Abstract
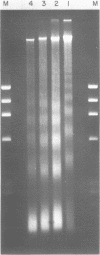
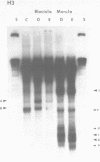
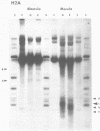
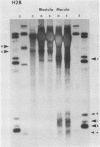
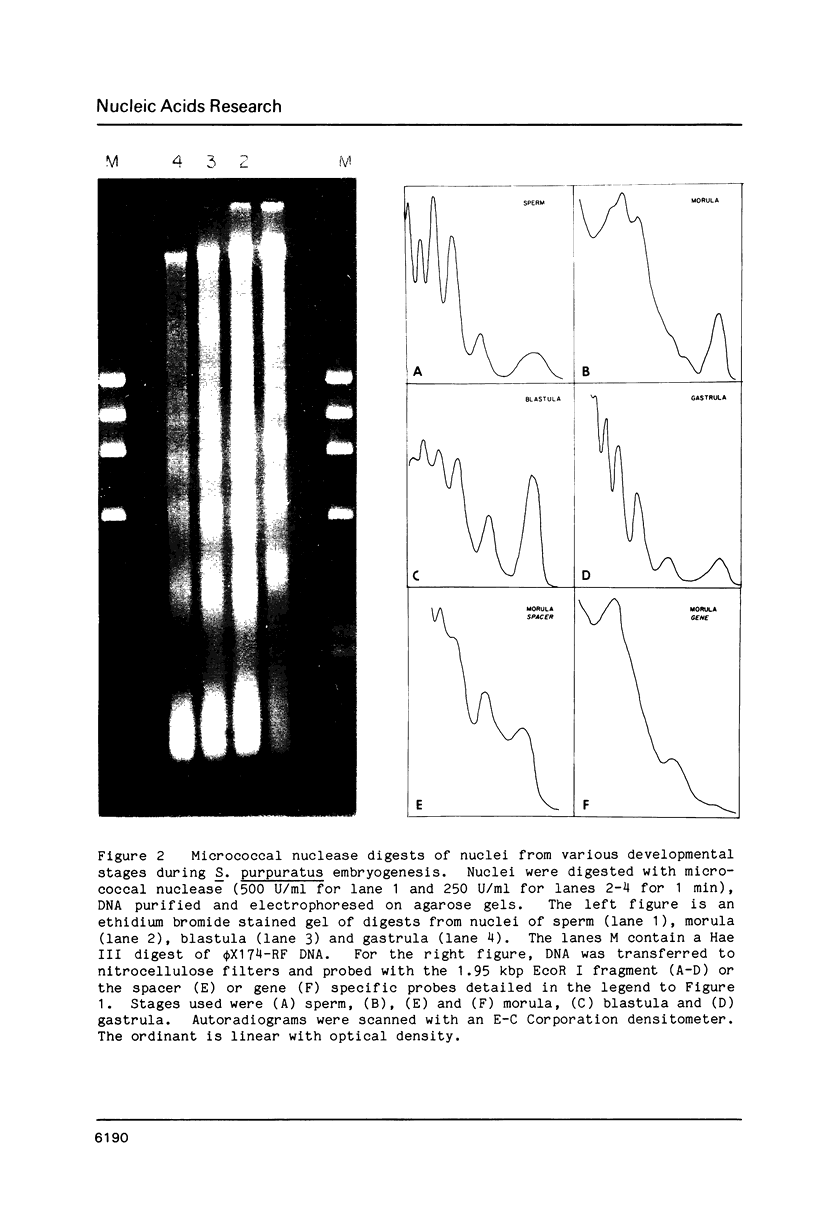
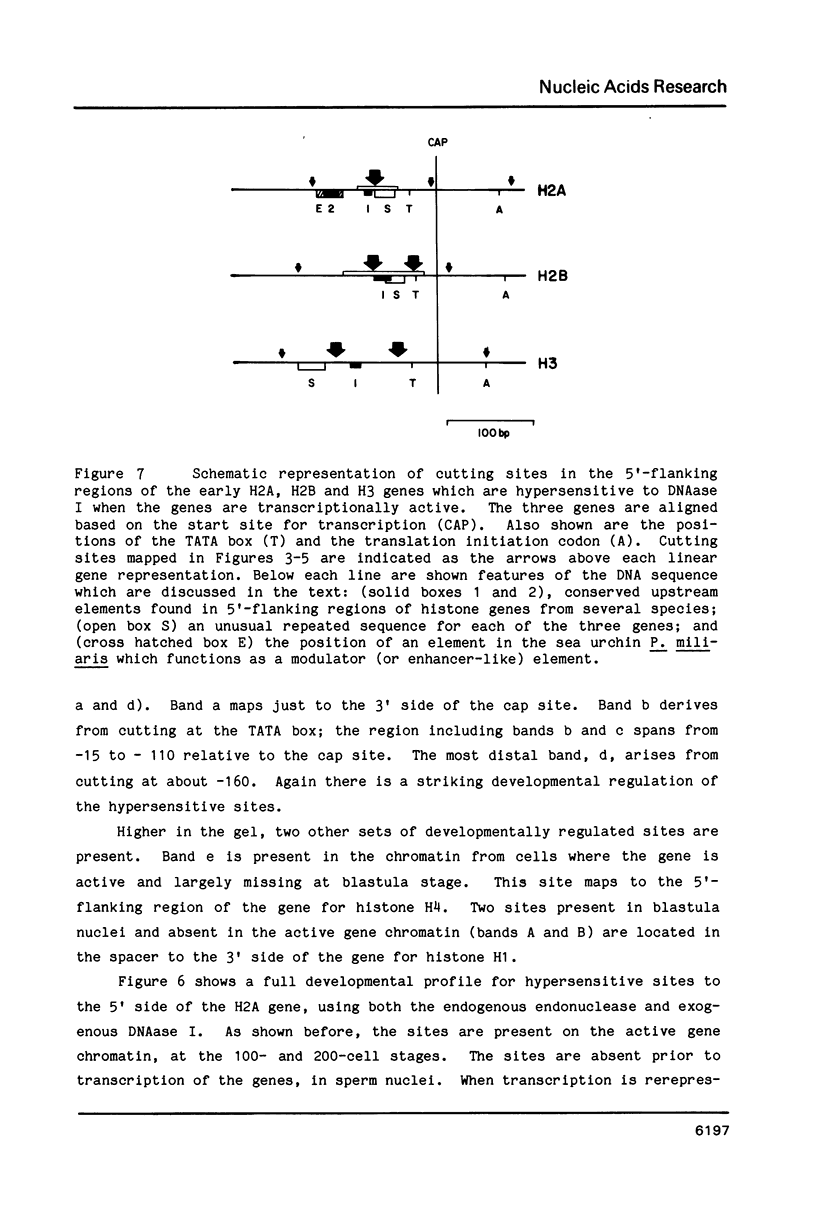
We describe features of the chromatin structure of the early histone gene family of Strongylocentrotus purpuratus during development. Before and after the histone genes are transcriptionally active, chromatin structure is quite similar with well-defined spaced nucleosomes and no major 5'-flanking sites hypersensitive to nucleases. During the period when the genes are active, marked changes in chromatin structure occur. Micrococcal nuclease digestion generates monomer nucleosomes and only trace amounts of higher multimers. Regions hypersensitive to an endogenous nuclease and DNAase I appear in the 5'-flanking regions of genes for H2A, H2B and H3. Each region consists of four sites spanning a DNA length of 200-250 base pairs. In each case, one major cutting site is near the TATA box; the bulk of the sensitive region is in the nontranscribed spacer. Other sites, in 3'-flanking regions of the genes, are sensitive to nucleases only when the histone genes are no longer transcribed.
Full text
PDF
















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bergman L. W., Kramer R. A. Modulation of chromatin structure associated with derepression of the acid phosphatase gene of Saccharomyces cerevisiae. J Biol Chem. 1983 Jun 10;258(11):7223–7227. [PubMed] [Google Scholar]
- Bryan P. N., Olah J., Birnstiel M. L. Major changes in the 5' and 3' chromatin structure of sea urchin histone genes accompany their activation and inactivation in development. Cell. 1983 Jul;33(3):843–848. doi: 10.1016/0092-8674(83)90026-0. [DOI] [PubMed] [Google Scholar]
- Cartwright I. L., Abmayr S. M., Fleischmann G., Lowenhaupt K., Elgin S. C., Keene M. A., Howard G. C. Chromatin structure and gene activity: the role of nonhistone chromosomal proteins. CRC Crit Rev Biochem. 1982;13(1):1–86. doi: 10.3109/10409238209108709. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E. Base sequence and helix structure variation in B and A DNA. J Mol Biol. 1983 May 25;166(3):419–441. doi: 10.1016/s0022-2836(83)80093-x. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Spacer DNA sequences upstream of the T-A-T-A-A-A-T-A sequence are essential for promotion of H2A histone gene transcription in vivo. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7102–7106. doi: 10.1073/pnas.77.12.7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosschedl R., Mächler M., Rohrer U., Birnstiel M. L. A functional component of the sea urchin H2A gene modulator contains an extended sequence homology to a viral enhancer. Nucleic Acids Res. 1983 Dec 10;11(23):8123–8136. doi: 10.1093/nar/11.23.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groudine M., Weintraub H. Propagation of globin DNAase I-hypersensitive sites in absence of factors required for induction: a possible mechanism for determination. Cell. 1982 Aug;30(1):131–139. doi: 10.1016/0092-8674(82)90019-8. [DOI] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Karpov V. L., Preobrazhenskaya O. V., Mirzabekov A. D. Chromatin structure of hsp 70 genes, activated by heat shock: selective removal of histones from the coding region and their absence from the 5' region. Cell. 1984 Feb;36(2):423–431. doi: 10.1016/0092-8674(84)90235-6. [DOI] [PubMed] [Google Scholar]
- Kedes L. H., Cohn R. H., Lowry J. C., Chang A. C., Cohen S. N. The organization of sea urchin histone genes. Cell. 1975 Nov;6(3):359–369. doi: 10.1016/0092-8674(75)90185-3. [DOI] [PubMed] [Google Scholar]
- Kedes L. H. Histone genes and histone messengers. Annu Rev Biochem. 1979;48:837–870. doi: 10.1146/annurev.bi.48.070179.004201. [DOI] [PubMed] [Google Scholar]
- Keichline L. D., Wassarman P. M. Developmental study of the structure of sea urchin embryo and sperm chromatin using micrococcal nuclease. Biochim Biophys Acta. 1977 Mar 2;475(1):139–151. doi: 10.1016/0005-2787(77)90348-3. [DOI] [PubMed] [Google Scholar]
- Khoury G., Gruss P. Enhancer elements. Cell. 1983 Jun;33(2):313–314. doi: 10.1016/0092-8674(83)90410-5. [DOI] [PubMed] [Google Scholar]
- Maxson R. E., Jr, Wilt F. H. The rate of synthesis of histone mRNA during the development of sea urchin embryos (Strongylocentrotus purpuratus). Dev Biol. 1981 Apr 30;83(2):380–386. doi: 10.1016/0012-1606(81)90485-1. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. A. The regulation of yeast mating-type chromatin structure by SIR: an action at a distance affecting both transcription and transposition. Cell. 1982 Sep;30(2):567–578. doi: 10.1016/0092-8674(82)90253-7. [DOI] [PubMed] [Google Scholar]
- Nedospasov S. A., Georgiev G. P. Non-random cleavage of SV40 DNA in the compact minichromosome and free in solution by micrococcal nuclease. Biochem Biophys Res Commun. 1980 Jan 29;92(2):532–539. doi: 10.1016/0006-291x(80)90366-6. [DOI] [PubMed] [Google Scholar]
- Overton G. C., Weinberg E. S. Length and sequence heterogeneity of the histone gene repeat unit of the sea urchin, S. purpuratus. Cell. 1978 Jun;14(2):247–257. doi: 10.1016/0092-8674(78)90111-3. [DOI] [PubMed] [Google Scholar]
- Shermoen A. W., Beckendorf S. K. A complex of interacting DNAase I-hypersensitive sites near the Drosophila glue protein gene, Sgs4. Cell. 1982 Jun;29(2):601–607. doi: 10.1016/0092-8674(82)90176-3. [DOI] [PubMed] [Google Scholar]
- Smith G. E., Summers M. D. The bidirectional transfer of DNA and RNA to nitrocellulose or diazobenzyloxymethyl-paper. Anal Biochem. 1980 Nov 15;109(1):123–129. doi: 10.1016/0003-2697(80)90019-6. [DOI] [PubMed] [Google Scholar]
- Struhl K. Promoter elements, regulatory elements, and chromatin structure of the yeast his3 gene. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):901–910. doi: 10.1101/sqb.1983.047.01.104. [DOI] [PubMed] [Google Scholar]
- Sures I., Lowry J., Kedes L. H. The DNA sequence of sea urchin (S. purpuratus) H2A, H2B and H3 histone coding and spacer regions. Cell. 1978 Nov;15(3):1033–1044. doi: 10.1016/0092-8674(78)90287-8. [DOI] [PubMed] [Google Scholar]
- Thoma F., Bergman L. W., Simpson R. T. Nuclease digestion of circular TRP1ARS1 chromatin reveals positioned nucleosomes separated by nuclease-sensitive regions. J Mol Biol. 1984 Aug 25;177(4):715–733. doi: 10.1016/0022-2836(84)90046-9. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]