Abstract
Streptococcal inhibitor of complement (SIC) is a highly polymorphic extracellular protein and putative virulence factor secreted by M1 and M57 strains of group A Streptococcus (GAS). The sic gene is highly upregulated in invasive M1T1 GAS isolates following selection of mutations in the covR/S regulatory locus in vivo. Previous work has shown that SIC (allelic form 1.01) binds to and inactivates complement C5b67 and human cathelicidin LL-37. We examined the contribution of SIC to innate immune resistance phenotypes of GAS in the intact organism, using (1) targeted deletion of sic in wild-type and animal-passaged (covS mutant) M1T1 GAS harboring the sic 1.84 allele and (2) heterologous expression of sic in M49 GAS, which does not possess the sic genein its genome. We find that M1T1 SIC production is strongly upregulated upon covS mutation but that the sic gene is not required for generation and selection of covS mutants in vivo. SIC 1.84 bound both human and murine cathelicidins and was necessary and sufficient to promote covS mutant M1T1 GAS resistance to LL-37, growth in human whole blood and virulence in a murine model of systemic infection. Finally, the sic knockout mutant M1T1 GAS strain was deficient in growth in human serum and intracellular macrophage survival. We conclude that SIC contributes to M1T1 GAS immune resistance and virulence phenotypes.
Key Words: Antimicrobial peptides, Complement system, Innate immunity, Streptococcus, Virulence factors, Cathelicidin
Introduction
Group A Streptococcus (GAS) is a leading human bacterial pathogen producing illnesses ranging from mild pharyngitis or impetigo to severe necrotizing fasciitis and streptococcal toxic shock syndrome. Over the last 30 years, an increase in severe invasive GAS infections has been documented, largely attributable to the global dissemination and persistence of one clonal lineage of the M1T1 serotype [1, 2, 3, 4]. In total, GAS is estimated to cause 650,000 invasive infections resulting in 150,000 deaths annually worldwide [5].
The medical significance of the M1T1 GAS clone has stimulated research attempting to identify specific virulence factors which could explain the strain's propensity for severe invasive infections. In their global transcriptional analysis of clonal M1T1 GAS strains associated with mucosal versus invasive human infections, Sumby et al. [6] made the critical discovery that characteristic frameshift mutations in the covS gene were present among the invasive isolates. CovS is the sensor kinase of the two-component regulator CovR/S, which regulates the expression of 10–15% of genes in the GAS genome [7]. Mutations in covS result in increased expression of hyaluronic capsule and several other known or hypothesized GAS virulence proteins [6, 8]. Simultaneously, covS mutation downregulates expression of the broad-spectrum cysteine protease, SpeB, that otherwise degrades many of the important GAS virulence determinants [9, 10]. The genetic and phenotypic switch to hypervirulence can be recapitulated in the murine model, where M1T1 GAS covS mutations are selected in vivo and lead to SpeB inactivation, neutrophil resistance and systemic dissemination [6, 11].
One of the most highly upregulated genes upon covS mutation and initiation of invasive M1T1 GAS infection is sic, encoding the streptococcal inhibitor of complement. SIC is a secreted 31-kDa protein first discovered because of its location in the Mga virulence regulon [12]. By biochemical assays, SIC was shown to bind C5b67 of the membrane attack complex, thus impairing terminal complement function [12, 13]. A screen of emm types revealed that sic is present only in M1 and M57 strains [12]. Later it was discovered that another protein with similarity to the C-terminal proline-rich region of SIC, termed DRS (distantly related to SIC), is expressed by M12 and M55 strains [12, 14]. With nearly 300 alleles discovered, sic is the most polymorphic gene in the M1 GAS genome, suggesting it is under strong immune selective pressure [15].
In more recent studies, purified SIC has been shown to bind several antimicrobial proteins important in host innate defense, including lysozyme, secretory leukocyte protease inhibitor, human α-defensin-1 and human cathelicidin (LL-37) [16, 17, 18]. SIC is also important for GAS epithelial cell adherence [19] and mucosal colonization [20]; however, the role of SIC in invasive M1T1 GAS infection has not been studied. The goal of the present study was to analyze the effect of SIC on M1T1 GAS resistance to cathelicidin antimicrobial peptides, growth in human serum and whole blood, and virulence and phenotype switching in a murine infection model. Work in our model invasive M1T1 GAS clone examines the sic 1.84 allele, in contrast to the sic 1.01 allele that has been the basis of most published studies using purified SIC protein. Our analysis is performed using intact organisms, pairing targeted mutagenesis of sic in M1T1 GAS (loss of function) with heterologous expression of sic in an M49 GAS strain that does not contain the sic gene (gain of function).
Materials and Methods
Bacterial Strains and Generation of SIC Mutants
The wild-type (WT) GAS M1T1 strain (5448) was originally isolated from a patient with necrotizing fasciitis and toxic shock syndrome [1]. Polymerase chain reaction (PCR) was used to amplify DNA fragments directly upstream and downstream of sic from the M1T1 GAS genome. Primer sets used were: (1) upF (5′-cttcgtcgtgacttggacgc-3′) + upR (5′-tagtattctctccttaata-3′ with 30-bp 5′ extension matching the 5′ end of the cat gene) and (2) downF (5′-agggatgcgataggaatag-3′ with 30-bp 5′ extension matching the 3′ end of the cat gene) + downR (5′-gcggattccggctataacg-3′). Fusion PCR was performed using the respective upstream and downstream fragments and an amplicon of the cat gene to create a new DNA fragment in which the sic gene was precisely replaced by cat in chromosomal context. The fusion construct was TA cloned into pCR2.1-TOPO (Invitrogen) and then subcloned into pHY304, a temperature-sensitive plasmid bearing erythromycin resistance. The knockout plasmid was transformed into the M1T1 GAS parent strain, and single recombination events were selected for by plating on Todd-Hewitt Agar (THA; Difco), supplemented with 2 μg/ml erythromycin, at 37°C. Single crossovers were relaxed at 30°C, and double crossovers were screened for sensitivity to erythromycin. The knockout, herein referred to as M1T1 GAS Δsic, was verified by PCR.
An animal-passaged (AP) version of the M1T1 GAS parent strain (5448AP) has been previously published and has been shown to contain a single inactivating adenine insertion at the 877-bp position of covS [10, 21]. The animal-passaged version of the M1T1 GAS Δsic was created by injecting 12-week-old female C57BL/6J mice (Charles River Laboratories) subcutaneously with 2 × 108 colony-forming units (CFU) of mid-log phase M1T1 GAS Δsic in 100 μl Dulbecco's phosphate-buffered saline (DPBS). After 72 h, mice were sacrificed, lesions excised and homogenized, and bacteria were plated. Bacteria were subsequently screened using a cysteine protease assay to detect SpeB activity, and the covR/S genes of SpeB-negative colonies were sequenced directly. A single base pair mutation (C → T) in covS was discovered at the 1,261-bp position, producing a premature stop codon. Hereforeward, the animal-passaged SIC mutant strain is designated M1T1 GAS (AP) Δsic.
Heterologous Expression of SIC
The sic gene was amplified from the M1T1 WT genome by PCR using primers sicF (5′-ggagagaatactaatg-3′) and sicR (5′-ttacgttgctgatggtgt-3′). The PCR product was TA cloned into pCR2.1-TOPO (Invitrogen) and then subcloned into shuttle expression vector pDCerm [22]. The resulting plasmid was introduced via electroporation into M49 GASstrain NZ131 [23], resulting in heterologous expression of SIC.
Western Immunoblot Analysis
Bacterial supernatants from late-log phase (SIC expression) or overnight cultures (SpeB expression, SIC cleavage) were collected after centrifugation at 3,220 g for 10 min. For SIC cleavage, cultures were grown with or without 10 μg/ml of E-64, a cysteine protease inhibitor. Supernatants were filtered using a 0.22-μm filter (Millipore) and then left unconcentrated (for SpeB blot) or concentrated 50-fold (for SIC blots) using a Vacufuge (Eppendorf). Samples were mixed with 4× sample buffer and 10× reducing agent (Invitrogen), boiled for 10 min, loaded onto a 10% Bis-Tris gel (Invitrogen) and run in MOPS running buffer at 140 V. The gels were then transferred onto a nitrocellulose membrane using a semi-dry apparatus (Bio-Rad) at 12 V for 45 min. After transfer, the membranes were blocked for 1 h at room temperature (RT) in phosphate-buffered saline (PBS)-0.03% Tween + 5% nonfat milk. For SIC Western blots, primary anti-SIC antibody was incubated at RT for 1 h at a 1:200 dilution in blocking buffer. For the SpeB blot, primary anti-SpeB antibody (Toxin Technology Inc.) was incubated at RT for 1 h at a 1:1,000 dilution in blocking buffer. After 3 washes with PBS-Tween, the membranes were incubated with a goat anti-rabbit HRP secondary antibody (Bio-Rad) at a 1:20,000 dilution for 1 h at RT. The membranes were subsequently washed 3× with PBS-Tween and developed using chemiluminescence.
Minimum Inhibitory Concentration Testing
Bacterial cultures were grown to mid-log phase in DPBS + 20% Todd-Hewitt Broth (THB; Difco). Bacteria were resuspended to 105 CFU/ml in the same buffer, and 90 μl of bacteria was added to 10 μl of decreasing concentrations of LL-37 or CRAMP in a 96-well plate. After incubation at 37°C for 24 h, 25 μl was removed from each well, diluted and plated for enumeration of bacteria to determine the minimum inhibitory concentration (MIC, inhibited bacterial growth) and minimum bactericidal concentration (MBC, no bacterial growth).
LL-37 Killing Kinetics
GAS cultures were grown to mid-log phase in DPBS + 20% THB. Bacteria were resuspended to 105 CFU/ml in the same buffer, and 450 μl of this suspension was added to 50 μl of 320 μM LL-37 (for a final concentration of 32 μM) in siliconized tubes. Tubes were placed on a rotisserie at 37°C, and 25 μl was removed at each time point to be diluted and plated for enumeration of CFU.
Co-Immunoprecipitation
One milliliter of supernatant from an overnight culture of M1T1 GAS (AP) was incubated with either 1 μM LL-37 or CRAMP at RT for 8 h. Twenty microliters of Protein A/G PLUS (Santa Cruz Biotechnology) beads was added to each tube and incubated while rotating at 4°C for 1 h to allow preclearing. Beads were centrifuged at 5,000 g for 5 min, and the supernatant was transferred to new tubes in 500-μl aliquots. For immunoprecipitation of LL-37, 1 μg of chicken anti-LL-37 was added while 1 μg IgY was added to a second tube as a negative control. Tubes were incubated, rotating, at 4°C overnight. The next day, goat anti-IgY was added to each tube and incubated, rotating, at 4°C for 2 h. Twenty microliters of Protein A/G PLUS beads was added to each tube and incubated at 4°C for 4 h. Beads were pelleted by centrifugation at 5,000 g for 5 min and subsequently washed 4 times with 500 μl ice-cold PBS. Each tube of beads was resuspended in 20 μl of PBS. Samples were then analyzed by Western blotting exactly as in the protocol above, except that subsequent to boiling, samples were spun down at 5,000 g for 5 min to remove beads. For immunoprecipitation of CRAMP, 1 μg of rabbit anti-CRAMP was added, while 1 μg rabbit IgG was added to a second tube as a negative control. Tubes were incubated, rotating, at 4°C overnight. The next day, Protein A/G PLUS beads were added, and the assay was carried out exactly as for LL-37.
Growth in Human Whole Blood
Blood was drawn from healthy donors after informed consent, and 2 × 103 CFU (M1T1) or 2 × 107 CFU (M49) of late-log phase bacteria in 100 μl PBS was added to 300 μl heparinized whole blood in siliconized tubes. Tubes were placed on a rotisserie at 37°C, and a 25-μl aliquot was removed at each time point to be diluted and plated for enumeration of CFU. Growth index was calculated as the ratio of surviving CFU after incubation versus the initial inoculum.
Growth in Human Serum
Blood was drawn from healthy human volunteers after informed consent. Normal human serum was prepared by pooling sera of 6 different donors and stored at −80°C. Overnight bacterial cultures grown in THB were diluted 1:100,000 in RPMI 1640 tissue culture medium (Invitrogen). Fifty microliters of bacteria was mixed with 50 μl of serum in RPMI 1640 in 96-well plates. Plates were incubated for 6 h at 37°C, and surviving bacteria were enumerated by plating serial dilutions on THA. Growth index was calculated as the ratio of surviving CFU after incubation versus the initial inoculum.
Macrophage Intracellular Survival
Eight-week-old CD-1 male mice (Charles River Laboratories) were injected intraperitoneally with 3 ml of 3% thioglycolate to stimulate macrophage induction. Three days after infection, mice were sacrificed and macrophages were harvested by rinsing the peritoneal cavity twice with 8 ml DPBS. Macrophages were counted, resuspended in RPMI + 10% FBS, seeded at 7 × 105 cells/well in a 24-well plate and allowed to adhere overnight. The next day, bacteria were grown to late-log phase and resuspended in RPMI + 2% FBS. An aliquot of 500 μl bacteria, representing a multiplicity of infection of 10:1 (GAS:macrophage), was added to wells after media was removed, and plates were centrifuged at 500 g for 5 min to ensure contact of the bacteria with the macrophages. Plates were incubated at 37°C + 5% CO2 for 1 h. Media was then removed and cells were washed 3 times with DPBS. Media containing 10 μg/ml penicillin and 100 μg/ml gentamicin was added to kill extracellular bacteria and the plates were incubated for an additional 1 h. Media was then removed, cells were washed 3 times with DPBS, and 500 μl of 0.025% Triton-X was added to lyse cells. Cells were removed by tituration and dilutions plated to enumerate bacteria; this represented the 1-hour time point. To wells of later time points, fresh media was added, and at the indicated time points, cells were harvested as described. Relative survival was calculated by comparing surviving CFU to the inoculum and normalizing all time points to the M1T1 GAS (AP) 1-hour time point.
Murine Infection Model
Eight-week-old male CD-1 mice were injected intraperitoneally with 2.5 × 106 CFU of mid-log phase M1T1 GAS (AP) or M1T1 GAS (AP) Δsic or alternatively, 1.3 × 107 CFU of mid-log M49 GAS or M49 GAS expressing SIC, suspended in 200 μl of DPBS + 5% gastric mucin. Survival was monitored for 4 (M1T1) or 7 (M49) days, and time of death was recorded for each mouse.
In vivo Phenotype Switching Assay
To study the influence of SIC on the capacity for in vivo phase-switching (covS mutation and SpeB inactivation), 8-week-old female C57BL/6J mice or outbred male CD-1 mice (Charles River Laboratories) were subcutaneously inoculated with a nonlethal dose (approx. 108 CFU/100 μl) of mid-log phase M1T1 GAS or M1T1 GAS Δsic in DPBS. For the C57BL/6J experiment, separate groups of 10 mice were injected with each strain; for the CD-1 experiment, each of 10 mice was injected on the left flank with the M1 GAS parent strain and on the right flank with the M1T1 GAS Δsic mutant. Three days after infection, mice were euthanized by CO2 asphyxiation, skin lesions excised and homogenized in 1 ml DPBS + 1 mm zirconia/silica beads using a Mini-Beadbeater (Biospec Products) and 2 × 1 min homogenization bursts. Homogenates were serially diluted in PBS, plated onto THA and 50 individual lesion-derived colonies (n = 50) were then assayed for SpeB protease by an azocaseinolytic assay [24] scaled to a 96-well plate format. Isolates were scored SpeB-negative following negative results in consecutive assays; for all assays, WT M1T1 GAS and its isogenic ΔspeB knockout mutant [10] were included as positive and negative controls, respectively.
Statistical Analyses
LL-37 kinetics, whole blood, serum, intracellular macrophage survival assays and in vivo covS switching were compared using the Student's t test. Mouse intraperitoneal challenge assay was assessed using the log-rank (Mantel Cox) test. All statistical tests were performed using GraphPad Prism version 5.0 (GraphPad Software Inc.). p values < 0.05 were considered statistically significant.
Ethics Permissions
Permission to collect human blood under informed consent was approved by the UCSD Human Research Protections Program. All animal experiments were conducted according to the guidelines approved by the UCSD Institutional Animal Use and Care Committee.
Results
Creation of Bacterial Reagents for SIC Analysis
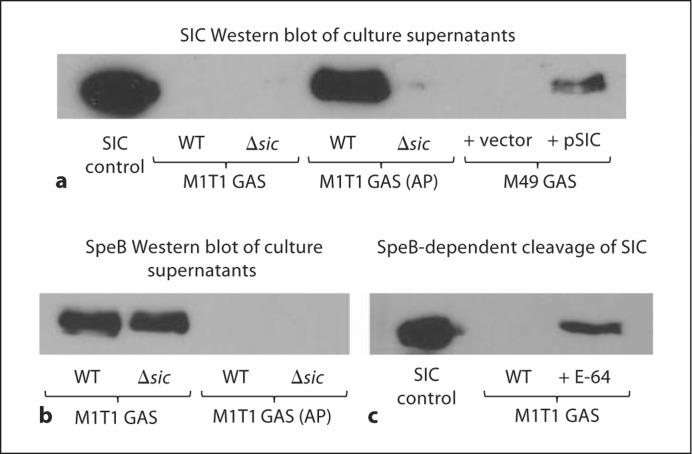
To study the role of SIC in the context of the living pathogen, we created a precise in-frame allelic replacement of the sic gene in M1T1 GAS with a cassette encoding chloramphenicol acetyltransferase (cat). The mutant was then animal passaged (AP) by murine subcutaneous passage, and identified SpeB-negative colonies were shown to possess an inactivating mutation within covS. For gain of function analyses, SIC was heterologously expressed on a plasmid in M49 GAS. To confirm the presence or lack of SIC expression in the generated strains, cell-free culture supernatants were concentrated 50-fold and subjected to Western blot analysis (fig. 1a). As expected, no detectable SIC was identified in supernatants from the two Δsic mutants, and heterologous expression in M49 GAS was successful. Even at a 50-fold concentration, however, the M1T1 GAS strain did not produce sufficient amounts of SIC for detection by Western blot, whereas this protein was highly abundant in the M1T1 GAS strain following covS mutation and associated transcriptional upregulation. Purified SpeB has been reported to cleave purified SIC [25], so we examined whether this was occurring in the context of the M1T1 GAS strain, resulting in the absence of SIC in the supernatant. We first looked at SpeB expression in the four M1T1 strains and found that it was expressed in the M1T1 GAS and M1T1 GAS Δsic strains but not in the M1T1 GAS (AP) or M1T1 GAS (AP) Δsic strains as expected (fig. 1b). To demonstrate cleavage of SIC by SpeB in M1T1 GAS, overnight cultures were grown with or without E-64, a cysteine protease inhibitor, concentrated 50-fold and subjected to Western blot. When SpeB was inhibited by E-64, full-length SIC was present in the supernatant (fig. 1c). Thus, in this representative strain of the invasive M1T1 GAS clone expressing the sic 1.84 allele, only very small amounts of the putative virulence factor may be present, due to cleavage by SpeB, until the in vivo selection event of covS mutation that leads to drastically reduced levels of SpeB and increased levels of SIC. Consequently, for many of the ensuing comparative studies of SIC function, the M1T1 GAS (AP) strain and the M1T1 GAS (AP) Δsic mutant were employed. Despite several attempts, we were technically unable to achieve complementation of the M1T1 GAS (AP) Δsic mutant with the expression vector used successfully in M49 GAS. We hypothesize these results derive from either (1) the marked hyperencapsulation of animal-passaged covS mutant GAS strains interfering with transformation or (2) potential toxicities of protein overexpression when the sic gene is present on a multicopy number plasmid and simultaneously subject to strong transcriptional upregulation.
Fig. 1.
Generation of bacterial reagents with altered SIC expression. a Western blot analysis of SIC expression following targeted mutagenesis and/or animal-passage in M1T1 GAS strains and heterologous expression in M49 GAS. Concentrated (50-fold) cell-free culture supernatants were probed using polyclonal anti- SIC antisera. Western blot analysis of SpeB expression (b) and SIC cleavage (c) by SpeB in M1T1 GAS strains. SIC control lanes contain 0.5 μg of purified SIC protein.
SIC 1.84 Binds to Human and Murine Cathelicidins
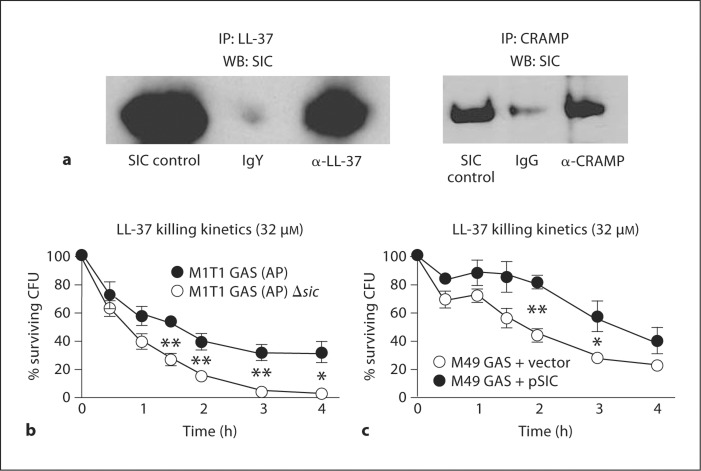
While previous studies of SIC protein interaction with human cathelicidin LL-37 were performed with the protein encoded by the sic 1.01 allele, our M1T1 GAS strain harbors the sic 1.84 allele. The alleles possess 89.6% sequence identity (online supplementary figure 1, www.karger.com/doi/10.1159/000317672). To determine whether a cathelicidin-binding phenotype was retained in the SIC 1.84 protein, we incubated M1T1 GAS (AP) supernatants with either 1 μM LL-37 or the related murine cathelicidin peptide CRAMP, then performed a co-immunoprecipitation by pulling down cathelicidin and immunoblotting for SIC. Increased amounts of SIC were seen in the anti-cathelicidin lanes, compared to the isotype control lanes, demonstrating that our particular sic variant binds both LL-37 and CRAMP (fig. 2a). To our knowledge, this is the first time SIC has been shown to bind murine cathelicidin.
Fig. 2.
SIC interferes with cathelicidin activity by sequestration. a SIC 1.84 allelic variant binds cathelicidins LL-37 (human) and CRAMP (murine) as demonstrated by co-immunoprecipitation after incubation of M1T1 GAS (AP) supernatant with 1 μM of the indicated cathelicidin. Samples were immunoprecipitated (IP) for cathelicidin and Western blotted (WB) for SIC. M1T1 GAS (AP) (b) and M49 GAS (c) strains lacking SIC were more rapidly killed over time by LL-37 in a kinetic killing assay. Experiments were performed in triplicate and repeated 3 times with similar results. Percent survival was calculated by dividing the surviving CFU by the initial inoculums and multiplying by 100. A representative experiment is shown with the mean ± SEM. * p < 0.05; ** p < 0.01, according to Student's unpaired t test.
SIC and Bacterial Resistance to Cathelicidin Killing
Finding that SIC binds both LL-37 and CRAMP, we sought to determine whether SIC expression influences bacterial resistance to killing by these host defense peptides. Strains were incubated with LL-37 or CRAMP, and bacteria were plated and enumerated at 24 h to determine the MICs and MBCs. It is important to recognize that other virulence factors implicated in GAS cathelicidin resistance are also altered upon covS mutation, including the loss of SpeB protease, capable of degrading LL-37 [25], and preservation of surface-anchored M1 protein, which is capable of binding the peptides via its N-terminus [26]. The presence of sic was found to promote GAS resistance to LL-37 in both the pre- and post-animal passage background (table 1), with the greatest relative difference in MIC observed upon direct comparison of the M1T1 GAS (AP) strain to the M1T1 GAS (AP) Δsic mutant (16 vs. 4 μM). Further functional linkage was provided by the observation that expression of SIC in M49 GAS led to increased resistance to LL-37 (table 1). In contrast, effects of loss or gain of SIC expression on GAS resistance to the murine cathelicidin CRAMP, if any, were insufficient to discriminate by in vitro MIC or MBC assays. Here we acknowledge the limitations of short-term in vitro analyses of a secreted protein, which may not be produced de novo in sufficient quantities from the freshly prepared bacterial inoculum to replicate levels that could accumulate over longer periods at foci of infection in vivo.
Table 1.
Activity of human and murine cathelicidin antimicrobial peptides against bacterial strains with altered SIC production
| MIC | MBC | |
|---|---|---|
| LL-37 (human) | ||
| M1T1 GAS | 14 μM | 24 μM |
| M1T1 GAS δsic | 10 μM | 14 μM |
| M1T1 GAS (AP) | 16 μM | 28 μM |
| M1T1 GAS (AP) δsic | 4 μM | 10 μM |
| M49 GAS + vector | 8 μM | 16 μM |
| M49 GAS + pSIC | 16 μM | 32 μM |
| CRAMP (mouse) | ||
| M1T1 GAS | 4 μM | 8 μM |
| M1T1 GAS δsic | 4 μM | 8 μM |
| M1T1 GAS (AP) | 4 μM | 16 μM |
| M1T1 GAS (AP) δsic | 4 μM | 16 μM |
| M49 GAS + vector | 4 μM | 16 μM |
| M49 GAS + pSIC | 4 μM | 16 μM |
To further investigate the role of SIC in resistance to LL-37, we examined the kinetics of bacterial killing. To test this, M1T1 GAS (AP) and M1T1 GAS (AP) Δsic were incubated with 32 μM LL-37 and time points were taken over a period of 4 h to allow for enumeration of bacteria. We found that M1T1 GAS (AP) Δsic was killed significantly faster than M1T1 GAS (AP) (fig. 2b). In addition, we observed that M49 GAS expressing SIC exhibited significantly more resistance to killing by LL-37 over time (fig. 2c). Thus, we conclude that SIC is necessary and sufficient to promote resistance to the human cathelicidin, LL-37.
SIC Promotes GAS Growth in Human Whole Blood and Human Serum
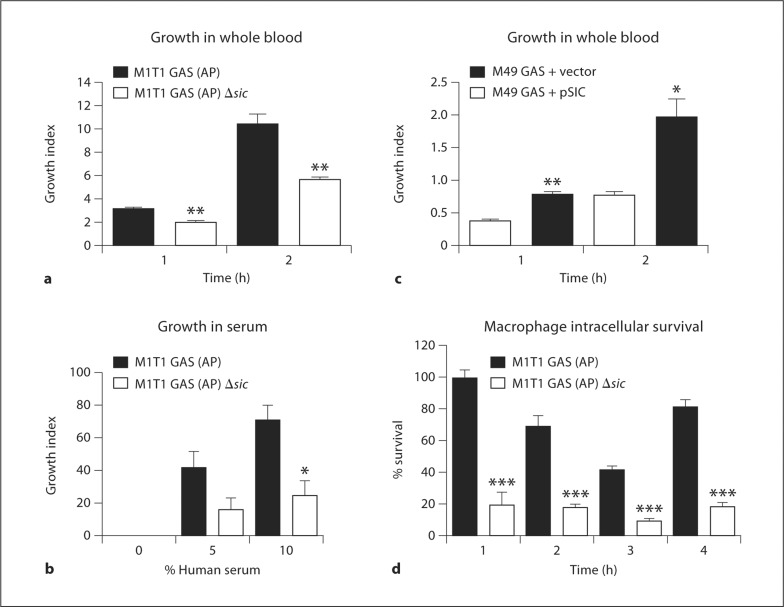
The in vivo selection of GAS M1T1 covS mutants is associated with systemic dissemination and enhanced resistance to phagocyte killing [6, 11, 21]. Here, we examined whether SIC is an important contributor to GAS proliferation in human blood and serum. Compared to the M1T1 GAS (AP) strain, the isogenic M1T1 GAS (AP) Δsic mutant grew significantly less efficiently in freshly isolated human whole blood (fig. 3a) and human serum (fig. 3b). Expression of SIC in the M49 GAS background significantly enhanced growth in human whole blood (fig. 3c). We conclude that SIC is both necessary and sufficient to support effective bacterial growth in human blood and necessary for growth in human serum.
Fig. 3.
SIC is necessary and sufficient to promote bacterial growth in human whole blood and necessary for growth in normal human serum and intracellular survival in primary murine macrophages. Absence of SIC expression in the M1T1 GAS (AP) background is associated with decreased survival in whole blood (a) and normal serum (b). c Heterologous expression of SIC in M49 GAS promotes bacterial growth in whole blood. For whole blood, experiments were performed in triplicate and repeated five times with similar results. Growth index was calculated by dividing the surviving CFU by the initial inoculum. A representative experiment is shown with the mean ± SEM. For serum, experiments were performed in duplicate and repeated 4 times with similar results. Mean ± SEM of 4 experiments is shown. d Increased survival within murine macrophages of GAS M1T1 (AP) vs. GAS M1T1 (AP) δ sic mutant. Experiment was performed with 6 replicates, then repeated 3 times with similar results. Relative survival was calculated by comparing surviving CFU to the inoculum and normalizing all time points to the GAS M1T1 (AP) 1-hour time point. * p < 0.05; ** p < 0.01; *** p < 0.001 according to Student's unpaired t test.
SIC Promotes GAS Survival in Murine Intraperitoneal Macrophages
To begin to investigate whether a murine model of infection might prove useful for assessing the contribution of SIC to GAS systemic virulence, we compared intracellular survival of the M1T1 GAS (AP) and M1T1 GAS (AP) Δsic in thioglycolate-elicited primary murine peritoneal macrophages. GAS were added to the macrophages at a multiplicity of infection of 10:1, and plates centrifuged to bring bacteria into close contact with the macrophages. After 1 h of incubation, aliquots were removed and plated to ensure similar rates of phagocytosis (data not shown), and antibiotic media was added to kill remaining extracellular bacteria. Cells were harvested at the indicated time points and wells were plated for enumeration of bacteria. Absence of SIC in the animal-passaged covS mutant GAS strain was associated with a significant intracellular survival defect (fig. 3d).
SIC Accelerates GAS-Induced Mortality in a Murine Systemic Model of Infection
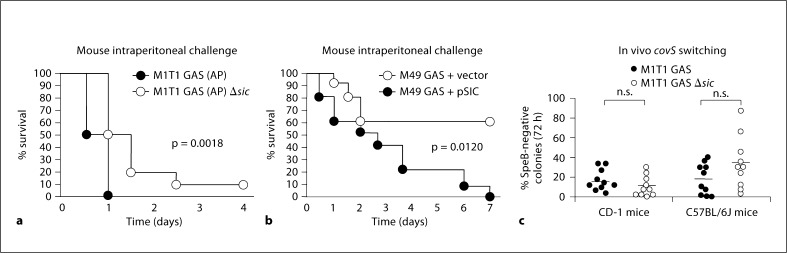
To determine the effect of SIC expression to the course of systemic GAS infection, 8-week-old CD-1 mice were administered intraperitoneal injections of M1 GAS (AP) or M1 GAS (AP) Δsic or for heterologous expression studies, M49 GAS or M49 GAS expressing SIC, and the kinetics of their survival was monitored over time. Mice injected with the Δsic mutant survived significantly longer than those challenged with the SIC-expressing strain (50 vs. 100% mortality at 24 h), suggesting a significant contribution of SIC to the overall virulence of the hyperinvasive covS mutant form of M1T1 GAS (fig. 4a). In addition, mice injected with M49 GAS survived significantly longer than those infected with M49 GAS expressing SIC, corroborating a contribution of SIC in invasive GAS infection (fig. 4b). We next wondered whether preservation of SIC, which itself is a target of SpeB-mediated proteolytic degradation [10], might be a selective force favoring in vivo covS mutations to eliminate speB expression. Two strains of mice (CD-1 and C57BL/6J) were injected subcutaneously with either the M1T1 GAS strain or its isogenic Δsic mutant (10 animals in each of the 4 groups). After 72 h, resulting lesions were excised to identify SpeB-negative colonies indicative of phase-switching to the covS mutant phenotype (fig. 4c). In both genetic backgrounds, SpeB-negative mutants were readily identified among the 50 bacterial colonies isolated from individual mice infected with the M1T1 GAS Δsic mutant, and no statistically significant difference in the frequency of covS phase-switching was found comparing the Δsic mutant to the parent M1T1 GAS strain. We conclude that in contrast to findings reported for the bacteriophage-encoded M1T1 GAS DNase Sda1 [21], presence of SIC is not required to maintain the selection pressure favoring covR/S mutation in vivo.
Fig. 4.
SIC promotes GAS virulence in murine models. Eightweek- old male CD-1 mice were injected intraperitoneally with 2.5 × 106 CFU of M1T1 GAS (AP) or M1T1 GAS (AP) δsic + 5% mucin (a) or 1.3 × 107 CFU of M49 GAS or M49 GAS expressing SIC + 5% mucin (b); n = 10. Survival was monitored for 4 or 7 days. Statistics were performed using the log-rank (Mantel Cox) test. c In vivo covS switching of GAS M1T1 vs. GAS M1T1 δsic as assessed by loss of SpeB activity 72 h after subcutaneous injection; no significant difference between WT and δsic mutant in either the CD-1 or C57BL/6J mouse background.
Discussion
We used targeted mutagenesis and heterologous expression to demonstrate that SIC contributes to the human cathelicidin antimicrobial peptide, whole blood and serum resistance phenotypes of invasive M1T1 GAS. These virulence phenotypes occur upon strong transcriptional upregulation of the sic gene following covS mutations selected in vivo during the transition to invasive infection. Concurrently, the reduction of SpeB expression upon covS mutation protects the highly expressed SIC protein from proteolytic degradation. Ultimately, sic can be functionally validated to be part of a critical suite of virulence factor genes that are upregulated during covS mutation and invasive M1T1 GAS infection, including those encoding hyaluronic acid capsule [28], DNase Sda1 [21], streptolysin O and interleukin-8 protease SpyCEP, that together promote innate immune resistance and systemic dissemination [4, 6].
SIC promoted GAS M1T1 survival in murine macrophages and enhanced the progression of invasive infection during mouse systemic infection. These results suggest at least some limited utility of murine models for further analyzing SIC virulence functions or evaluating therapeutic interventions directed against SIC. We observe that the 1.84 allelic variant of SIC binds both human and murine cathelicidin, although in the intact organism its influence on LL-37 resistance is clearly more pronounced than any potential influence on CRAMP resistance. Theoretically, a virulence contribution of SIC binding to CRAMP could be manifest in vivo upon greater accumulation of the secreted virulence factor in established foci of infection or by effects upon other pro-inflammatory or immunostimulatory functions of the cathelicidin peptide [29, 30]. The observed SIC contribution to mouse virulence could also derive from enhanced survival in macrophages, enhanced serum resistance, or other yet to be determined pathogenic effects.
Cathelicidin is a critical aspect of host defense against invasive GAS infection [27], and serotype M1 GAS strains show higher intrinsic resistance levels to LL-37 killing than GAS of other serotypes uncommonly associated with invasive infection [26]. The expression of two LL-37 binding/sequestering molecules, namely SIC and the N-terminal domain of M1 protein [26], pose a significant challenge for our innate immune system attempting to restrict the spread of M1T1 GAS infection. The unusually high allelic variation of SIC in M1 GAS may parallel that of M protein across all serotype strains – evidence of an ongoing evolutionary battle of pathogen and host, with M1T1 GAS seeking to preserve pro-survival functions of the SIC peptide while escaping host-neutralizing antibody responses.
Supplementary Material
Acknowledgements
The authors thank Lars Björck of Lund University for providing anti-SIC antisera and purified SIC protein. This work was supported by National Institutes of Health grants AI77780 (V.N.) and AI48176 (R.L.G). M.A.P. is supported in part by the UCSD Genetics Training Program, NIH/NIGMS T32 GM008666, S.H.M.R. by an EMBO Postdoctoral Fellowship and the Netherlands Organization of Scientific Research, J.N.C. by a National Health and Medical Research Council of Australia grant (514639) and A.H. by a Department of Employment Science and Technology (Australia) International Sciences Linkages grant (CG001105).
References
- 1.Chatellier S, Ihendyane N, Kansal RG, Khambaty F, Basma H, Norrby-Teglund A, Low DE, McGeer A, Kotb M. Genetic relatedness and superantigen expression in group A Streptococcus serotype M1 isolates from patients with severe and nonsevere invasive diseases. Infect Immun. 2000;68:3523–3534. doi: 10.1128/iai.68.6.3523-3534.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cleary PP, LaPenta D, Vessela R, Lam H, Cue D. A globally-disseminated M1 subclone of group A streptococci differs from other subclones by 70 kilobases of prophage DNA and capacity for high-frequency intracellular invasion. Infect Immun. 1998;66:5592–5597. doi: 10.1128/iai.66.11.5592-5597.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cockerill FR, 3rd, MacDonald KL, Thompson RL, Roberson F, Kohner PC, Besser-Wiek J, Manahan JM, Musser JM, Schlievert PM, Talbot J, Frankfort B, Steckelberg JM, Wilson WR, Osterholm MT. An outbreak of invasive group A streptococcal disease associated with high carriage rates of the invasive clone among school-aged children. JAMA. 1997;277:38–43. [PubMed] [Google Scholar]
- 4.Aziz RK, Kotb M. Rise and persistence of global M1T1 clone of Streptococcus pyogenes. Emerg Infect Dis. 2008;14:1511–1517. doi: 10.3201/eid1410.071660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis. 2005;5:685–694. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 6.Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-wide analysis of group A streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2006;2:41–49. doi: 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graham MR, Smoot LM, Migliaccio CA, Virtaneva K, Sturdevant DE, Porcella SF, Federle MJ, Adams GJ, Scott JR, Musser JM. Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc Natl Acad Sci USA. 2002;99:13855–13860. doi: 10.1073/pnas.202353699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engleberg NC, Heath A, Miller A, Rivera C, DiRita VJ. Spontaneous mutations in the CsrRS two-component regulatory system of Streptococcus pyogenes result in enhanced virulence in a murine model of skin and soft tissue infection. J Infect Dis. 2001;183:1043–1054. doi: 10.1086/319291. [DOI] [PubMed] [Google Scholar]
- 9.Raeder R, Harokopakis E, Hollingshead S, Boyle MD. Absence of SpeB production in virulent large capsular forms of group A streptococcal strain 64. Infect Immun. 2000;68:744–751. doi: 10.1128/iai.68.2.744-751.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aziz RK, Pabst MJ, Jeng A, Kansal R, Low DE, Nizet V, Kotb M. Invasive M1T1 group A Streptococcus undergoes a phase-shift in vivo to prevent proteolytic degradation of multiple virulence factors by SpeB. Mol Microbiol. 2004;51:123–134. doi: 10.1046/j.1365-2958.2003.03797.x. [DOI] [PubMed] [Google Scholar]
- 11.Cole JN, McArthur JD, McKay FC, Sanderson-Smith ML, Cork AJ, Ranson M, Rohde M, Itzek A, Sun H, Ginsburg D, Kotb M, Nizet V, Chhatwal GS, Walker MJ. Trigger for group A streptococcal M1T1 invasive disease. FASEB J. 2006;20:1745–1747. doi: 10.1096/fj.06-5804fje. [DOI] [PubMed] [Google Scholar]
- 12.Akesson P, Sjöholm AG, Björck L. Protein SIC, a novel extracellular protein of Streptococcus pyogenes interfering with complement function. J Biol Chem. 1996;271:1081–1088. doi: 10.1074/jbc.271.2.1081. [DOI] [PubMed] [Google Scholar]
- 13.Fernie-King BA, Seilly DJ, Willers C, Wurzner R, Davies A, Lachmann PJ. Streptococcal inhibitor of complement (SIC) inhibits the membrane attack complex by preventing uptake of C567 onto cell membranes. Immunology. 2001;103:390–398. doi: 10.1046/j.1365-2567.2001.01249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartas J, Sriprakash KS. Streptococcus pyogenes strains containing emm12 and emm55 possess a novel gene coding for distantly related sic protein. Microb Pathog. 1999;26:25–33. doi: 10.1006/mpat.1998.0244. [DOI] [PubMed] [Google Scholar]
- 15.Hoe NP, Vuopio-Varkila J, Vaara M, Grigsby D, De Lorenzo D, Fu YX, Dou SJ, Pan X, Nakashima K, Musser JM. Distribution of streptococcal inhibitor of complement variants in pharyngitis and invasive isolates in an epidemic of serotype M1 group A Streptococcus infection. J Infect Dis. 2001;183:633–639. doi: 10.1086/318543. [DOI] [PubMed] [Google Scholar]
- 16.Frick IM, Akesson P, Rasmussen M, Schmidtchen A, Björck L. SIC, a secreted protein of Streptococcus pyogenes that inactivates antibacterial peptides. J Biol Chem. 2003;278:16561–16566. doi: 10.1074/jbc.M301995200. [DOI] [PubMed] [Google Scholar]
- 17.Fernie-King BA, Seilly DJ, Davies A, Lachmann PJ. Streptococcal inhibitor of complement inhibits two additional components of the mucosal innate immune system: secretory leukocyte proteinase inhibitor and lysozyme. Infect Immun. 2002;70:4908–4916. doi: 10.1128/IAI.70.9.4908-4916.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernie-King BA, Seilly DJ, Lachmann PJ. The interaction of streptococcal inhibitor of complement (SIC) and its proteolytic fragments with the human β defensins. Immunology. 2004;111:444–452. doi: 10.1111/j.0019-2805.2004.01837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoe NP, Ireland RM, DeLeo FR, Gowen BB, Dorward DW, Voyich JM, Liu M, Burns EH, Jr, Culnan DM, Bretscher A, Musser JM. Insight into the molecular basis of pathogen abundance: group A Streptococcus inhibitor of complement inhibits bacterial adherence and internalization into human cells. Proc Natl Acad Sci USA. 2002;99:7646–7651. doi: 10.1073/pnas.112039899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lukomski S, Hoe NP, Abdi I, Rurangirwa J, Kordari P, Liu M, Dou SJ, Adams GG, Musser JM. Nonpolar inactivation of the hypervariable streptococcal inhibitor of complement gene (SIC) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonization. Infect Immun. 2000;68:535–542. doi: 10.1128/iai.68.2.535-542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker MJ, Hollands A, Sanderson-Smith ML, Cole JN, Kirk JK, Henningham A, McArthur JD, Dinkla K, Aziz RK, Kansal RG, Simpson AJ, Buchanan JT, Chhatwal GS, Kotb M, Nizet V. DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. Nat Med. 2007;13:981–985. doi: 10.1038/nm1612. [DOI] [PubMed] [Google Scholar]
- 22.Jeng A, Sakota V, Li Z, Datta V, Beall B, Nizet V. Molecular genetic analysis of a group A Streptococcus operon encoding serum opacity factor and a novel fibronectin-binding protein, Sfbx. J Bacteriol. 2003;185:1208–1217. doi: 10.1128/JB.185.4.1208-1217.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simon D, Ferretti JJ. Electrotransformation of Streptococcus pyogenes with plasmid and linear DNA. FEMS Microbiol Lett. 1991;66:219–224. doi: 10.1016/0378-1097(91)90336-9. [DOI] [PubMed] [Google Scholar]
- 24.Collin M, Olsén A. Generation of a mature streptococcal cysteine proteinase is dependent on cell wall-anchored M1 protein. Mol Microbiol. 2000;36:1306–1318. doi: 10.1046/j.1365-2958.2000.01942.x. [DOI] [PubMed] [Google Scholar]
- 25.Hoe NP, Kordari P, Cole R, Liu M, Palzkill T, Huang W, McLellan D, Adams CJ, Hu M, Vuopio-Varkila J, Cate TR, Pichichero ME, Edwards KM, Eskola J, Low DE, Musser JM. Human immune response to streptococcal inhibitor of complement, a serotype M1 group A Streptococcus extracellular protein involved in epidemics. J Infect Dis. 2000;182:1425–1436. doi: 10.1086/315882. [DOI] [PubMed] [Google Scholar]
- 26.Johansson L, Thulin P, Sendi P, Hertzén E, Linder A, Akesson P, Low DE, Agerberth B, Norrby-Teglund A. Cathelicidin LL-37 in severe Streptococcus pyogenes soft tissue infections in humans. Infect Immun. 2008;76:3399–3404. doi: 10.1128/IAI.01392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lauth X, von Köckritz-Blickwede M, McNamara CW, Myskowski S, Zinkernagel AS, Beall B, Ghosh P, Gallo RL, Nizet V. M1 protein allows group A streptococcal survival in phagocyte extracellular traps through cathelicidin inhibition. J Innate Immun. 2009;1:202–214. doi: 10.1159/000203645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nizet V, Ohtake T, Lauth X, Trowbridge J, Rudisill J, Dorschner RA, Pestonjamasp V, Piraino J, Huttner K, Gallo RL. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature. 2001;414:454–457. doi: 10.1038/35106587. [DOI] [PubMed] [Google Scholar]
- 29.Ravins M, Jaffe J, Hanski E, Shetzigovski I, Natanson-Yaron S, Moses AE. Characterization of a mouse-passaged, highly encapsulated variant of group A Streptococcus in in vitro and in vivo studies. J Infect Dis. 2000;182:1702–1711. doi: 10.1086/317635. [DOI] [PubMed] [Google Scholar]
- 30.Lai Y, Gallo RL. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009;30:131–141. doi: 10.1016/j.it.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Radek K, Gallo RL. Antimicrobial peptides: natural effectors of the innate immune system. Semin Immunopathol. 2007;29:27–43. doi: 10.1007/s00281-007-0064-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.