Abstract
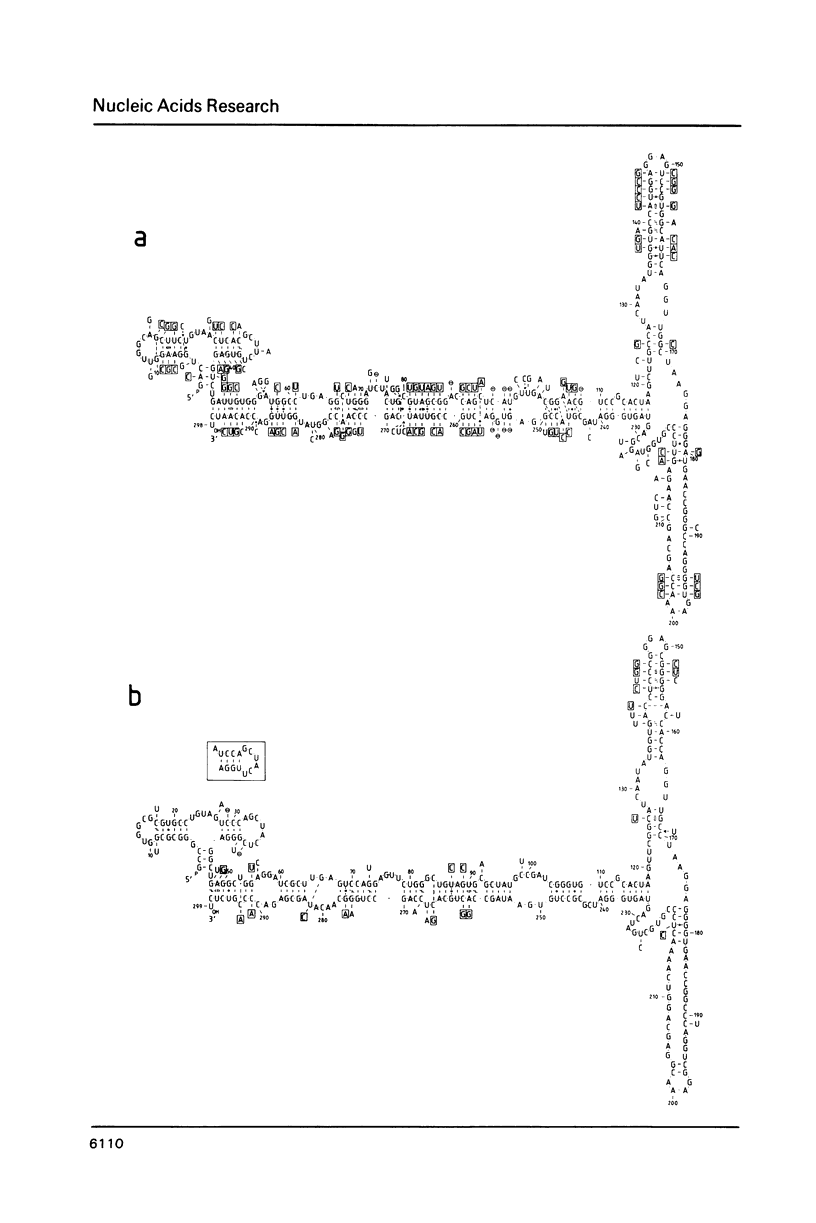
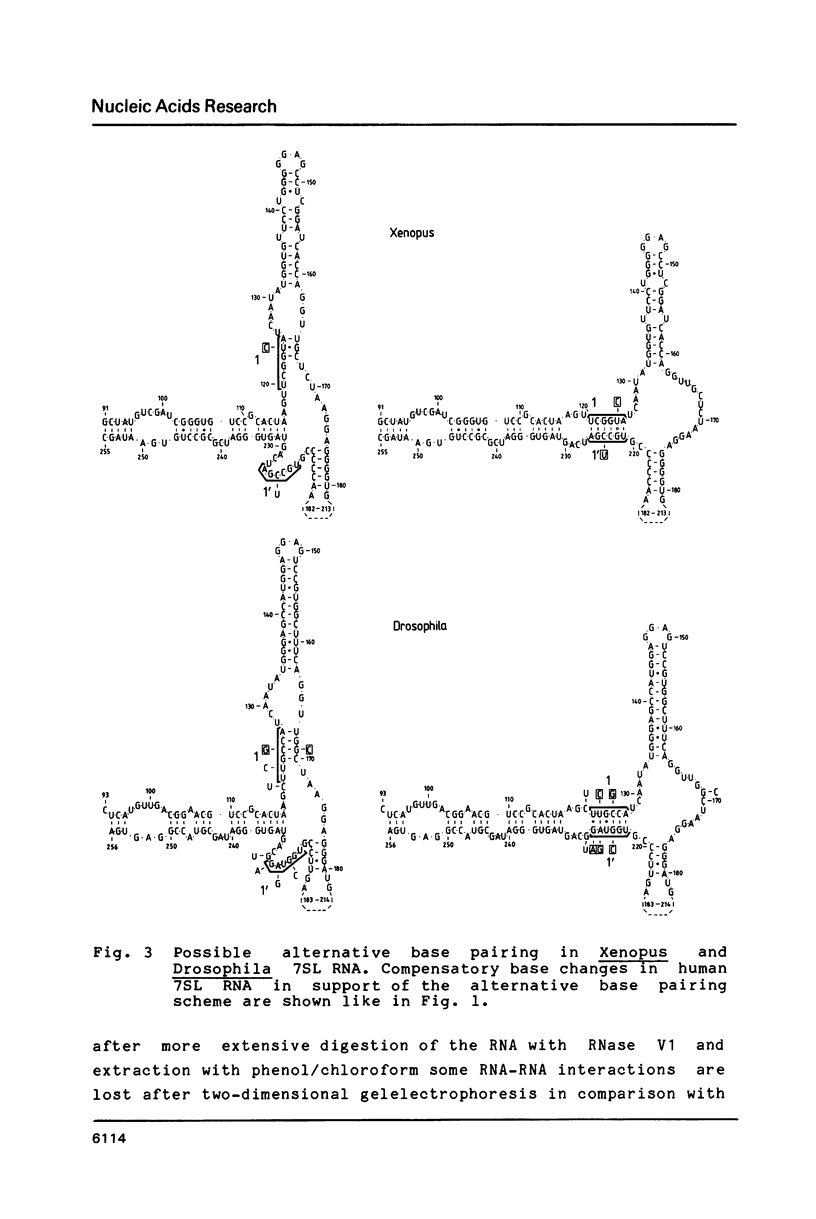
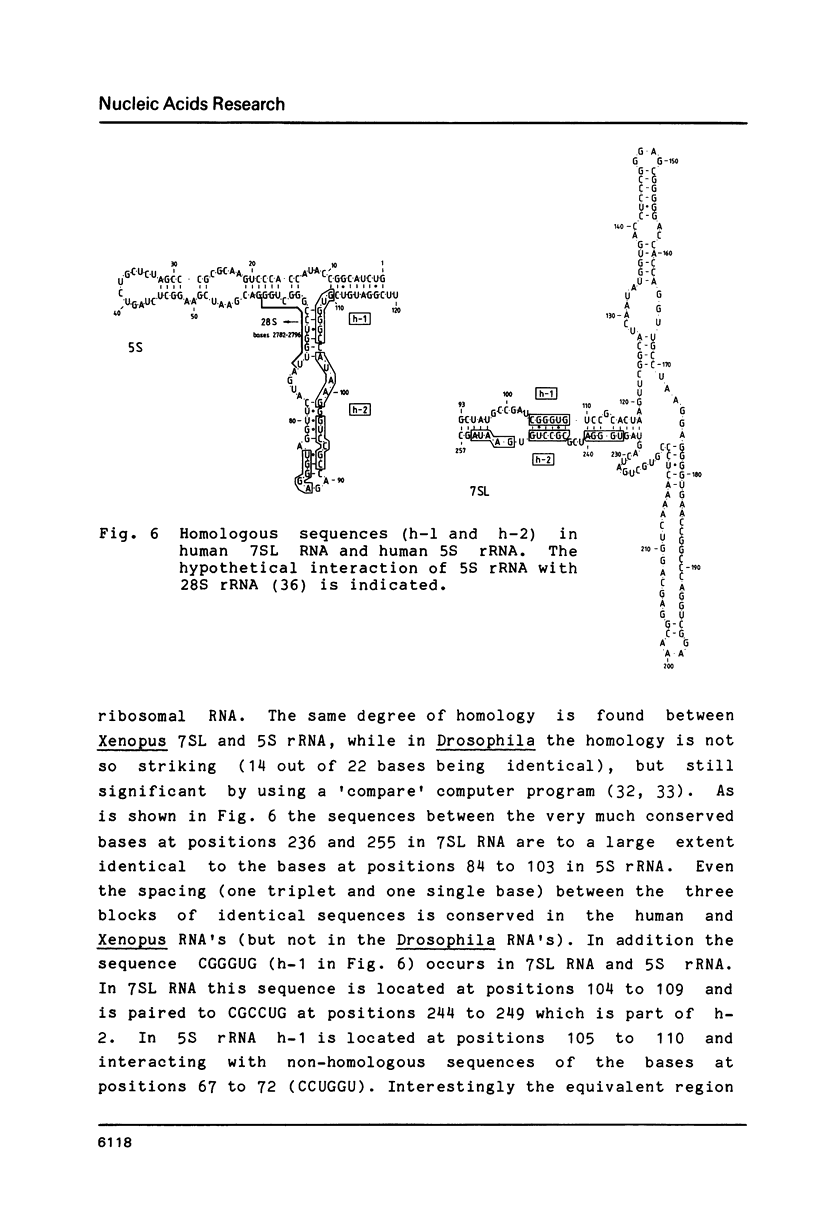
The secondary structure of the 7SL RNA in the signal recognition particle was determined by applying both a theoretical and an experimental approach. The compensatory base change approach was taken comparing the published sequences of human, Drosophila and Xenopus 7SL RNA's. The deduced secondary structure was confirmed by post-labeling of an RNase V1-nicked dog SRP with P32-pCp and RNA-ligase and analysis of the labeled RNA-fragments by non-denaturing/denaturing 2D polyacrylamide gel electrophoresis. Two interesting features in the secondary structure were revealed: Firstly, bases at positions 122 to 127 of the human 7SL RNA are not only able to pair with bases at positions 167 to 170, but also with a single-stranded region of the bases at positions 223 to 228, suggesting an alternative base pairing scheme for the 7SL RNA in all three organisms. In agreement with this finding, four different conformations were identified after transcription of the 7SL RNA from the genomic human clone. The involvement of the particular basepairing interaction postulated was confirmed by the analysis of a 7SL RNA deletion mutant (Sma1-409). Secondly, a significant sequence homology of the paired bases at positions 236 to 255 and 104 to 109 in 7SL RNA with bases in 5S ribosomal RNA at the positions 84 to 110 was noticed, suggesting that 5S ribosomal and 7SL RNA interact with the same target during protein biosynthesis. These findings are summarized by proposing a mechanism for the translational arrest of protein synthesis by the signal recognition particle using specific sequences and an alternative configuration in the 7SL RNA.
Full text
PDF
















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews D. W., Walter P., Ottensmeyer F. P. Structure of the signal recognition particle by electron microscopy. Proc Natl Acad Sci U S A. 1985 Feb;82(3):785–789. doi: 10.1073/pnas.82.3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansorge W., Barker R. System for DNA sequencing with resolution of up to 600 base pairs. J Biochem Biophys Methods. 1984 Mar;9(1):33–47. doi: 10.1016/0165-022x(84)90064-2. [DOI] [PubMed] [Google Scholar]
- Brimacombe R., Maly P., Zwieb C. The structure of ribosomal RNA and its organization relative to ribosomal protein. Prog Nucleic Acid Res Mol Biol. 1983;28:1–48. doi: 10.1016/s0079-6603(08)60081-1. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Tanner N. K., Tinoco I., Jr, Weir B. R., Zuker M., Perlman P. S. Secondary structure of the Tetrahymena ribosomal RNA intervening sequence: structural homology with fungal mitochondrial intervening sequences. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3903–3907. doi: 10.1073/pnas.80.13.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark C. G., Tague B. W., Ware V. C., Gerbi S. A. Xenopus laevis 28S ribosomal RNA: a secondary structure model and its evolutionary and functional implications. Nucleic Acids Res. 1984 Aug 10;12(15):6197–6220. doi: 10.1093/nar/12.15.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- Dahlberg J. E., Kintner C., Lund E. Specific binding of tRNAMet to 23S rRNA of Escherichia coli. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1071–1075. doi: 10.1073/pnas.75.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingman C. W., Peacock A. C. Analytical studies on nuclear ribonucleic acid using polyacrylamide gel electrophoresis. Biochemistry. 1968 Feb;7(2):659–668. doi: 10.1021/bi00842a022. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann V. A., Wolters J., Huysmans E., Vandenberghe A., De Wachter R. Collection of published 5S and 5.8S ribosomal RNA sequences. Nucleic Acids Res. 1984;12 (Suppl):r133–r166. doi: 10.1093/nar/12.suppl.r133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox G. E., Woese C. R. 5S RNA secondary structure. Nature. 1975 Aug 7;256(5517):505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- Gilmore R., Blobel G., Walter P. Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J Cell Biol. 1982 Nov;95(2 Pt 1):463–469. doi: 10.1083/jcb.95.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotz C., Zwieb C., Brimacombe R., Edwards K., Kössel H. Secondary structure of the large subunit ribosomal RNA from Escherichia coli, Zea mays chloroplast, and human and mouse mitochondrial ribosomes. Nucleic Acids Res. 1981 Jul 24;9(14):3287–3306. doi: 10.1093/nar/9.14.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier-Takada C., Altman S. Catalytic activity of an RNA molecule prepared by transcription in vitro. Science. 1984 Jan 20;223(4633):285–286. doi: 10.1126/science.6199841. [DOI] [PubMed] [Google Scholar]
- Gundelfinger E. D., Di Carlo M., Zopf D., Melli M. Structure and evolution of the 7SL RNA component of the signal recognition particle. EMBO J. 1984 Oct;3(10):2325–2332. doi: 10.1002/j.1460-2075.1984.tb02134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundelfinger E. D., Krause E., Melli M., Dobberstein B. The organization of the 7SL RNA in the signal recognition particle. Nucleic Acids Res. 1983 Nov 11;11(21):7363–7374. doi: 10.1093/nar/11.21.7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinek W. R., Schmid C. W. Repetitive sequences in eukaryotic DNA and their expression. Annu Rev Biochem. 1982;51:813–844. doi: 10.1146/annurev.bi.51.070182.004121. [DOI] [PubMed] [Google Scholar]
- Maizel J. V., Jr, Lenk R. P. Enhanced graphic matrix analysis of nucleic acid and protein sequences. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7665–7669. doi: 10.1073/pnas.78.12.7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D. I., Dobberstein B. A membrane component essential for vectorial translocation of nascent proteins across the endoplasmic reticulum: requirements for its extraction and reassociation with the membrane. J Cell Biol. 1980 Nov;87(2 Pt 1):498–502. doi: 10.1083/jcb.87.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D. I., Krause E., Dobberstein B. Secretory protein translocation across membranes-the role of the "docking protein'. Nature. 1982 Jun 24;297(5868):647–650. doi: 10.1038/297647a0. [DOI] [PubMed] [Google Scholar]
- Noller H. F. Topography of 16S RNA in 30S ribosomal subunits. Nucleotide sequences and location of sites of reaction with kethoxal. Biochemistry. 1974 Nov 5;13(23):4694–4703. doi: 10.1021/bi00720a003. [DOI] [PubMed] [Google Scholar]
- Ross A., Brimacombe R. Experimental determination of interacting sequences in ribosomal RNA. Nature. 1979 Sep 27;281(5729):271–276. doi: 10.1038/281271a0. [DOI] [PubMed] [Google Scholar]
- Ullu E., Murphy S., Melli M. Human 7SL RNA consists of a 140 nucleotide middle-repetitive sequence inserted in an alu sequence. Cell. 1982 May;29(1):195–202. doi: 10.1016/0092-8674(82)90103-9. [DOI] [PubMed] [Google Scholar]
- Ullu E., Tschudi C. Alu sequences are processed 7SL RNA genes. Nature. 1984 Nov 8;312(5990):171–172. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- Ullu E., Weiner A. M. Human genes and pseudogenes for the 7SL RNA component of signal recognition particle. EMBO J. 1984 Dec 20;3(13):3303–3310. doi: 10.1002/j.1460-2075.1984.tb02294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilenko S. K., Carbon P., Ebel J. P., Ehresmann C. Topography of 16 S RNA in 30 S subunits and 70 S ribosomes accessibility to cobra venom ribonuclease. J Mol Biol. 1981 Nov 15;152(4):699–721. doi: 10.1016/0022-2836(81)90123-6. [DOI] [PubMed] [Google Scholar]
- Volckaert G., Fiers W. Micro thin-layer techniques for rapid sequence analysis of 32P-labeled RNA: double digestion and pancreatic ribonuclease analyses. Anal Biochem. 1977 Nov;83(1):228–239. doi: 10.1016/0003-2697(77)90531-0. [DOI] [PubMed] [Google Scholar]
- Walter P., Blobel G. Purification of a membrane-associated protein complex required for protein translocation across the endoplasmic reticulum. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7112–7116. doi: 10.1073/pnas.77.12.7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., Blobel G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature. 1982 Oct 21;299(5885):691–698. doi: 10.1038/299691a0. [DOI] [PubMed] [Google Scholar]
- Walter P., Blobel G. Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J Cell Biol. 1981 Nov;91(2 Pt 1):557–561. doi: 10.1083/jcb.91.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., Gilmore R., Blobel G. Protein translocation across the endoplasmic reticulum. Cell. 1984 Aug;38(1):5–8. doi: 10.1016/0092-8674(84)90520-8. [DOI] [PubMed] [Google Scholar]
- Walter P., Ibrahimi I., Blobel G. Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein. J Cell Biol. 1981 Nov;91(2 Pt 1):545–550. doi: 10.1083/jcb.91.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G., Dobberstein B. Protein transfer across microsomal membranes reassembled from separated membrane components. Nature. 1978 Jun 15;273(5663):569–571. doi: 10.1038/273569a0. [DOI] [PubMed] [Google Scholar]
- Zieve G., Penman S. Small RNA species of the HeLa cell: metabolism and subcellular localization. Cell. 1976 May;8(1):19–31. doi: 10.1016/0092-8674(76)90181-1. [DOI] [PubMed] [Google Scholar]
- Zweib C., Dahlberg A. E. Structural and functional analysis of Escherichia coli ribosomes containing small deletions around position 1760 in the 23S ribosomal RNA. Nucleic Acids Res. 1984 Sep 25;12(18):7135–7152. doi: 10.1093/nar/12.18.7135. [DOI] [PMC free article] [PubMed] [Google Scholar]