Abstract
Transcriptional regulation of the delta-aminolevulinic acid synthetase gene of Rhizobium meliloti was investigated under conditions of normal vegetative growth and during symbiosis with the legume host alfalfa. S1 nuclease mapping and DNA sequence analysis indicated that transcription originates from two sites separated by 238 base pairs. A deletion analysis of the putative promoter regions P1 and P2, corresponding to the proximal and distal RNA start sites, was carried out with Bal-31 nuclease. Promoter function was monitored as beta-galactosidase activity after fusing the deletions to lac Z and introducing them into Rhizobium on a broad host range plasmid. The data obtained suggest that both regions function equivalently as promoters. The DNA sequences of P1 and P2 show considerable homology in the region between -35 and the start of transcription. Both contain a -35 region that is analogous to the consensus E. coli promoter sequence, while the -10 region is dissimilar. No resemblance was found between either P1 or P2 and the promoter regions of genes under general nitrogen control.
Full text
PDF

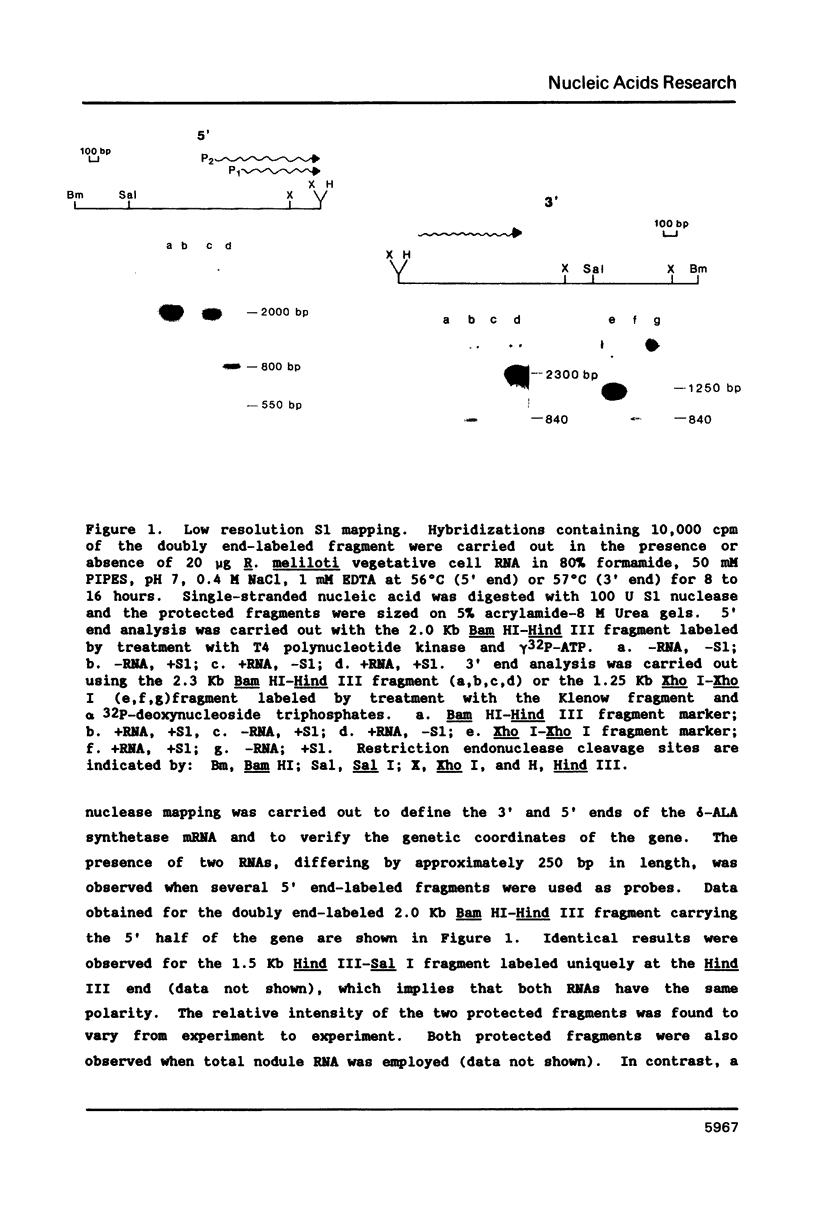


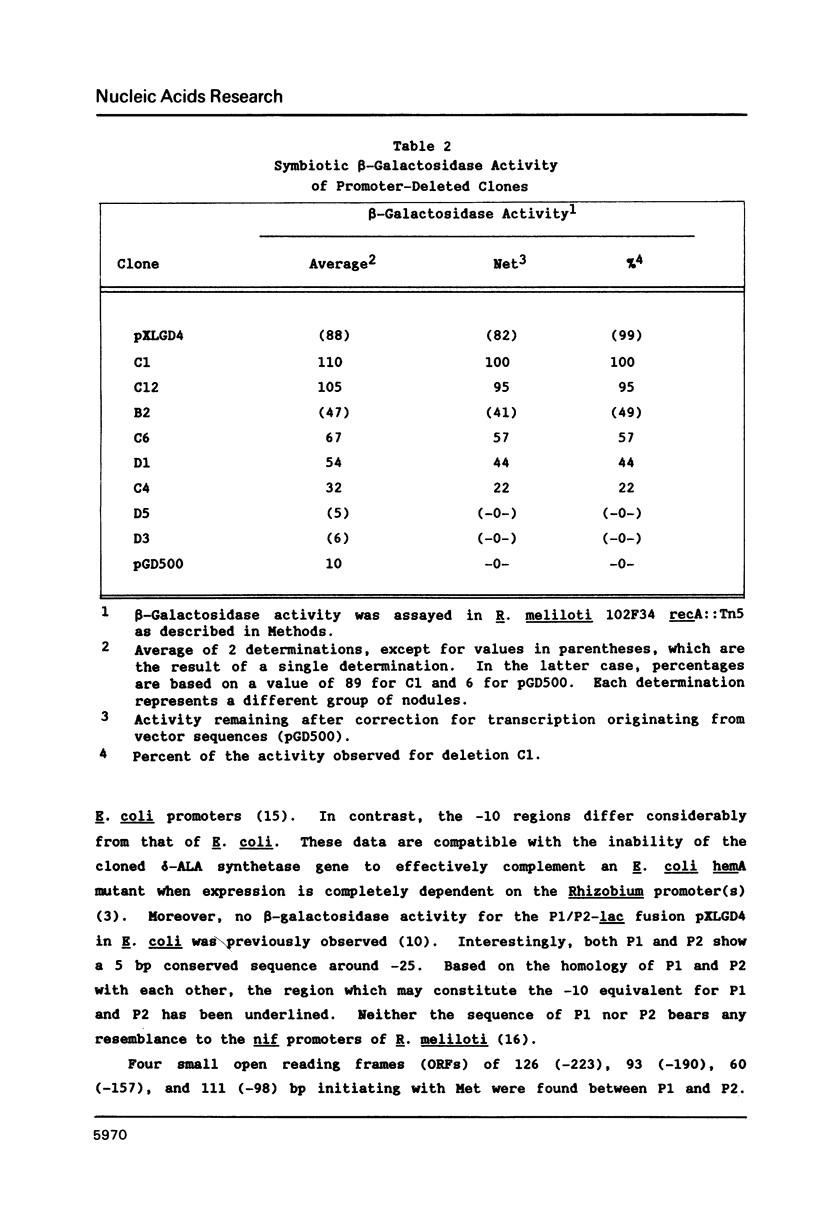
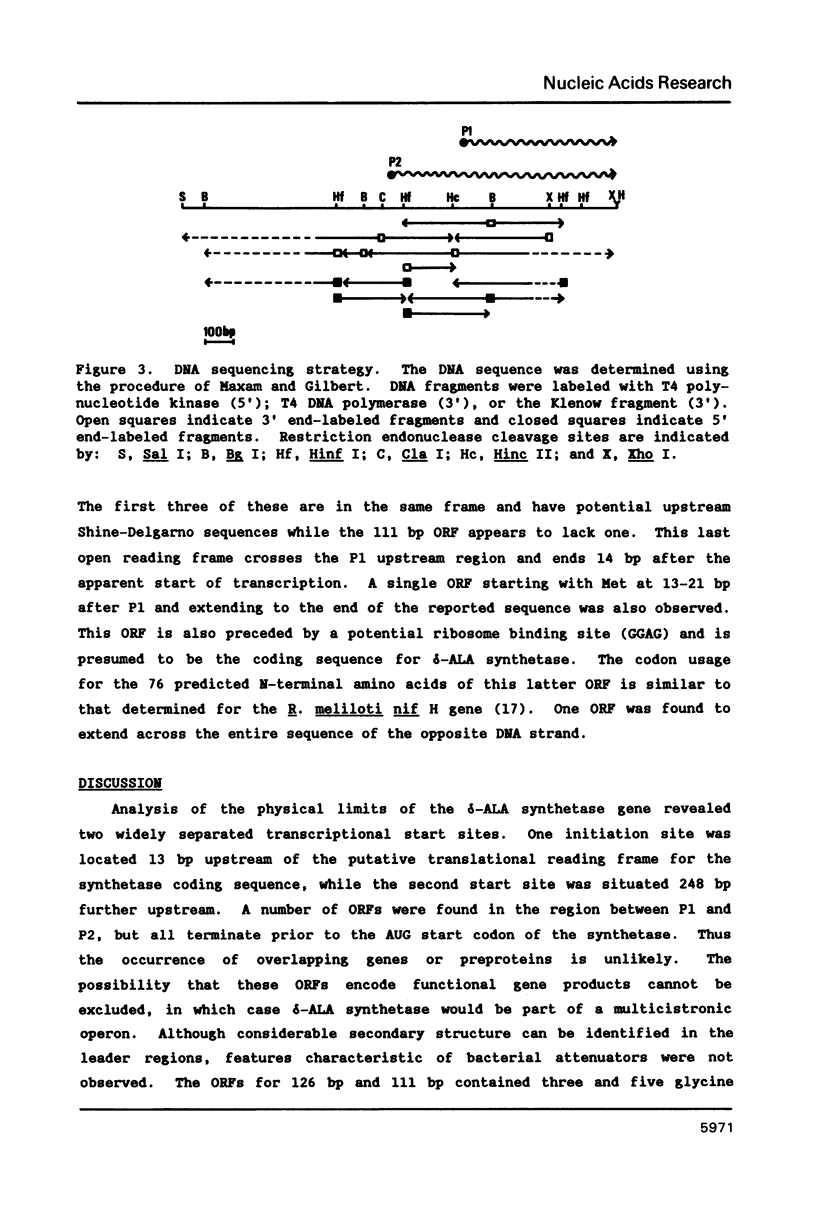

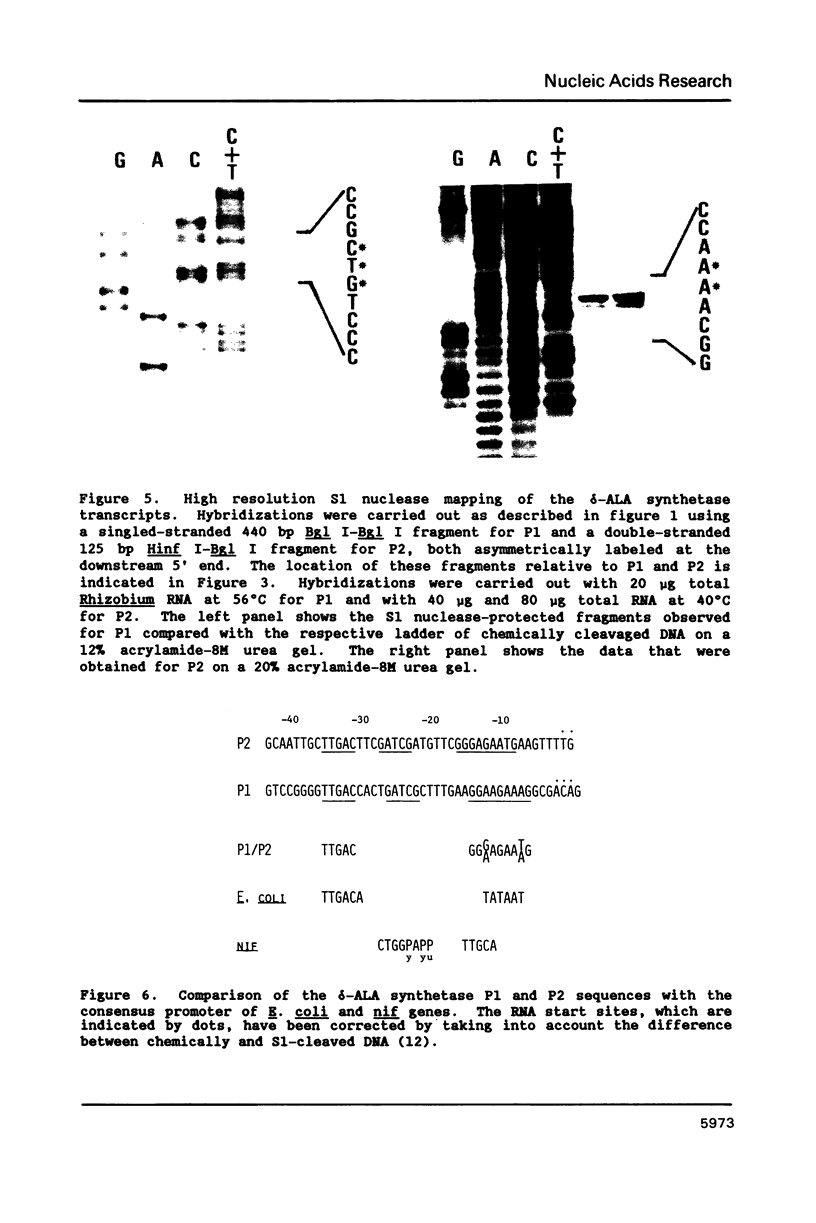



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allan M., Lanyon W. G., Paul J. Multiple origins of transcription in the 4.5 Kb upstream of the epsilon-globin gene. Cell. 1983 Nov;35(1):187–197. doi: 10.1016/0092-8674(83)90221-0. [DOI] [PubMed] [Google Scholar]
- Benyajati C., Spoerel N., Haymerle H., Ashburner M. The messenger RNA for alcohol dehydrogenase in Drosophila melanogaster differs in its 5' end in different developmental stages. Cell. 1983 May;33(1):125–133. doi: 10.1016/0092-8674(83)90341-0. [DOI] [PubMed] [Google Scholar]
- Better M., Helinski D. R. Isolation and characterization of the recA gene of Rhizobium meliloti. J Bacteriol. 1983 Jul;155(1):311–316. doi: 10.1128/jb.155.1.311-316.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman A. R., Fromm M., Berg P. Complex regulation of simian virus 40 early-region transcription from different overlapping promoters. Mol Cell Biol. 1984 Sep;4(9):1900–1914. doi: 10.1128/mcb.4.9.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton Z. F., Gross C. A., Watanabe K. K., Burgess R. R. The operon that encodes the sigma subunit of RNA polymerase also encodes ribosomal protein S21 and DNA primase in E. coli K12. Cell. 1983 Feb;32(2):335–349. doi: 10.1016/0092-8674(83)90453-1. [DOI] [PubMed] [Google Scholar]
- Close T. J., Rodriguez R. L. Construction and characterization of the chloramphenicol-resistance gene cartridge: a new approach to the transcriptional mapping of extrachromosomal elements. Gene. 1982 Dec;20(2):305–316. doi: 10.1016/0378-1119(82)90048-8. [DOI] [PubMed] [Google Scholar]
- Corbin D., Barran L., Ditta G. Organization and expression of Rhizobium meliloti nitrogen fixation genes. Proc Natl Acad Sci U S A. 1983 May;80(10):3005–3009. doi: 10.1073/pnas.80.10.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Schmidhauser T., Yakobson E., Lu P., Liang X. W., Finlay D. R., Guiney D., Helinski D. R. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid. 1985 Mar;13(2):149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R. Tandem promoters determine regulation of the Klebsiella pneumoniae glutamine synthetase (glnA) gene. Nucleic Acids Res. 1984 Oct 25;12(20):7811–7830. doi: 10.1093/nar/12.20.7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R., Roeder R. G. Definition of a novel promoter for the major adenovirus-associated virus mRNA. Cell. 1980 Nov;22(1 Pt 1):231–242. doi: 10.1016/0092-8674(80)90171-3. [DOI] [PubMed] [Google Scholar]
- Kassavetis G. A., Geiduschek E. P. Bacteriophage T4 late promoters: mapping 5' ends of T4 gene 23 mRNAs. EMBO J. 1982;1(1):107–114. doi: 10.1002/j.1460-2075.1982.tb01132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong S. A., Ditta G. S., Helinski D. R. Heme biosynthesis in Rhizobium. Identification of a cloned gene coding for delta-aminolevulinic acid synthetase from Rhizobium meliloti. J Biol Chem. 1982 Aug 10;257(15):8724–8730. [PubMed] [Google Scholar]
- Ley T. J., Nienhuis A. W. A weak upstream promoter gives rise to long human beta-globin RNA molecules. Biochem Biophys Res Commun. 1983 May 16;112(3):1041–1048. doi: 10.1016/0006-291x(83)91723-0. [DOI] [PubMed] [Google Scholar]
- Majumdar A., Adhya S. Demonstration of two operator elements in gal: in vitro repressor binding studies. Proc Natl Acad Sci U S A. 1984 Oct;81(19):6100–6104. doi: 10.1073/pnas.81.19.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Rose M., Botstein D. Structure and function of the yeast URA3 gene. Differentially regulated expression of hybrid beta-galactosidase from overlapping coding sequences in yeast. J Mol Biol. 1983 Nov 15;170(4):883–904. doi: 10.1016/s0022-2836(83)80193-4. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sarokin L., Carlson M. Upstream region required for regulated expression of the glucose-repressible SUC2 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Dec;4(12):2750–2757. doi: 10.1128/mcb.4.12.2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G. C., Bott K. F. DNA sequence of the tandem ribosomal RNA promoter for B. subtilis operon rrnB. Nucleic Acids Res. 1983 Sep 24;11(18):6289–6300. doi: 10.1093/nar/11.18.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Török I., Kondorosi A. Nucleotide sequence of the R.meliloti nitrogenase reductase (nifH) gene. Nucleic Acids Res. 1981 Nov 11;9(21):5711–5723. doi: 10.1093/nar/9.21.5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wong H. C., Schnepf H. E., Whiteley H. R. Transcriptional and translational start sites for the Bacillus thuringiensis crystal protein gene. J Biol Chem. 1983 Feb 10;258(3):1960–1967. [PubMed] [Google Scholar]
- Young R. A., Hagenbüchle O., Schibler U. A single mouse alpha-amylase gene specifies two different tissue-specific mRNAs. Cell. 1981 Feb;23(2):451–458. doi: 10.1016/0092-8674(81)90140-9. [DOI] [PubMed] [Google Scholar]




