Yokoyama et al. solved the crystal structure of UTX, an H3K27me3/2-specific demethylase bound with an H3 peptide. The authors found that both the catalytic jumonji domain and a novel zinc-binding domain bind their targets in a sequence-specific manner. Therefore, the structure reported here demonstrates that UTX uses two lock-and-key sets to discriminate between H3K27me3 and H3K9me3 in histones.
Keywords: epigenetics, UTX/KDM6A, histone demethylase, jumonji domain, crystal structure, H3K27 methylation
Abstract
Tri- and dimethylations of histone H3K9 (H3K9me3/2) and H3K27 (H3K27me3/2), both situated in the “A-R-Kme-S” sequence motif, mediate transcriptional repression of distinct genomic regions. H3K9me3/2 mainly governs constitutive heterochromatin formation, while H3K27me3/2 represses key developmental genes. The mechanisms by which histone-modifying enzymes selectively regulate the methylation states of H3K9 and H3K27 are poorly understood. Here we report the crystal structures of the catalytic fragment of UTX/KDM6A, an H3K27me3/2-specific demethylase, in the free and H3 peptide-bound forms. The catalytic jumonji domain binds H3 residues 25–33, recognizing H3R26, H3A29, and H3P30 in a sequence-specific manner, in addition to H3K27me3 in the catalytic pocket. A novel zinc-binding domain, conserved within the KDM6 family, binds residues 17–21 of H3. The zinc-binding domain changes its conformation upon H3 binding, and thereby recognizes the H3L20 side chain via a hydrophobic patch on its surface, which is inaccessible in the H3-free form. Mutational analyses showed that H3R17, H3L20, H3R26, H3A29, H3P30, and H3T32 are each important for demethylation. No other methyllysines in the histone tails have the same set of residues at the corresponding positions. Thus, we clarified how UTX discriminates H3K27me3/2 from the other methyllysines with distinct roles, including the near-cognate H3K9me3/2, in histones.
Histones are subjected to a variety of post-translational modifications, such as acetylation of lysine; methylation of lysine and arginine; phosphorylation of serine, threonine, and tyrosine; and ubiquitination of lysine. These modifications regulate the structure and function of chromatin, mainly by modulating the histone•DNA interactions and the binding of effector proteins that recognize specific modified/unmodified states of histone residues (Jenuwein and Allis 2001).
Histone lysine methylations, found in at least five sites in H3 and one site in H4, have different biological consequences, depending on their locations and methylation states (non-, mono-, di-, and trimethylations) (Kouzarides 2007). For example, the tri- and dimethylations of H3K4 and H3K36 (H3K4me3/2 and H3K36me3/2, respectively) are associated with transcriptional activation. On the other hand, both H3K9me3/2 and H3K27me3/2 mediate transcriptional repression. Nevertheless, they are thought to operate through distinct mechanisms and to target different genomic regions. H3K9me3, which recruits heterochromatin protein 1 (HP1) and multiple repressor complexes associated with it, is mainly found in pericentric and telomeric regions and repeat elements. In contrast, H3K27me3 cooperates with polycomb group proteins to repress developmental genes. In multipotent stem cells, most of the genes encoding developmental regulators are repressed by H3K27me3 (Bernstein et al. 2006; Boyer et al. 2006). As the cells differentiate, a particular set of the genes loses H3K27me3 to start developmental programs, depending on the specific cell lineage to which they have committed (Mikkelsen et al. 2007).
So far, two related H3K27me3 demethylases, both belonging to the jumonji family, have been identified (Agger et al. 2007; De Santa et al. 2007; Hong et al. 2007; Lan et al. 2007; Lee et al. 2007b; Smith et al. 2008). UTX, also designated as KDM6A, regulates the expression of HOX genes by demethylating H3K27me3/2 and thereby antagonizing the repression mediated by polycomb proteins (Agger et al. 2007; Lan et al. 2007; Lee et al. 2007b). The other known H3K27me3/2 demethylase, JMJD3 (also designated as KDM6B), is induced in macrophages in response to inflammatory signals and also activates polycomb target genes during macrophage activation (De Santa et al. 2007). Notably, no activity has been detected for UTX and JMJD3 toward H3K9, although H3K27 and H3K9 reside in remarkably similar amino acid sequences, including the identical “A-R-Kme-S” motif.
Aberrant histone modifications often result in developmental defects and human diseases, underscoring the importance of their strict regulation (Bhaumik et al. 2007; Chi et al. 2010; Pedersen and Helin 2010). Inactivating mutations of the UTX gene are found in multiple tumor types (van Haaften et al. 2009), suggesting that it acts as a tumor suppressor, possibly by regulating the expression of cell cycle regulators, such as retinoblastoma-binding proteins (Herz et al. 2010; Wang et al. 2010). On the other hand, the decreased level of H3K9 methylation leads to chromosome instability, abnormal telomere elongation, impaired animal viability, and an increased tumor risk (Peters et al. 2001; García-Cao et al. 2004). Thus, the H3K27 demethylation activity of the KDM6 proteins, as well as their ability to discriminate H3K27 from H3K9, is important for normal cell functions.
Unlike the KDM6 proteins, several jumonji demethylases act on multiple histone lysine residues. Functional and structural analyses have provided insights into the multiple site specificity of these enzymes (Hou and Yu 2010). JMJD2A/JHDM3A/KDM4A, an H3K9me3/2 and H3K36me3/2 demethylase (Klose et al. 2006; Whetstine et al. 2006), is the first jumonji demethylase for which a crystal structure was reported (Chen et al. 2006). The JmjC and JmjN domains, which are the signature motifs of the jumonji family, tightly associate with each other and with other regions to form a single larger domain (hereafter collectively called the jumonji domain) (Horton et al. 2010). The catalytic jumonji domain binds and demethylates methyllysine in an Fe(II)- and α-ketoglutarate (αKG)-dependent manner. JMJD2A primarily interacts with the main chain of H3 and does not use the sequence-specific recognition of the H3 side chains (Chen et al. 2007; Couture et al. 2007; Ng et al. 2007). PHF8 and KDM7A are H3K9me2/1 and H3K27me2/1 demethylases belonging to the PHF cluster (Horton et al. 2010; Yang et al. 2010). Their jumonji domains are thought to bind H3K9 and H3K27 by recognizing the identical set of H3 residues near the substrate methyllysine in the “A-R-Kme-S” motif, whereas the second methyllysine (H3K4me3) modulates the specificity of the enzymes.
On the other hand, the structures and the H3K27-specific recognition mechanism of the KDM6 proteins have remained unclear. It is not known how they discriminate H3K27 from H3K9 and whether any domain other than the catalytic jumonji domain is relevant to the site specificity. To address these questions, we solved the crystal structures of the C-terminal catalytic fragment of UTX in the H3-free and H3-bound forms. The structures revealed that the catalytic jumonji domain cooperates with a novel zinc-binding domain in order to recognize two separate segments of H3. UTX thereby recognizes H3K27me3 in a highly sequence-specific manner. Structure-based mutational analyses confirmed that the H3 residues recognized by UTX are essential for demethylation. These analyses have revealed the molecular basis for the high specificity of UTX, which can discriminate H3K27me3 even from the near-cognate H3K9me3.
Results
Overall structures
The C-terminal region of UTX (residues 880–1401), containing the jumonji domain, is highly conserved between UTX and JMJD3 (Fig. 1). To gain insight into the sequence-specific demethylation mechanism of UTX, we prepared the fragment corresponding to the C-terminal region of UTX, which actually exhibits demethylase activity against H3K27me3 (see below). The crystal structures of the fragment were solved in two forms—(1) the complex with N-oxyalylglycine (NOG; a nonreactive analog of αKG) and Ni(II), which coordinates in the Fe(II)-binding site but does not support catalysis, and (2) the complex with NOG, Ni(II), and an H3K27me3 peptide (residues 17–38 of H3)—at 1.85 and 1.80 Å resolutions, respectively (Fig. 2; Table 1). Clear electron density is visible for residues 17–33 of the bound H3 peptide, with the exception of the side chains of H3R17, H3K18, and H3K23, which are less ordered (Supplemental Fig. 1).
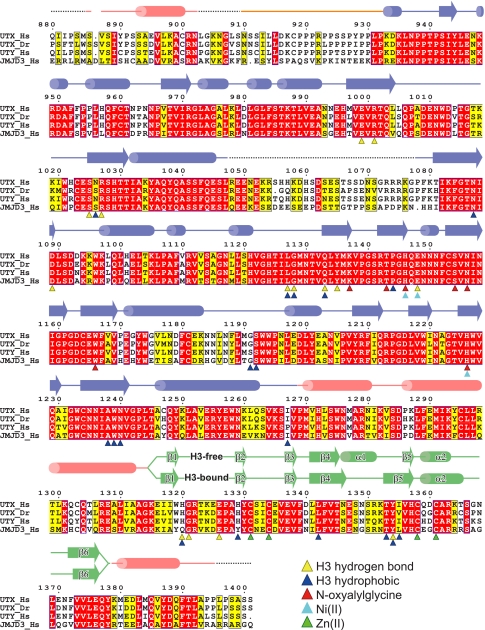
Figure 1.
Sequence alignment of KDM6 proteins. Alignment of the amino acid sequences of human and zebrafish UTX, human UTY, and human JMJD3. The helical domain, the linker region, the jumonji domain, and the zinc-binding domain are colored pink, orange, blue, and green, respectively. Disordered regions are shown with dashed lines. Secondary structural elements (cylinders for helices and arrows for β strands) of the H3-free and H3-bound structures are shown separately. Yellow and blue triangles show residues interacting with H3 via hydrogen bonds and hydrophobic interactions, respectively. Red, cyan, and green triangles show residues interacting with NOG, Ni(II), and Zn(II), respectively. UTY also conserves the residues involved in H3 and cofactor binding.
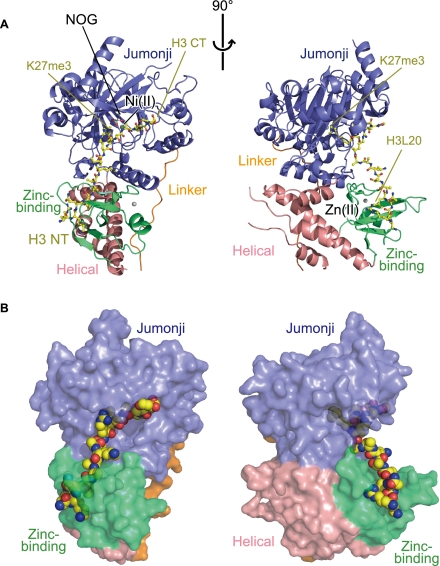
Figure 2.
Overall structure of the UTX•H3 complex. (A) Orthogonal views of the overall structure of the UTX•H3 complex. UTX is shown in a ribbon representation. The bound H3 peptide (yellow), NOG (white), Ni(II) (pale green), and Zn(II) (silver) are shown in a ball-and-stick representation. The N and C termini of the H3 peptide are labeled with “H3 NT” and “H3 CT,” respectively. (B) Same as A, but UTX and the H3 peptide are shown in surface and space-filling representations, respectively.
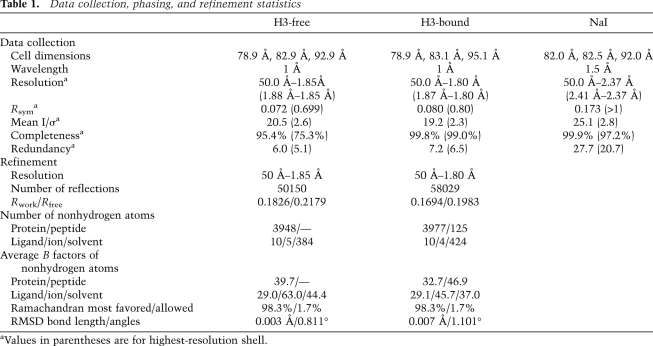
Table 1.
Data collection, phasing, and refinement statistics
As shown in Figures 1 and 2, the UTX structure consists of four regions: the helical domain (residues 886–902, 1269–1314, and 1379–1395, shown in pink), the linker region (residues 910–932, orange), the jumonji domain (residues 933–1047 and 1078–1268, blue), and the zinc-binding domain (residues 1315–1378, green). The helical domain is composed of five helices that reside on both sides of the jumonji domain in the amino acid sequence. The jumonji domain assumes a jellyroll fold with the central β barrel flanked by α helices, similar to other oxygenases belonging to the cupin superfamily (Clissold and Ponting 2001). The helical domain is tightly associated with the other two domains via hydrophobic interactions, thereby connecting them together and stabilizing the overall architecture. The zinc-binding domain is inserted in the helical domain in the amino acid sequence. The bound H3 peptide assumes an extended conformation without intramolecular hydrogen bonds, and its N-terminal and C-terminal parts interact with the zinc-binding and jumonji domains, respectively.
H3K27me3 binding and catalysis
The jumonji domain accommodates the H3K27me3 side chain in the central cavity of its β barrel (Figs. 2, 3A,D). No significant structural change of this domain is observed upon H3 binding. The main chain amino and carbonyl groups of H3K27me3 interact with the L1127 carbonyl group and the Q1133 side chain of UTX, respectively. As a result, the trimethyllysine moiety is oriented toward the center of the cavity, where NOG and Ni(II) are also bound. The three methyl groups are recognized by characteristic CH–O hydrogen bonds, as in the cases of the SET domain methyltransferases (Xiao et al. 2003) and the JMJD2A demethylase (Chen et al. 2007; Couture et al. 2007; Ng et al. 2007). Y1135 and G1128 (corresponding to Y177 and G170 of JMJD2A, respectively) each form bifurcated CH–O hydrogen bonds with two methyl groups of H3K27me3. The side chain of E1148 (corresponding to JMJD2A E190) also hydrogen bonds with one of the methyl groups and interacts electrostatically with the positively charged methylammonium group. These interactions fix the orientation of one of the methyl groups so that it can be subjected to demethylation. One difference between the trimethyllysine recognition modes of UTX and JMJD2A is that JMJD2A forms an additional CH–O hydrogen bond with a methyl ammonium group via its S288 residue. This residue is frequently substituted by Ala in other enzymes, such as JMJD2D and UTX (A1238) (Fig. 3A). The substitution of S288 in JMJD2A with Ala was shown to enhance its activity, especially on dimethylated substrates, suggesting its role in the determination of the methylation state specificity (Chen et al. 2006; Couture et al. 2007).
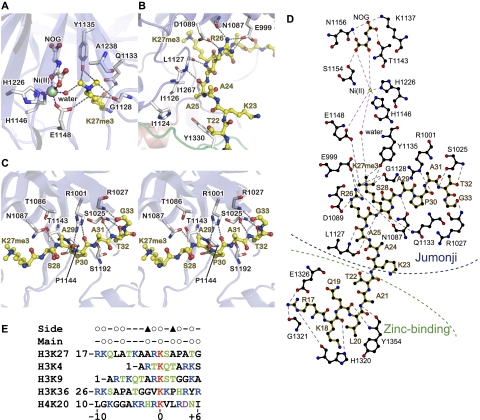
Figure 3.
H3 binding. (A) Binding of H3K27me3, NOG, and Ni(II). Purple and gray dashed lines represent bonds coordinating Ni(II) and hydrogen bonds, respectively. (B) Binding of H3R26 and surrounding residues. (C) Binding of the C-terminal region of the H3 peptide (stereo view). (D) A schematic of the interactions between UTX, the H3 peptide, NOG, and Ni(II), created by ligplot (Wallace et al. 1995). Hydrophobic interactions are omitted for clarity. (E) Sequence alignment of the methylation sites in the N termini of histones H3 and H4. Open circles indicate the residues around H3K27 with a main chain or side chain that interacts with UTX in the present structure. Filled triangles indicate the residues whose substitutions with large amino acids may abolish UTX binding.
NOG is also accommodated in the central cavity of the barrel, where it hydrogen bonds with K1137, T1143, S1154, and N1156 (Fig. 3A,D). The bound Ni(II) ion is octahedrally coordinated to two oxygen atoms of NOG; the side chains of H1146, E1148, and H1226; and one water molecule. In the H3 peptide complex structure, this water is located in close proximity to one of the methyl groups of the trimethylated H3K27. Similar enzyme–cofactor interactions have been observed in the structures of JMJD2A–H3 complexes (Chen et al. 2007; Couture et al. 2007; Ng et al. 2007).
Jumonji–H3 interaction
The jumonji domain binds residues 25–33 of H3 by hydrogen bonding and hydrophobic interactions via the side chains as well as the main chain of H3 (Fig. 3B–E). In particular, H3R26, which resides at position −1 relative to the methyllysine substrate, is extensively recognized by three hydrogen bonds with the side chains of E999 and D1089 and the N1087 main chain carbonyl group. The H3P30 side chain also forms hydrophobic interactions with P1144, and the H3T32 side chain hydrogen bonds with the S1025 main chain amide group. Moreover, UTX hydrogen bonds with the main chain carbonyl groups of H3A25, H3A29, H3P30, and H3G33 (via the main chain amide groups of L1127 and R1027 and the side chain of R1001) and the main chain amide groups of H3A29 and H3T32 (via the side chain of N1087 and the main chain carbonyl group of S1025). These interactions fix the conformation of the bound peptide so that the side chains of H3A25 (position −2), H3A29 (position +2), and H3T32 (position +5) protrude toward small pockets on the surface of UTX (Fig. 3B,C). Molecular modeling suggests that the substitution of these residues with larger ones may interfere with peptide binding (Supplemental Fig. 2). A valine, glutamate, or larger residue at position −2; a threonine or larger residue at position +2; and an asparagine or larger residue at position +5 may sterically clash with UTX. On the other hand, the side chains of H3T22, H3K23, H3A24, H3S28, and H3A31 protrude toward the solvent region and do not contact UTX. Taken together, these observations indicate that the residues at positions −2, −1, +2, +3, and +5 may contribute to the specific binding and demethylation by UTX.
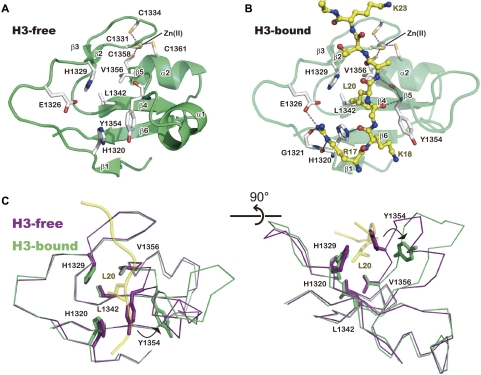
The zinc-binding domain changes its conformation to accommodate H3
In the H3-free structure, the zinc-binding domain is composed of six β strands that constitute two sheets (β1–β6–β4–β5 and β2–β3) and two α helices (Fig. 4A). The Zn(II) ion is coordinated by four cysteine residues, which are conserved among the KDM6 family members (C1331, C1334, C1358, and C1361) (see Fig. 1), stabilizing the overall structure. An analysis using the Dali sever (Holm and Rosenström 2010) revealed that this domain adopts a novel fold; the closest structural neighbor, with a Z-score of 4.1 (Protein Data Bank [PDB] ID: 2BAY; U-box of the splicing factor Prp19), does not bind zinc. H1320, H1329, L1342, Y1354, and V1356 form hydrophobic interactions with each other at the center of the domain (Fig. 4A).
Figure 4.
Conformational change of the zinc-binding domain to accommodate the H3 peptide. (A) Structure of the zinc-binding domain in the H3-free structure. Secondary elements are labeled as in Figure 1. Purple dashed lines represent bonds coordinating Zn(II). (B) Structure of the zinc-binding domain bound with the H3 peptide. (C) Orthogonal views of the H3-free (purple) and H3-bound (green) structures of the zinc-binding domain. The H3 main chain (ribbon) and the L20 side chain (ball and stick) in the H3-bound structure are shown in yellow.
Unexpectedly, H3 binding induces a significant structural change of the zinc-binding domain (Fig. 4). Residues 1353–1355 are located in the loop connecting α1 and β5 in the H3-free structure, while these residues newly form hydrogen bonds with the adjacent β4 strand to elongate the β5 strand in the H3-bound structure. This conformational change is accompanied by the deformation of the α1 helix (Fig. 4B). Consequently, Y1354 is displaced from the above-mentioned hydrophobic interactions, which exposes a hydrophobic patch consisting of H1320, H1329, L1342, and V1356 on the surface (Fig. 4A,B). This patch binds the hydrophobic side chain of H3L20, which is located at position −7 relative to the methyllysine substrate. The main chain of H3Q19 and H3L20 occupies the space where Y1354 is located in the H3-free structure (Fig. 3D, 4B,C). Moreover, the main chain amide and carbonyl groups of Y1354 hydrogen bond with the H3Q19 carboxyl group and the H3A21 amide group, respectively, forming an intermolecular β-sheet-like interaction. The displaced Y1354 side chain also stacks with the H3K18 side chain, and the H3R17 guanidium group is located within hydrogen-bonding distance from E1326 (Fig. 3D, 4B). However, we note that the electron density for the side chain of H3R17 (the most N-terminal residue in the H3 peptide used in this study) is poor (Supplemental Fig. 1), suggesting its structural flexibility under the present crystallization conditions. A sequence alignment analysis showed that most of the residues of the zinc-binding domain, including those involved in the structural integrity and H3 binding, of UTX are shared by JMJD3 (Fig. 1). Collectively, these results suggest that the zinc-binding domain is important for the site specificity of the KDM6 family.
H3R17, H3L20, H3R26, H3A29, H3P30, and H3T32 are important for the substrate specificity of UTX
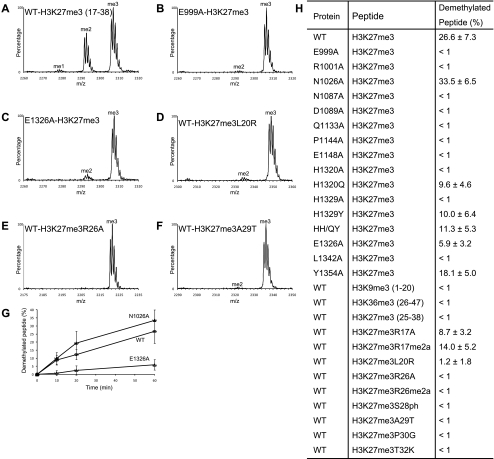
To determine whether the interactions seen in the structure are important for demethylation, we created UTX fragments (residues 880–1402) possessing the E999A, R1001A, N1087A, D1089A, Q1133A, P1144A, E1148A, H1320A, H1329A, E1326A, L1342A, or Y1354A substitution and measured their activities using the H3K27me3 peptide (residues 17–38) (summarized in Fig. 5H). As a control, we also created the alanine mutation of N1026, whose main chain atoms, but not side chain atoms, interact with H3. As expected, the N1026A mutant retained activity comparable with that of the wild type (Fig. 5A,G,H). The E1148A mutant lacked detectable activity after a 60-min reaction, which is consistent with its essential role in catalysis (Fig. 5H). The E999A, D1089A, R1001A, N1087A, and P1144A mutants also lacked detectable activity (Fig. 5B,H). As described, E999, D1089, and P1144 interact with the H3 side chains, and R1001 and N1087 bind the H3 main chain (Fig. 3B–D). These results demonstrated the essential roles of the jumonji domain residues that recognize the H3 main chain and side chains in demethylation. Moreover, the activities of the H1320A, H1329A, and L1342A mutants were also undetectable, whereas those of the E1326A and Y1354A mutants exhibited large and moderate decreases, respectively (Fig. 5C,H). Furthermore, the short H3K27me3 peptide spanning residues 25–38, which is able to contact only the jumonji domain, is a very poor substrate of UTX (Fig. 5H). These results indicate that the recognition of both H3R17 (via E1326) and H3L20 (via H1320, H1329, L1342, and Y1354) by the zinc-binding domain is indeed important for demethylation.
Figure 5.
Demethylation assay. (A–F) Representative MALDI-TOF mass spectra of the 60-min demethylation reactions. (A) The wild-type UTX. (B) The E999A mutant. (C) The E1326A mutant. (D) The wild-type UTX and the H3 peptide with the L20R substitution. (E) The wild-type UTX and the H3 peptide with the R26A substitution. (F) The wild-type UTX and the H3 peptide with the A29T substitution. (G) Time course of demethylation reactions by the wild-type, N1026A, and E1326A proteins. Mean values with standard deviations are plotted. (H) Summary of the demethylation assay. The percentages of demethylated peptide after a 60-min reaction (mean values with standard deviations) are shown. Unless otherwise noted, the peptide substrates contain residues 17–38 of H3.
UTX H1320 and H1329, which interact with H3L20, are replaced by glutamine and tyrosine, respectively, in JMJD3 (Fig. 1). Thus, we also examined the activity of the UTX fragments carrying the single H1320Q or H1329Y substitution as well as the H1320Q/H1329Y double mutant (designated as HH/QY). H1320Q, H1329Y, and HH/QY exhibited reduced, but still significant, activity (Fig. 5H), showing that these substitutions decrease, but do not abolish, the recognition of H3L20 by the zinc-binding domain.
Next, we sought to examine how UTX discriminates H3K27 from other methylated lysine residues in histones H3 and H4, especially the near-cognate H3K9. We first confirmed that neither H3K9me3 (residues 1–20) nor H3K36me3 (residues 26–47) can be demethylated by UTX (Fig. 5H), consistent with previous reports identifying UTX as the H3K27-specific demethylase (Agger et al. 2007; Hong et al. 2007; Lan et al. 2007; Lee et al. 2007b). H3K27, H3K9, and H4K20 share an arginine residue at position −1. No demethylation activity was detected when the H3 peptide with the R26A substitution was used (Fig. 5E,H), further confirming its essential role in the substrate recognition. We also substituted H3L20 (position −7), H3A29 (position +2), H3P30 (position +3), and H3T32 (position +5) with the corresponding residues in the sequence around H3K9 (H3L20R, H3A29T, H3P30G, and H3T32K, respectively), and H3R17 (position −10; no corresponding residue exists for H3K9) with alanine (H3R17A). No demethylation activity was detected for the H3A29T, H3P30G, and H3T32K variants, whereas very weak activity was observed for H3R17A and H3L20R (Fig. 5D,F,H). These results, taken together with the mutational analyses of UTX, show that the residues at positions −10, −7, −1, +2, +3, and +5 (H3R17, H3L20, H3R26, H3A29, H3G30, and H3T32, respectively) predominantly contribute to the specificity.
Finally, we examined the effects of the modification of H3 residues on the demethylation activity of UTX. H3R17 and H3R26, both recognized by UTX, are asymmetrically dimethylated by CARM1/PRMT4 (Wolf 2009). The demethylation activity was decreased greatly and slightly by the H3R26 and H3R17 asymmetrical dimethylation (R26me2a and R17me2a), respectively (Fig. 5H). H3S28 is phosphorylated (H3S28ph) by kinases such as MSK1 and MSK2 (Gehani et al. 2010). Although H3S28 is not directly recognized by UTX in the present structure, the phosphorylation of H3S28 abolished the demethylation activity of UTX (Fig. 5H). These results point to possible mechanisms by which cross-talk between histone modifications regulates the state of H3K27 methylation.
Discussion
The jumonji and zinc-binding domains of UTX recognize two separate segments of histone H3 to achieve the site specificity for H3K27
Our structural and biochemical analyses presented here provide the molecular basis for the UTX-specific demethylation of H3K27 and the discrimination against the other methylated lysine residues, including the near-cognate H3K9, in histones H3 and H4. The jumonji domain of UTX accommodates H3K27 and its surrounding residues, from 25 to 33, by specific side chain and main chain interactions (Fig. 3). The most intensively recognized residue is H3R26 (Figs. 3B, 5E), while H3K9 also has an arginine residue at position −1. On the other hand, H3P30 at position +3 forms a hydrophobic interaction with UTX, while the +3 residue of H3K9 is glycine (H3G12), which does not have a large hydrophobic surface. Furthermore, H3T11 and H3K14, located at positions +2 and +5 of H3K9, hinder demethylation by UTX, as they are too large to fit in the small pockets on the surface of UTX that binds H3A29 and H3T32 (Supplemental Fig. 2). Therefore, the jumonji domain is able to recognize H3K27 preferably over H3K9. Moreover, the zinc-binding domain of UTX recognizes the upstream residues, especially H3L20 and H3R17 at positions −7 and −10 (Figs. 4B, 5C,D). As H3K9 has an arginine residue at position −7 and lacks position −10, the zinc-binding domain of UTX has a preference for H3K27 over H3K9. Thus, the two domains of UTX cooperatively discriminate H3K27 from H3K9, H3K4, H3K36, and H4K20 because none of them has the same residues at positions −10, −7, −1, +2, +3, and +5 as those of H3K27. In addition, the pocket for H3A25 at position −2 may also be involved in determining the site specificity, as a valine, glutamate, or larger residue cannot be accommodated in it (Supplemental Fig. 2). We examined whether transplantation of the six H3K27 characteristic residues could convert H3K9me3 into a substrate of UTX. However, UTX was unable to demethylate the H3K9me3 peptide with residue changes at the −10, −9, −7, +2, +3, and +5 positions (sequence RKALTKQTARKme3SAPGTAPRKQL; added or substituted residues are underlined) (data not shown). One possible explanation for this phenomenon is that Thr at position −6 (H3T3), but not Ala (H3A21), sterically clashes with the zinc-binding domain (data not shown). Collectively, these observations reveal the very strict substrate specificity of UTX and may be helpful for identifying its new substrates.
The mechanism of substrate recognition by UTX revealed in this study is remarkably different from those employed by other jumonji proteins that possess multiple site specificity. JMJD2A forms few interactions with the H3 side chains, thereby exhibiting “plasticity” in the recognition of distinct methylation sites (Chen et al. 2007; Couture et al. 2007; Ng et al. 2007). The jumonji domains of PHF8 and KDM7A themselves do not discriminate H3K9 and H3K27 (Horton et al. 2010; Yang et al. 2010). In vitro, human KDM7A even demethylates H3K36me2/1, which lacks the “A-R-Kme-S” motif (Fig. 3E). The N-terminal PHD domains of these enzymes, both of which specifically bind H3K4me3, adopt different orientations relative to the jumonji domain. As a result, the second methylation (H3K4me3) in the same histone molecule facilitates the H3K9me2 demethylation by PHF8 through cooperative binding, while it decreases the activity of KDM7A toward H3K9me2 by inhibiting the access of the substrate methyllysine to the catalytic center. Thus, the PHD–H3K4me3 interactions regulate these enzymes by “reading” the modification patterns (Horton et al. 2010). In contrast, the jumonji and zinc-binding domains of UTX do not require additional histone modifications and exhibit the strict specificity for H3K27me3/2 by themselves. It is intriguing that these enzymes, all of which belong to the jumonji superfamily, achieve and regulate their site specificities in such different manners.
Our database search revealed that only the other members of the KDM6 family, UTY and JMJD3/KDM6B, show sequence homology with the zinc-binding domain of UTX/KDM6A (data not shown). Still, it is possible that other histone-binding proteins employ additional domains that are structurally and/or functionally homologous to the zinc-binding domain to achieve their site specificities. In this regard, H3L20 could serve as a functional “tag” to be recognized by the cognate domain because of the relatively rare occurrence of hydrophobic residues in the histone tails. We expect more examples in which the recognition of hydrophobic residues—not only H3L20, but also H4L10 and H4L22—plays a role in determining the protein's specificity.
Effects of other modifications on the activity of UTX
Growing evidence suggests that cross-talk occurs between histone modifications (Lee et al. 2010). In some instances, one modification can affect the creation of the second on the same histone molecule by regulating the activity of the enzyme responsible for it. For example, the acetylation of H3K14 by GCN5 is activated by the prior phosphorylation of H3S10 (Cheung et al. 2000; Lo et al. 2000), while the asymmetrical methylation of H3R2 abrogates the trimethylation of H3K4 by Set1-like methyltransferases (Guccione et al. 2007; Kirmizis et al. 2007). On the other hand, little is known about whether the enzymatic activity of UTX is regulated by any second modification of H3.
Our biochemical results indicated that the modifications of H3R17, H3R26, and H3S28 might regulate the demethylation activity of UTX and thus target gene expression. It is surprising that, although H3S28 is not recognized, its phosphorylation inhibits H3K27 demethylation by UTX. The phosphate group of H3S28ph should be in close proximity to the H3R28 side chain, which intimately interacts with E999, N1087, and D1089. We speculate that H3S28ph interacts with the H3R26 side chain, thus interfering with the interaction between H3R26 and UTX. MSK1 and MSK2 reportedly phosphorylate S28 of the K27-trimethylated histone H3, which leads to the displacement of the polycomb proteins from chromatin and the subsequent gene activation (Gehani et al. 2010). Thus, H3K27me3S28ph represents the state to be neither “read” nor “erased” by the proteins acting on H3K27me3. As previously proposed (Gehani et al. 2010), this doubly modified state may function as a transient activation mark, which can readily be turned off by the H3S28 dephosphorylation and is resistant to constitutive gene activation due to the H3K27 demethylation. A similar on–off switch reportedly operates in the H3K9me–H3S10ph double mark, where H3S10 phosphorylation dissociates HP1 proteins from the H3K9me3 chromatin during M phase (Fischle et al. 2005; Hirota et al. 2005).
Structural comparison with other jumonji demethylases
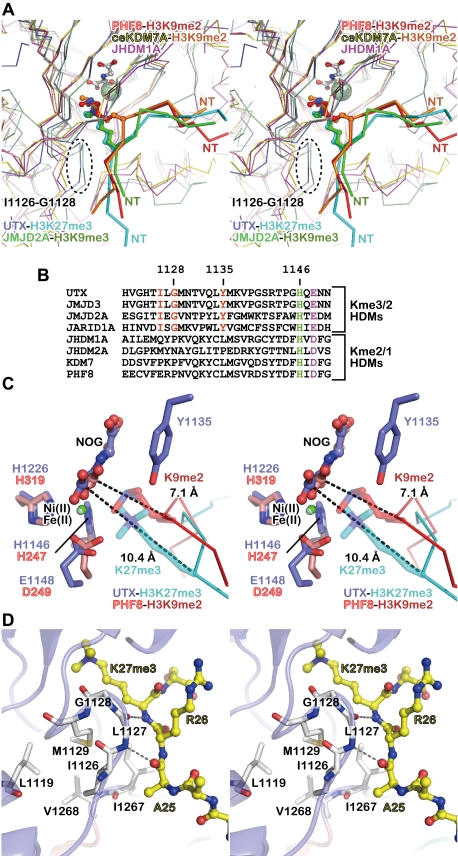
Interestingly, PHF8 and KDM7A bind an H3 peptide in the orientation opposite to that of JMJD2A (Chen et al. 2007; Couture et al. 2007; Ng et al. 2007; Horton et al. 2010; Yang et al. 2010). In the present UTX–H3 structure, the orientation of the H3 peptide is the same as that in the JMJD2 complex (and opposite to that in the PHF8 and KDM7A complexes). Moreover, the location of the substrate methyllysine in the UTX complex is similar to that in the JMJD2A complex, but quite different from those in the PHF8 and KDM7A complexes. PHF8 and KDM7A have no space for the “third” methyl group (Horton et al. 2010; Yang et al. 2010), which is consistent with their inability to demethylate trimethyllysine. Compared with the structures of PHF8 and KDM7A (Fig. 6), both JMJD2A and UTX (which can demethylate trimethyllysine), contain the trimethyllysine residue farther from the catalytic center, thus leaving sufficient space for all three methyl groups (Chen et al. 2007; Couture et al. 2007; Ng et al. 2007).
Figure 6.
Structural comparison with other jumonji demethylases. (A) Stereo view of the superposed structures of the UTX•H3 complex (blue and cyan), the JMJD2A•H3K9me3 peptide complex (pale green and green; PDB ID: 2OQ6), the PHF8•H3K9me2 peptide complex (pink and red; PDB ID: 3KV4), the Caenorhabditis elegans KDM7A•H3K9me2 peptide complex (yellow and orange; PDB ID: 3N9N), and the substrate-free JHDM1A (purple; PDB ID: 2YU2). The N-terminal side of each histone peptide is indicated as “NT.” (B) Alignment of the sequences near the Fe(II)-binding motif (shown in green and purple) from eight jumonji demethylases (four tri/didemethylases and four di/mono-demethylases). The residues conserved only among tri/didemethylases are shown in red. (C) Stereo view of the methyllysine-binding sites. The structures of the UTX•H3 (blue and cyan) and PHF8•H3K9me2 (pink and red) complexes are superposed. The distances between the methyllysine Cα atom and the NOG C1 atom are indicated (dashed lines). Note that Y1135 of UTX and the H3K9me2 side chain of the PHF8 structure are close to each other. (D) Stereo view of the L-X-G-M/V motif.
We noticed two structural features that may account for the different substrate-binding modes. First, as described above, Y1135 of UTX participates in the recognition of one of the three methyl groups of trimethyllysine in the present structure. This tyrosine residue is strictly conserved in the KDM6 and JMJD2 proteins, but is absent in the PHF and JHDM1/KDM2 proteins (H3K36me2/1 demethylases) (Fig. 6B). In the superposed structures, Y1135 of UTX is located too close to the H3K9me2 side chain of the PHF8 complex, suggesting that UTX cannot bind the methyllysine in a manner similar to that of PHF8 (Fig. 6C). Second, the conserved I-X-G-M/V sequence (residues 1126–1129 of UTX; X denotes any amino acid) is found 24–21 residues upstream of the Fe(II)-binding H-X-D/E motif (H1146 and E1148 of UTX) in the KDM6 and JMJD2 proteins (Fig. 6B). In the structures of UTX and JMJD2A, this segment protrudes near the entrance of the methyllysine-binding pocket (Fig. 6A) and, as described above, hydrogen bonds with the main chain of H3. In the structure of UTX, the substitution of G1128 with larger amino acids would cause a serious clash with the K27me3 side chain (data not shown). I1126 and M1129 participate in the network of hydrophobic interactions with L1119, I1267, and V1268, thus stabilizing the local conformation of the segment (Fig. 6D). PHF8 and KDM7A, lacking the protrusion, cannot bind H3 in the same manner (Fig. 6A,B). The H3-free structure of JHDM1A (an H3K36me2/1 demethylase) (Han et al. 2007) also lacks the protrusion (Fig. 6A,B).
A sequence alignment analysis revealed that the proteins from the JARID1/KDM5 family (H3K4me3/2 demethylases) (Christensen et al. 2007; Iwase et al. 2007; Klose et al. 2007; Lee et al. 2007a; Tahiliani et al. 2007; Yamane et al. 2007) also have the conserved tyrosine residue and the L-X-G-M/V motif (Fig. 6B), suggesting that they may bind H3 in a manner similar to that of UTX. On the other hand, NO66, a distantly related H3K4me3/2 and H3K36me3/2 demethylase (Sinha et al. 2010), lacks both of the two motifs at the same positions in its sequence (data not shown). Collectively, these observations have revealed the sequence and structural features that seem to characterize several jumonji tridemethylases. The elucidation of the substrate-binding mechanisms of other tridemethylases awaits the determination of their structures.
Materials and methods
Protein purification and crystallization
The C-terminal fragment of UTX (residues 880–1401) was cloned into pET47b (Merck) and expressed in Rosetta 2(DE3)pLysS cells (Merck) as the N-terminally 6xHis-tagged protein. The protein was purified by Ni Sepharose (GE Healthcare) chromatography, followed by tag cleavage with 6xHis-tagged HRV3C protease, a second round of Ni Sepharose chromatography to remove the tag and the protease, and Superdex 200 pg chromatography (GE Healthcare). The purified protein was concentrated to 10–12 mg/mL in 20 mM Tris-HCl buffer (pH 7.5) containing 200 mM NaCl and 1 mM DTT, frozen in liquid nitrogen, and stored at −80°C. Mutant proteins were expressed and purified in the same manner as the wild-type protein. Initial crystals were grown by vapor diffusion at 4°C against 0.1 M Tris-HCl buffer (pH 8.5), 0.2 M Li2SO4, and 25% (w/v) PEG 3350. Using microseeding, the best crystals were obtained in 0.1 M Tris-HCl buffer (pH 8.5), 0.2 M Li2SO4, and 18%–20% PEG 3350. The crystals belonging to the space group P212121 were harvested by serial transfer into the reservoir buffer supplemented with 1 mM NiCl2, 1 mM NOG, and 10% or 20% (v/v) ethylene glycol and flash-cooled in liquid nitrogen. NaI-soaked crystals were prepared by using harvest buffer containing 1 M NaI instead of 0.2 M Li2SO4. H3-bound UTX crystals were prepared by soaking the crystals in harvest buffer containing 5 mg/mL H3 peptide (residues 17–38, containing K27me3) for 3 d at 4°C.
Structure determination
X-ray diffraction data were collected at the SPring-8 BL41XU beamline and processed with the HKL2000 program suite. The structure of H3-free UTX was solved by the single-wavelength anomalous dispersion method using the data set from the NaI-soaked crystal (Dauter et al. 2000) collected with X-rays at 1.5 Å wavelength to enhance the anomalous signal from the iodine atoms. Initially, 42 iodine sites were located with the program Shelx (Sheldrick 2008) and then used to calculate the initial phases with the program Phaser (McCoy et al. 2007). Most of the model was automatically created by the program Phenix (Adams et al. 2010) and then manually corrected and refined by using the programs Coot (Emsley et al. 2010) and Phenix. The UTX–H3 complex structure was solved with the program Phaser by the molecular replacement method, using the H3-free structure as a search model. Structural superposition was performed with Coot. Figures were prepared with the program Pymol (Schrödinger, LLC.).
MALDI-TOF analysis of demethylation activity
We found that the purified proteins tended to precipitate during the incubation. Thus, arginine and glutamate were added to prevent precipitation. The UTX proteins (10 μM) were incubated with histone H3 peptides (80 μM) in 50 mM Tris-HCl buffer (pH 7.5) containing 30 mM NaCl, 5% glycerol, 1 mM αKG, 80 μM Fe(NH4)2 (SO4)2, 2 mM ascorbic acid, 50 mM arginine, and 50 mM glutamate at 25°C. At 10, 20, and 60 min, 1 μL of the reaction solution was removed and mixed with 9 μL of 50% (v/v) acetonitrile and 0.3% (v/v) trifluoroacetic acid. For the MALDI-TOF analysis, 0.5 μL of the stopped reaction solution was mixed with 0.5 μL of 10 mg/mL α-cyano-4-hydroxycinnamic acid in 50% (v/v) acetonitrile and 0.3% (v/v) trifluoroacetic acid and analyzed on a Voyager-DE Biospectrometry Workstation MALDI-TOF mass spectrometer (Applied Biosystems). For quantification, mass spectra were recorded from triplicate reactions, each with 10 different points in the sample spots. The resultant 30 mass spectra were processed by the Data Explorer software and averaged.
Accession codes
The atomic coordinates and structure factors have been deposited in the PDB (IDs 3AVS and 3AVR for the H3-free and H3-bound structures, respectively).
Acknowledgments
We thank the staff of SPring-8 BL41XU (Harima, Japan) and Y. Fujii for assistance with our data collection. We also thank A. Ishii, T. Nakayama, and T. Kosuge for assistance in manuscript preparation; R. Akasaka for technical assistance; and M. Shirouzu and Y. Muto for advice and comments. This work was supported by the Targeted Proteins Research Program (TPRP); the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan; and the Uehara Foundation.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.172296.111.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K 2007. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449: 731–734 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie XH, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Smith E, Shilatifard A 2007. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol 14: 1008–1016 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Chen ZZ, Zang JY, Whetstine J, Hong X, Davrazou F, Kutateladze TG, Simpson M, Mao QL, Pan CH, Dai SD, et al. 2006. Structural insights into histone demethylation by JMJD2 family members. Cell 125: 691–702 [DOI] [PubMed] [Google Scholar]
- Chen ZZ, Zang JY, Kappler J, Hong X, Crawford F, Wang Q, Lan F, Jiang CY, Whetstine J, Dai S, et al. 2007. Structural basis of the recognition of a methylated histone tail by JMJD2A. Proc Natl Acad Sci 104: 10818–10823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD 2000. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell 5: 905–915 [DOI] [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG 2010. Covalent histone modifications–miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer 10: 457–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J, Agger K, Cloos PA, Pasini D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE, Helin K 2007. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell 128: 1063–1076 [DOI] [PubMed] [Google Scholar]
- Clissold PM, Ponting CP 2001. JmjC: cupin metalloenzyme-like domains in jumonji, hairless and phospholipase A(2)β. Trends Biochem Sci 26: 7–9 [DOI] [PubMed] [Google Scholar]
- Couture JF, Collazo E, Ortiz-Tello PA, Brunzelle JS, Trievel RC 2007. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat Struct Mol Biol 14: 689–695 [DOI] [PubMed] [Google Scholar]
- Dauter Z, Dauter M, Rajashankar KR 2000. Novel approach to phasing proteins: derivatization by short cryo-soaking with halides. Acta Crystallogr D Biol Crystallogr 56: 232–237 [DOI] [PubMed] [Google Scholar]
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G 2007. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130: 1083–1094 [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD 2005. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438: 1116–1122 [DOI] [PubMed] [Google Scholar]
- García-Cao M, O'Sullivan R, Peters AH, Jenuwein T, Blasco MA 2004. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet 36: 94–99 [DOI] [PubMed] [Google Scholar]
- Gehani SS, Agrawal-Singh S, Dietrich N, Christophersen NS, Helin K, Hansen K 2010. Polycomb group protein displacement and gene activation through MSK-dependent H3K27me3S28 phosphorylation. Mol Cell 39: 886–900 [DOI] [PubMed] [Google Scholar]
- Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, Schuchlautz H, Lüscher B, Amati B 2007. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 449: 933–937 [DOI] [PubMed] [Google Scholar]
- Han Z, Liu P, Gu L, Zhang Y, Li H, Chen S, Chai J 2007. Structural basis for histone demethylation by JHDM1. Frontier Science 1: 52–67 [Google Scholar]
- Herz HM, Madden LD, Chen Z, Bolduc C, Buff E, Gupta R, Davuluri R, Shilatifard A, Hariharan IK, Bergmann A 2010. The H3K27me3 demethylase dUTX is a suppressor of Notch- and Rb-dependent tumors in Drosophila. Mol Cell Biol 30: 2485–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Lipp JJ, Toh BH, Peters JM 2005. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 438: 1176–1180 [DOI] [PubMed] [Google Scholar]
- Holm L, Rosenström P 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res 38: W545–W549 doi: 10.1093/nar/gkq366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K 2007. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci 104: 18439–18444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JR, Upadhyay AK, Qi HH, Zhang X, Shi Y, Cheng XD 2010. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat Struct Mol Biol 17: 38–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H, Yu H 2010. Structural insights into histone lysine demethylation. Curr Opin Struct Biol 20: 739–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y 2007. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 128: 1077–1088 [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD 2001. Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, Bähler J, Green RD, Kouzarides T 2007. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature 449: 928–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Yamane K, Bae YJ, Zhang DZ, Erdjument-Bromage H, Tempst P, Wong JM, Zhang Y 2006. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442: 312–316 [DOI] [PubMed] [Google Scholar]
- Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument-Bromage H, Tempst P, Gilliland DG, Zhang Y, Kaelin WG 2007. The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell 128: 889–900 [DOI] [PubMed] [Google Scholar]
- Kouzarides T 2007. Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen SZ, Iwase S, Alpatov R, Issaeva I, Canaani E, et al. 2007. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449: 689–694 [DOI] [PubMed] [Google Scholar]
- Lee MG, Norman J, Shilatifard A, Shiekhattar R 2007a. Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomb-like protein. Cell 128: 877–887 [DOI] [PubMed] [Google Scholar]
- Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di Croce L, Shiekhattar R 2007b. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 318: 447–450 [DOI] [PubMed] [Google Scholar]
- Lee JS, Smith E, Shilatifard A 2010. The language of histone crosstalk. Cell 142: 682–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo WS, Trievel RC, Rojas JR, Duggan L, Hsu JY, Allis CD, Marmorstein R, Berger SL 2000. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol Cell 5: 917–926 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ 2007. Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku MC, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SS, Kavanagh KL, McDonough MA, Butler D, Pilka ES, Lienard BM, Bray JE, Savitsky P, Gileadi O, von Delft F, et al. 2007. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature 448: 87–91 [DOI] [PubMed] [Google Scholar]
- Pedersen MT, Helin K 2010. Histone demethylases in development and disease. Trends Cell Biol 20: 662–671 [DOI] [PubMed] [Google Scholar]
- Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schöfer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107: 323–337 [DOI] [PubMed] [Google Scholar]
- Sheldrick GM 2008. A short history of SHELX. Acta Crystallogr A 64: 112–122 [DOI] [PubMed] [Google Scholar]
- Sinha KM, Yasuda H, Coombes MM, Dent SY, de Crombrugghe B 2010. Regulation of the osteoblast-specific transcription factor Osterix by NO66, a Jumonji family histone demethylase. EMBO J 29: 68–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ER, Lee MG, Winter B, Droz NM, Eissenberg JC, Shiekhattar R, Shilatifard A 2008. Drosophila UTX is a histone H3 Lys27 demethylase that colocalizes with the elongating form of RNA polymerase II. Mol Cell Biol 28: 1041–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Mei P, Fang R, Leonor T, Rutenberg M, Shimizu F, Li J, Rao A, Shi Y 2007. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature 447: 601–605 [DOI] [PubMed] [Google Scholar]
- van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, Edkins S, Hardy C, O'Meara S, Teague J, et al. 2009. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet 41: 521–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace AC, Laskowski RA, Thornton JM 1995. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng 8: 127–134 [DOI] [PubMed] [Google Scholar]
- Wang JK, Tsai MC, Poulin G, Adler AS, Chen SZ, Liu HL, Shi Y, Chang HY 2010. The histone demethylase UTX enables RB-dependent cell fate control. Genes Dev 24: 327–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, et al. 2006. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125: 467–481 [DOI] [PubMed] [Google Scholar]
- Wolf SS 2009. The protein arginine methyltransferase family: an update about function, new perspectives and the physiological role in humans. Cell Mol Life Sci 66: 2109–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Wilson JR, Gamblin SJ 2003. SET domains and histone methylation. Curr Opin Struct Biol 13: 699–705 [DOI] [PubMed] [Google Scholar]
- Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P, Zhang Y 2007. PLU-1 is an H3K4 dernethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell 25: 801–812 [DOI] [PubMed] [Google Scholar]
- Yang Y, Hu L, Wang P, Hou H, Lin Y, Liu Y, Li Z, Gong R, Feng X, Zhou L, et al. 2010. Structural insights into a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans. Cell Res 20: 886–898 [DOI] [PubMed] [Google Scholar]