google Scholar]
- Schulze J, Jackson J, Nakanishi S, Gardner J, Hentrich T, Haug J, Johnston M, Jaspersen S, Kobor M, Shilatifard A 2009. Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol Cell 35: 626–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A 2006. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem 75: 243–269 [DOI] [PubMed] [Google Scholar]
- Shukla A, Bhaumik SR 2007. H2B-K123 ubiquitination stimulates RNAPII elongation independent of H3-K4 methylation. Biochem Biophys Res Commun 359: 214–220 [DOI] [PubMed] [Google Scholar]
- Smith E, Shilatifard A 2009. Developmental biology. Histone cross-talk in stem cells. Science 323: 221–222 [DOI] [PubMed] [Google Scholar]
- Smith E, Shilatifard A 2010. The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol Cell 40: 689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Ahn SH 2010. A Bre1-associated protein, large 1 (Lge1), promotes H2B ubiquitylation during the early stages of transcription elongation. J Biol Chem 285: 2361–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi YH, Schulze JM, Jackson J, Hentrich T, Seidel C, Jaspersen SL, Kobor MS, Shilatifard A 2011. Dot1 and histone H3K79 methylation in natural telomeric and HM silencing. Mol Cell 42: 118–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny JC, Erdjument-Bromage H, Tempst P, Allis CD 2007. Ubiquitylation of histone H2B controls RNA polymerase II transcription elongation independently of histone H3 methylation. Genes Dev 21: 835–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Chiang JH, Lin CH, Arens CE, Saleem RA, Smith JJ, Aitchison JD 2010. Histone chaperone Chz1p regulates H2B ubiquitination and subtelomeric anti-silencing. Nucleic Acids Res 38: 1431–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A, Krogan N, Dover J, Schneider J, Heidt J, Boateng M, Dean K, Golshani A, Zhang Y, Greenblatt J, et al. 2003. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell 11: 267–274 [DOI] [PubMed] [Google Scholar]
- Wyce A, Xiao T, Whelan KA, Kosman C, Walter W, Eick D, Hughes TR, Krogan NJ, Strahl BD, Berger SL 2007. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol Cell 27: 275–288 [DOI] [PubMed] [Google Scholar]
- Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, Strahl BD 2005. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol Cell Biol 25: 637–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ma H, Pugh BF 2011. Stable and dynamic nucleosome states during a meiotic developmental process. Genome Res 21: 875–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Wyrick JJ, Reese JC 2010. Novel trans-tail regulation of H2B ubiquitylation and H3K4 methylation by the N terminus of histone H2A. Mol Cell Biol 30: 3635–3645 [DOI] [PMC free article] [PubMed] [Google Scholar]