Abstract
Group A Streptococcus (GAS) causes rare but life-threatening syndromes of necrotizing fasciitis and toxic shock-like syndrome in humans. The GAS serotype M1T1 clone has globally disseminated, and mutations in the control of virulence regulatory sensor kinase (covRS) operon correlate with severe invasive disease. Here, a cohort of non-M1 GAS was screened to determine whether mutation in covRS triggers systemic dissemination in divergent M serotypes. A GAS disease model defining parameters governing invasive propensity of differing M types is proposed. The vast majority of GAS infection is benign. Nonetheless, many divergent M types possess limited capacity to cause invasive infection. M1T1 GAS readily switch to a covRS mutant form that is neutrophil resistant and frequently associated with systemic infection. Whilst non-M1 GAS are shown in this study to less frequently accumulate covRS mutations in vivo, such mutants are isolated from invasive infections and exhibit neutrophil resistance and enhanced virulence. The reduced capacity of non-M1 GAS to switch to the hypervirulent covRS mutant form provides an explanation for the comparatively less frequent isolation of non-M1 serotypes from invasive human infections.
Key Words: Animal models, Bacteriology, Immunity, Innate, Neutrophils, Streptococcus, Virulence factors, Invasive infection
Introduction
Group A Streptococcus (GAS) causes 700 million infections each year, resulting in over 500,000 deaths. Invasive GAS infections account for >600,000 disease episodes, incurring a death rate of approximately 25% [1]. Over the past 30 years, a resurgence of life-threatening invasive GAS pathologies has been documented, in parallel with the emergence of the globally disseminated M1T1 clone [2, 3]. The M1T1 clone remains the most frequently isolated M serotype from cases of invasive GAS infection and also from simple pharyngitis [4]. Nonetheless, other GAS serotypes cause invasive infections in Western populations, and GAS invasive disease in indigenous populations, such as Aboriginal communities of Northern Australia, is often associated with multiple M serotypes [5].
Historically, it is documented that hyperencapsulated GAS isolates are associated with invasive human infection and increased virulence in murine models [6, 7, 8, 9, 10]. Similarly, an inverse correlation has been described between SpeB production and disease severity in both human clinical disease and in murine models [11, 12]. In the GAS M1T1 clone, mutations in the control of virulence regulatory sensor kinase (covRS; alternatively designated csrRS) operon are selected for in vivo, and result in upregulation of capsule, loss of SpeB expression, increased disease severity in murine infection models and are more frequently associated with severe human invasive disease [10, 13, 14, 15].
The GAS M1T1 clone is distinguished from closely related M1 strains by the acquisition of the bacteriophage-encoded DNase Sda1 and superantigen SpeA [16, 17]. Human neutrophil-mediated killing of GAS selects for the neutrophil-resistant covRS mutant form of M1T1. The acquisition of the bacteriophage-encoded sda1 gene provided M1T1 with enhanced capacity to switch to the covRS mutant form [14], as Sda1 mediates escape from neutrophil extracellular traps [18, 19]. The loss of SpeB-mediated proteolytic degradation in vivo, as a result of covRS mutation, preserves expression of Sda1 and other virulence factors [20], allowing GAS to recruit and activate the broad-spectrum human protease plasmin on the bacterial surface, resulting in extensive tissue destruction and triggering systemic dissemination [11, 14].
While significant advances in the understanding of GAS M1T1 invasive disease initiation have been made, parameters governing invasive propensity of other M types have not been elucidated. Recently, analysis of GAS isolates of varying M type documented an association between invasive clinical isolates and mutations in genes encoding GAS global gene regulators (covRS/csrRS and ropB/rgg)[21]. In this study, we have examined a set of non-M1 serotype GAS isolates to determine whether such mutations trigger systemic dissemination in divergent M types. A model describing the invasive potential of differing M types is proposed.
Materials and Methods
GAS Strains and Culture Conditions
Clinical GAS isolates examined in this study have been described previously (table 1). Routine culture of GAS was conducted in stasis at 37°C in Todd-Hewitt broth supplemented with 1% (w/v) yeast extract or on horse-blood agar. GAS cultures for use in microarray experiments were propagated in Todd-Hewitt broth supplemented with 1.5% (w/v) yeast extract.
Table 1.
Characteristics of GAS isolates and mutant strains utilized
| Isolate | emm type | Isolate origin | Clinical origin | Reference or source |
|---|---|---|---|---|
| 5448 | 1.0 | USA | invasive; STSS/NF | [20] |
| 5448 AP | 1.0 | animal passage | generated during murine passage | [20] |
| NS13 | 53 | Australia | invasive; blood | [37] |
| NS88.2 | 98.1 | Australia | invasive; blood | [37] |
| NS179 | 9.1 | Australia | invasive; blood; pustules on foot | [37] |
| NS210 | 22 | Australia | invasive; diabetic ulcer with fever | [37] |
| NS223 | 91 | Australia | invasive; blood | [37] |
| NS452 | 25 | Australia | invasive; cellulitis; wound | [37] |
| NS455 | 52 | Australia | invasive; blood | [37] |
| NS501 | 14 | Australia | invasive; blood | [37] |
| A20 | 23 | Japan | invasive; blood | [41] |
| NS730 | 90 | Australia | invasive; nf; pus from left hip | [37] |
| NS733 | 90 | Australia | invasive; nf; wrist aspirate | [37] |
| NS931 | 69 | Australia | invasive; nf; blood | [37] |
| NS1133 | 101 | Australia | invasive; blood | [37] |
| ALAB49 | 53 | USA | superficial; impetigo; skin lesion | [42] |
| NS10 | 53 | Australia | superficial; throat swab | [37] |
| NS14 | 102 | Australia | superficial; post-operative wound | [37] |
| NS32 | 101 | Australia | superficial; wound infection | [37] |
| NS50.1 | 108 | Australia | superficial; wound infection | [37] |
| NS53 | 71 | Australia | superficial; fever | [37] |
| NS59 | 53 | Australia | superficial; wound infection | [37] |
| NS236 | 77 | Australia | superficial; sore throat; throat swab | [37] |
| NS253 | 52 | Australia | superficial; wound infection | [37] |
| NS265 | 56 | Australia | superficial; wound infection | [37] |
| NS297 | 44/61 | Australia | superficial; skin sore | [37] |
| NS474 | 58 | Australia | superficial; wound infection | [37] |
| NS488 | 12 | Australia | superficial; sinusitis; pharyngeal pus | [37] |
| NS836 | ck249 | Australia | superficial; wound infection | [37] |
| NS88.2rep | 98.1 | NA | sogenic covS repaired NS88.2 strain | this study |
| NS88.2covS | 98.1 | NA | reverse complemented NS88.2 covS mutant | this study |
Clinical origin classified as invasive if infected tissue is normally sterile in a healthy host. STSS = Streptococcal toxic shock-like syndrome; NF = necrotising fasciitis; NA = not applicable.
SpeB Activity Assays
SpeB cysteine protease activity in cell-free stationary-phase supernatants was determined using the chromogenic substrate N-benzoyl-Pro-Phe-Arg-p-nitroanilide-hydrochloride (Sigma), according to the method of Hytönen et al. [22]. To screen large numbers of GAS colonies recovered following subcutaneous murine passage (n = 1,500), single colonies were transferred to designated grid locations on Columbia agar plates supplemented with 15% (v/v) commercial skim milk (Devondale) and assayed for secreted SpeB activity as described by Ashbaugh et al. [23].
Western Blot Analysis
Stationary-phase supernatant proteins were concentrated 37.5-fold in 100 mM Tris (pH 7.6) by precipitation with 10% trichloroacetic acid. SpeB protein was then detected using Western blot analysis essentially as previously described [11].
Quantification of Hyaluronic Acid Capsule Biomass
Overnight cultures in Todd-Hewitt broth supplemented with 1% (w/v) yeast extract were subinoculated 1:14 and grown to mid-logarithmic phase (OD600 0.6). Capsule extraction and quantification were conducted using the method of Ashbaugh and Wessels [24].
DNA Sequence Analysis of the covRS Locus
The method for mapping mutations in the GAS covRS operon was as described previously [14]. Genomic DNA was isolated using the QIAGEN DNeasy Blood and Tissue Kit (Qiagen) according to the manufacturer's guidelines. For each operon, sequences were assembled in Chromas Pro v1.33 (Technelysium Pty Ltd) and aligned with the intact covRS operon of the M1T1 isolate 5448 (BioEdit v7.0.9.0; Ibis Biosciences).
Microarray Design and Production
An oligonucleotide microarray with probes representing M1 core ORFeome in addition to ORFs representing various M1, M3, M18 and Streptococcus dysgalactiae prophages was used in this study. The microarray was an expansion of one described previously [25]. Oligomers (70mers) were obtained from Dr. Kevin McIver and Dr. June Scott, and printed in the Molecular Resource Center, University of Tennessee Health Science Center by the use of MicroGrid II (Genomic Solutions). Additional 70mers, representing M1T1-specific prophages and prophage 3396 of S. dysgalactiae, were designed in batch according to the same design criteria applied for the other oligomers (Oligo Wiz 2.0, http://www.cbs.dtu.dk/services/OligoWiz) and obtained from Integrated DNA Technologies.
DNA-DNA Microarray
DNA-DNA microarray experiments were conducted in dye-flipped biological triplicates for each GAS isolate, and all steps involving Alexa Fluor®dyes were conducted in the dark. RNA-free genomic DNA was extracted from overnight liquid cultures by a modified phenol-chloroform procedure [17] and randomly sheared into <1-kb fragments using a Misonix 3000 cup-horn sonicator and Branson Sonifier®250. Sheared DNA samples were fluorescently labeled with the BioPrime®Total Genomic Labeling System (Invitrogen) as described by the manufacturer. In each hybridization reaction, equal amounts of Alexa Fluor® 3-labeled and Alexa Fluor® 5-labeled samples from different pairs of isolates were combined with hybridization buffer (Genisphere) and applied to the microarray slide. Following incubation at 55°C for 16 h, glass slides were washed, dried via centrifugation and scanned using a GenePix 4000B scanner (Axon Instruments Inc.).
DNA Microarray Data Analysis
The GenePixPro 4.0 software (Axon Instruments Inc.) was used for primary analysis of the scanned GenePix files. The fluorescent intensities were then normalized to the median intensity for each channel. Data from all probes representing the same gene were averaged, and a mean hybridization score was calculated for each gene. An average threshold of 40 median-normalized fluorescence units was selected, under which a gene was called ‘absent’.
Transcriptional Microarray
Overnight GAS cultures were sub-inoculated 1:10 into fresh prewarmed media and grown to mid-logarithmic phase (OD600 0.4). Bacteria were concentrated 20-fold in Buffer RLT (Qiagen) containing β-mercaptoethanol, lysed by mechanical disruption in Lysing matrix B tubes (Q-Biogene) with a FastPrep FP120 Homogenizer (Q-Biogene) and flash frozen for storage at −80°C. Bacterial RNA was extracted using the RNeasy Mini Kit (Qiagen), treated with TURBO DNA-free™DNase to remove contaminating genomic DNA (Ambion), re-concentrated on RNeasy columns (Qiagen) and converted to dendrimer-labeled cDNA with the Genisphere 3DNA Array 900MPX Kit as described by the corresponding manufacturer. Dendrimer-labeled cDNA samples from different pairs of isolates were combined and hybridized to the microarray slide for 16 h at 55°C. Slides were washed to remove unbound cDNA and labeled with dendrimer-targeted Alexa Fluor 546 and Alexa Fluor 647 dyes for 5 h at 55°C. Following a final wash to remove excess fluorescent dye, slides were coated in DyeSaver 2 (Genisphere) to preserve the fluorescent signal, and subsequently polished with toluene/acetone (3:1, v/v) to minimize background fluorescence immediately prior to scanning in a GenePix 4000B scanner (Axon Instruments Inc.). Scanned images were processed with GenePixPro 4.0 software (Axon Instruments Inc.), and all transcriptional and statistical analyses undertaken in silico using GeneSpring GX 10 (Agilent Technologies). All transcriptional microarray data were submitted to the NCBI Gene Expression Omnibus (GEO) according to the MIAME standards (GEO accession No. GSE23825).
Neutrophil Killing Assays
The capacity of GAS isolates to survive during co-incubation with human neutrophils in vitro was determined as described by Hollands et al. [25]. Briefly, 2 × 104 colony-forming units (CFU) of mid-logarithmic-phase bacteria were incubated with 2 × 105 neutrophils in RPMI with 2% heat-inactivated plasma for 30 min at 37°C. The final percent survival was calculated following comparison to the same bacterial culture incubated under the same conditions in the absence of neutrophils.
In vivo Phase-Switching
To examine the capacity for a phenotypic phase switch through covRS mutation in vivo, sublethal doses of SpeB-positive GAS isolates, in the order of 107 CFU per dose, were subcutaneously administered in sterile 0.7% (w/v) NaCl to the right flank of C57BL/J6 mice less than 8 months of age (10 animals per isolate). On the third day after infection, mice were sacrificed by CO2 asphyxiation and the infected cutaneous lesions surgically removed. In vivo passaged bacteria were recovered from murine lesions on horse-blood agar and single colonies assayed for SpeB status as outlined above.
Transgenic Murine Infection Model
Humanized plasminogen transgenic (Tg+)AlbPLG1 mice, heterozygous for the human plasminogen gene [26], served as the animal model for determining GAS invasive potential as previously described [14]. GAS isolates were grown to logarithmic phase (OD600 0.6), washed with sterile 0.7% (w/v) NaCl and appropriately diluted to prepare the inoculum. Final dose of viable bacteria was confirmed using a plate-based serial dilution cultured on horse-blood agar. For each GAS isolate, a cohort of 10 humanized mice were subcutaneously challenged in the right flank and mortality was documented over a 10-day period.
Restoration of covS in NS88.2
The covS gene from NS88.2 was amplified using primers pHYcovSF (5′-gggggatccatggaaaatcagaaacaaaaacag-3′) and pHYcovSR (5′-ggggaattcctaactctctttagactgggcc-3′). The resulting amplicon was cloned into the temperature-sensitive vector pHY304 using BamHI/EcoRI restriction-enzyme digestion and ligation with T4 DNA ligase. Site-directed mutagenesis of the adenine nucleotide at position 581 to guanine was performed according to the method of Sanderson-Smith et al. [27], using primers pHYcovsa581gF (5′-gccaaataactcaacaactagtagccaaaacagcagtcatgc-3′) and pHYcovsa581gR (5′-gcaagactgctgttttggctactagttgttgagttatttggc-3′). The resulting plasmids (pHYcovS and pHYrep) were transformed into Escherichia coli MC1061 using standard electroporation procedures. Allelic replacement mutants were constructed as described previously [28]. The covRS operons of the isogenic mutants NS88.2rep and NS88.2covS were sequenced as outlined above to confirm both the presence of the desired mutation and the integrity of the covRS operon.
GAS Surface Plasmin Activity Assays
GAS were incubated in human plasma as described previously [11]. Overnight growth in Todd-Hewitt broth supplemented with 1% (w/v) yeast extract was diluted to OD600 0.5 and co-incubated with human plasma at 37°C for 3 h. GAS were twice washed with PBS, 0.01% gelatin and 0.01 M EDTA, prior to resuspension in PBS and 0.01% gelatin. Plasmin activity was determined using the chromogenic substrate Spectrozyme PL (American Diagnostica).
Results
Expression of SpeB, Capsule Production and covRS Mutation of Non-M1 GAS
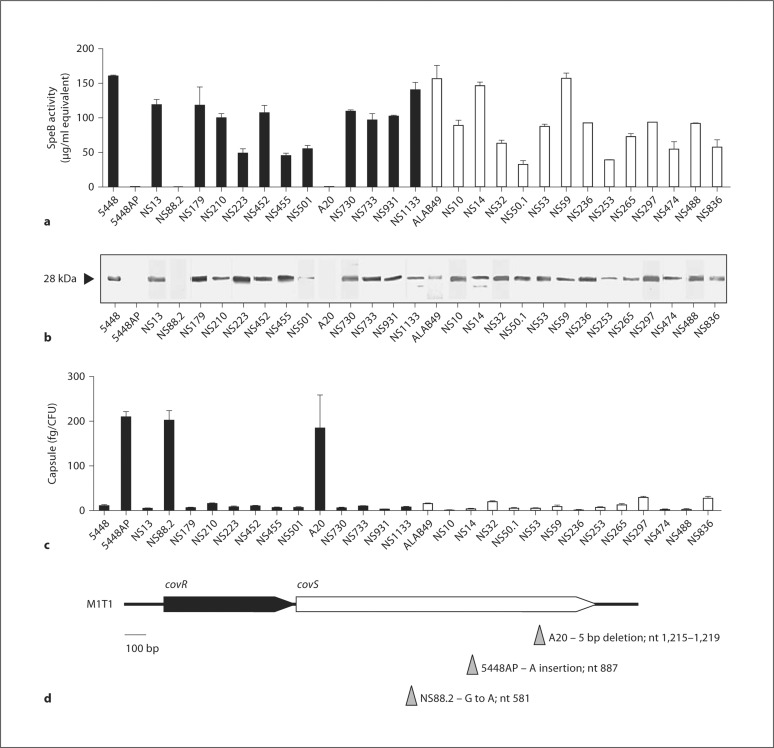
A range of GAS clinical isolates of differing emm types from invasive and benign infections were selected for this study (table 1). The well-characterized M1T1 clinical isolate 5448 and the natural isogenic covS mutant 5448AP [14] were included for comparison. With the exception of 3 strains, 5448AP, NS88.2 and A20, each of the GAS isolates described here expressed and secreted active SpeB at the stationary phase of growth(fig. 1a, b). An inverse correlation between SpeB expression and hyaluronic acid capsule production was observed, with 5448AP, NS88.2 and A20 hyperencapsulated with respect to all SpeB-positive GAS isolates (fig. 1c). GAS M1T1 strain 5448AP contains an adenine insertion at nucleotide position 887 in the ORF of the covS gene, resulting in the premature truncation of the translated CovS protein [14]. Correspondingly, DNA sequence analysis of the SpeB-negative hyperencapsulated strains NS88.2 and A20 revealed a guanine to adenine substitution at position 581 in the covS gene of NS88.2 and a 5-base pair deletion between positions 1,215 and 1,219 of the A20 covS gene (fig. 1d). Each mutation results in premature truncation of the translated CovS protein.
Fig. 1.
Molecular and phenotypic analyses of GAS isolates representing distinct M serotypes. a SpeB activity in cell-free stationary-phase supernatants of invasive (filled bars) and uncomplicated infection (open bars) GAS isolates. Isolate 5448 represents the globally disseminated M1T1 clone, while 5448AP is a hypervirulent animal passaged variant of 5448 [14]. b Western blot detection of SpeB in stationary-phase GAS supernatants. The 28-kDa mature SpeB protease is indicated with an arrowhead. c Hyaluronic acid capsule biomass of mid-logarithmic-phase invasive (filled bars) and uncomplicated infection (open bars) GAS isolates. d Schematic representation of the covRS operon. DNA sequence analysis confirmed the presence of inactivating covS mutations in the SpeB-deficient isolates A20, NS88.2 and 5448AP. The nature and nucleotide positions of the mutations in each isolate are indicated by the corresponding arrowheads.
DNA Microarray and Transcriptomic Analyses
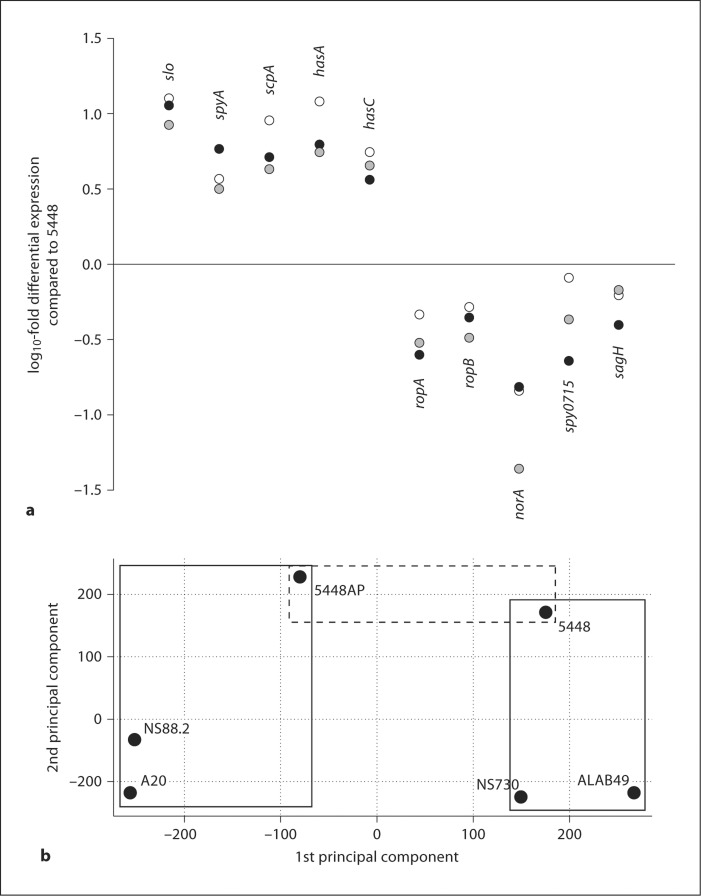
Three GAS strains with intact covRS (5448, ALAB49 and NS730) and 3 covS mutant forms (5448AP, NS88.2 and A20) were subjected to DNA microarray and transcriptomic analyses. DNA microarray identified 1,580 genes representing the core genome of the 6 GAS strains under examination (online suppl. table 1 and online suppl. fig. 1, www.karger.com/doi/10.1159/000317640). The bacteriophage-encoded sda1 gene, which confers on M1T1 GAS the capacity to switch to the covRS mutant form at high frequency [14], was not present in the genomes of the non-M1 GAS strains (online suppl. table 1 and online suppl. fig. 1). Only ubiquitous genes were included in subsequent transcriptomic analyses undertaken on GAS strains grown to mid-logarithmic phase. The virulence-related genes found to be strongly upregulated in the covS mutant strains, in comparison to the covRS intact strain 5448, include genes of the has operon (hasA, hasB and hasC; capsule biosynthesis), slo (streptolysin O) and the spyA exotoxin. Corroborating studies on M1T1 covS mutation [13], the positive regulators of SpeB activity and expression ropA and ropB [29, 30], in addition to genes of the sag operon (sagB, sagC and sagH), involved in streptolysin S production [31], were found to be downregulated in the covS mutant strains studied here (fig. 2a). No significant SpeB expression was detected at the mid-logarithmic phase of growth, in accordance with previous studies [32]. Principal component analysis of these transcriptomic data revealed 2 distinct clusters of covRS intact strains and covS mutant forms (fig. 2b). These data suggest that differing M types harboring covS mutations express related transcriptomic profiles.
Fig. 2.
In vitro mid-logarithmic phase transcriptional microarray analysis of covRS intact and covRS mutant non-M1 GAS. a log10-fold differential expression of virulence-associated and regulatory genes of the covS mutants 5448AP (filled circles), A20 (open circles) and NS88.2 (shaded circles) compared to the covRS intact M1T1 strain 5448. Selected genes are significantly differentially expressed in 5448AP with respect to 5448 (p < 0.05). b Principal component analysis on the non-M1 isolates ALAB49 (M53) and NS730 (M90), NS88.2 (M98.1) and A20 (M23), in addition to the M1T1 reference strains 5448 and 5448AP, revealed 2 distinct expression profiles in this 6-isolate strain set. Solid-line boxes placed in the plot area highlight strain clusters. The dashed-line box highlights the M1T1 strains 5448 and 5448AP, which, apart from a single base insertion in the 5448AP covS gene, harbor identical genomes.
Neutrophil Resistance, SpeB Switching and Virulence
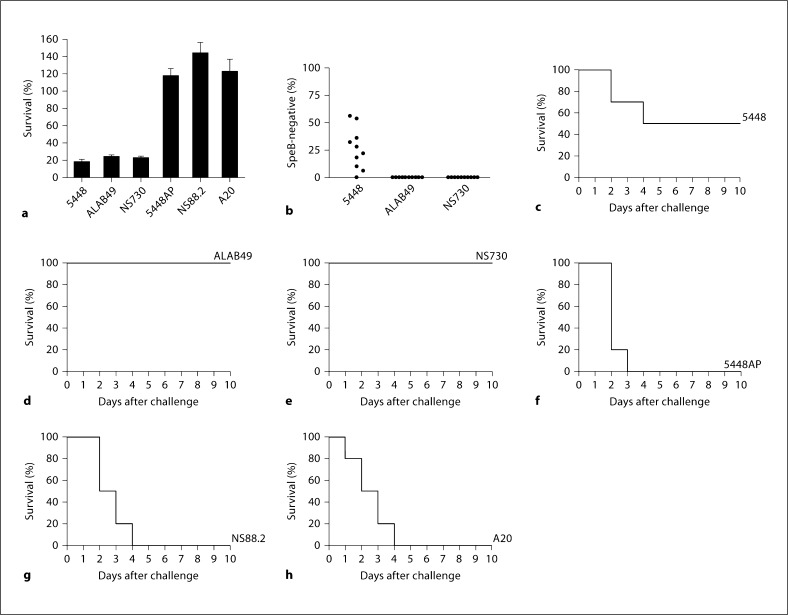
In comparison with the covRS intact strains 5448, NS730 and ALAB49, the hyperencapsulated covS mutant strains 5448AP, NS88.2 and A20 displayed enhanced resistance to human neutrophil killing (fig. 3a). Of the covRS intact strains, the M1T1 strain 5448 readily switched to the SpeB-negative covRS mutant form in vivo (fig. 3b), in accordance with previous studies [11, 14, 20]. However, the covRS intact non-M1 strains NS730 and ALAB49 only infrequently switch to a covRS mutant form in vivo (fig. 3b). Utilizing the humanized plasminogen transgenic mouse line AlbPLG1, we assessed the virulence of both covRS intact and mutant GAS strains. The M1T1 strain 5448, which has the capacity to switch at a high frequency to the covRS mutant form in vivo, was virulent in this mouse model (fig. 3c), corroborating previous work [11, 14]. The covRS intact strains NS730 and ALAB49, which infrequently switch to the covRS mutant form (fig. 3b), failed to establish a lethal infection (fig. 3d, e). Each of the covS mutant strains, 5448AP, NS88.2 and A20, were highly virulent (fig. 3f–h). These data suggest that while covRS mutation may occur only infrequently in non-M1 GAS, such covRS mutant forms are hypervirulent.
Fig. 3.
Characterization of the covRS intact isolates ALAB49 (M53) and NS730 (M90), and the covS mutant isolates NS88.2 (M98.1) and A20 (M23), in comparison with the M1T1 reference strains 5448 and 5448AP. a The percent survival of GAS isolates during co-culture with human neutrophils in vitro. b The capacity of SpeB-positive isolates to phase-switch to a SpeB-negative phenotype was assessed following a 3-day subcutaneous passage in C57BL/J6 mice. Each data point represents the percent of SpeBnegative covRS mutants recovered from the infection site of each mouse (n = 10 mice per strain). Subcutaneous infection of humanized plasminogen transgenic AlbPLG1 (n = 10) 5448 (3.9 × 107 CFU/dose) (c), ALAB49 (3.7 × 108 CFU/dose) (d), NS730 (2.2 × 108 CFU/dose) (e), 5448AP (5.1 × 107 CFU/dose) (f), NS88.2 (2.0 × 107 CFU/dose) (g) and A20 (1.2 × 108 CFU/dose) (h).
Repair of the covS Mutation in GAS Strain NS88.2 and Phenotypic Characterization
In order to investigate whether low-frequency switching of non-M1 GAS results in the hypervirulent covRS mutant form, the non-M1 GAS strain NS88.2 harboring a covS mutation was chosen. The adenine nucleotide point mutation at position 581 in the NS88.2 covS gene (fig. 1d) was converted to guanine by allelic replacement mutagenesis to construct strain NS88.2rep, with an intact or ‘repaired’ covS gene. Then, in order to fulfill Koch's molecular postulates [33], allelic replacement mutagenesis was undertaken on NS88.2rep to restore the original adenine nucleotide point mutation, resulting in strain NS88.2covS.
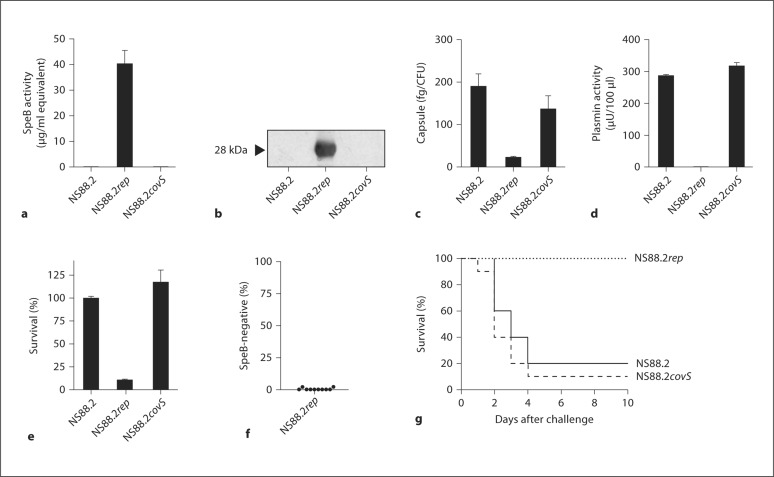
Repair of the covS mutation restored SpeB expression and activity in NS88.2rep, while NS88.2 and NS88.2covS remained SpeB-negative (fig. 4a, b). NS88.2 and NS88.2covS were hyperencapsulated in comparison to NS88.2rep (fig. 4c), and acquired substantial surface plasmin activity following incubation in human plasma (fig. 4d). Both NS88.2 and NS88.2covS also displayed enhanced resistance to killing by human neutrophils (fig. 4e). The covS intact NS88.2rep displayed only limited capacity to switch to the more virulent covRS mutant form (fig. 4f) and was not virulent in comparison to NS88.2 and NS88.2covS in AlbPLG1 mice (fig. 4g). These data support the hypothesis that non-M1 GAS serotypes switch less frequently to the hypervirulent covRS mutant form, providing an explanation for the comparatively less frequent isolation of non-M1 serotypes from invasive human infections.
Fig. 4.
Characterization of NS88.2 covS mutant (NS88.2 and NS88.2 covS) and intact (NS88.2 rep) isogenic strains. a SpeB activity in stationary-phase GAS supernatants. b Western blot detection of SpeB in stationary-phase supernatants. The mature SpeB protease is indicated with an arrowhead. c Hyaluronic acid capsule biomass of mid-logarithmic phase isogenic GAS strains. d Acquired surface plasmin activity following incubation in human plasma. e Percent survival following co-culture with human neutrophils in vitro. f The capacity of the SpeB-positive NS88.2 rep to phase-switch to a SpeB-negative phenotype following a 3-day subcutaneous passage in C57BL/J6 mice. g Subcutaneous infection of humanised plasminogen transgenic A lbPLG1 mice (n = 10 mice per strain) with NS88.2 (1.0 × 107 CFU/dose), NS88.2 rep (1.3 × 107 CFU/dose) and NS88.2 covS (9.8 × 106 CFU/dose).
Discussion
Severe group A streptococcal invasive disease progresses rapidly and results in high patient morbidity, with approximately one quarter of cases being fatal despite the susceptibility of the pathogen to antibiotic treatment [1, 34]. Host genetic factors [35] and the human fibrinolytic protease plasmin [26, 36] have both been documented as contributing to GAS invasive disease potential. Whilst progression of GAS disease from benign mucosal infections to invasive disease occurs infrequently, the M1T1 clone is clinically and epidemiologically associated with deep tissue infections in Western countries [4]. Mutations in the M1T1 covRS locus result in a hyperencapsulated, SpeB-negative, hypervirulent phenotype, and are correlated with invasive diagnosis in patients [10, 12, 13]. The capacity of M1T1 to switch to the covRS mutant form at high frequency may result from acquisition of the phage borne sda1 gene by M1T1 which confers resistance to neutrophil killing [14].
The contribution of similar mutations in covRS to invasive disease potential of non-M1 GAS remains largely unknown. Many differing M types have been associated with invasive infections [21, 34, 37, 38]. A naturally occurring mutation in the covR gene of an M3 isolate from a case of streptococcal toxic shock-like syndrome has been described. This covR mutation was associated with increased capsule expression and enhanced virulence [39]. Mutation in covR, associated with enhanced expression of the interleukin-8 cleaving protease SpyCEP, has also been observed in an M81 serotype GAS isolated from a lethal case of bacteremia and necrotizing fasciitis [40]. A correlation between mutation in global gene regulators (covRS and ropB) and invasive pathology has been documented in a range of M types. In comparison to mutations in ropB, inactivation of covS is clinically predominant and results in greater virulence in murine infection models [21].
In this study, a variety of non-M1 GAS strains from invasive disease episodes were examined, including isolates representing the emm sequence types14,22,23,25,52,53,69,90,91,98.1 and101. These included SpeB-positive isolates and SpeB-negative covS mutant forms. We propose that divergent M types with intact covRS possess an underlying capacity to cause invasive infection. In each serotype background, the covRS mutant form represents a more virulent state, which has greater propensity to cause invasive infection.
Using allelic replacement mutagenesis, we demonstrate that repair of the covS defect in the invasive emm98.1 GAS strain NS88.2 (covS mutant form) renders the isogenic covS intact strain NS88.2rep SpeB-positive, susceptible to neutrophil killing and less able to accumulate surface plasmin activity following growth in human plasma. The covS intact NS88.2rep strain was also found to be avirulent in the humanized plasminogen transgenic mouse model. In comparison to the M1T1 GAS covS intact strain 5448, we propose that the lack of virulence of NS88.2rep is due to the reduced capacity of this strain to switch to the invasive covRS mutant form in vivo. The lower frequency of switching to the covS mutant form limits the number of these invasive variants at the site of local infection. This lower frequency of switching may reflect the absence of the sda1 gene in this genetic background. The reduced capacity of non-M1 GAS serotypes to switch to the hypervirulent covRS mutant form may provide an explanation for the comparatively less frequent isolation of non-M1 serotypes from invasive human infections.
Supplementary Material
Acknowledgements
P.M. and A.H. are the recipients of Australian Postgraduate Awards. J.C. is the recipient of an Australian National Health and Medical Research Council Overseas Biomedical Training Fellowship (514639). This work was funded by NIH grant AI077780 (V.N., M.W.), National Health and Medical Research Council of Australia Grant 573401 (M.W.) and an Australian International Science Linkages grant CG110095 (M.W., M.K., V.N., A.H.).
References
- 1.Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis. 2005;5:685–694. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 2.Tart AH, Walker MJ, Musser JM. New understanding of the group A Streptococcus pathogenesis cycle. Trends Microbiol. 2007;15:318–325. doi: 10.1016/j.tim.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Musser JM, Kapur V, Szeto J, Pan X, Swanson DS, Martin DR. Genetic diversity and relationships among Streptococcuspyogenes strains expressing serotype M1 protein – recent intercontinental spread of a subclone causing episodes of invasive disease. Infect Immun. 1995;63:994–1003. doi: 10.1128/iai.63.3.994-1003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aziz RK, Kotb M. Rise and persistence of global M1T1 clone of Streptococcuspyogenes. Emerg Infect Dis. 2008;14:1511–1517. doi: 10.3201/eid1410.071660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hassell M, Fagan P, Carson P, Currie BJ. Streptococcal necrotising fasciitis from diverse strains of Streptococcus pyogenes in tropical northern Australia: case series and comparison with the literature. BMC Infect Dis. 2004;4:60. doi: 10.1186/1471-2334-4-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ravins M, Jaffe J, Hanski E, Shetzigovski I, Natanson-Yaron S, Moses AE. Characterization of a mouse-passaged, highly encapsulated variant of group A Streptococcus in in vitro and in vivo studies. J Infect Dis. 2000;182:1702–1711. doi: 10.1086/317635. [DOI] [PubMed] [Google Scholar]
- 7.Moses AE, Wessels MR, Zalcman K, Alberti S, Natanson-Yaron S, Menes T, Hanski E. Relative contributions of hyaluronic acid capsule and M protein to virulence in a mucoid strain of the group A Streptococcus. Infect Immun. 1997;65:64–71. doi: 10.1128/iai.65.1.64-71.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson DR, Stevens DL, Kaplan EL. Epidemiologic analysis of group A streptococcal serotypes associated with severe systemic infections, rheumatic fever, or uncomplicated pharyngitis. J Infect Dis. 1992;166:374–382. doi: 10.1093/infdis/166.2.374. [DOI] [PubMed] [Google Scholar]
- 9.Marcon MJ, Hribar MM, Hosier DM, Powell DA, Brady MT, Hamoudi AC, Kaplan EL. Occurrence of mucoid M-18 Streptococcuspyogenes in a central Ohio pediatric population. J Clin Microbiol. 1988;26:1539–1542. doi: 10.1128/jcm.26.8.1539-1542.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engleberg NC, Heath A, Miller A, Rivera C, DiRita VJ. Spontaneous mutations in the CsrRS two-component regulatory system of Streptococcus pyogenes result in enhanced virulence in a murine model of skin and soft tissue infection. J Infect Dis. 2001;183:1043–1054. doi: 10.1086/319291. [DOI] [PubMed] [Google Scholar]
- 11.Cole JN, McArthur JD, McKay FC, Sanderson-Smith ML, Cork AJ, Ranson M, Rohde M, Itzek A, Sun H, Ginsburg D, Kotb M, Nizet V, Chhatwal GS, Walker MJ. Trigger for group A streptococcal M1T1 invasive disease. FASEB J. 2006;20:1745–1747. doi: 10.1096/fj.06-5804fje. [DOI] [PubMed] [Google Scholar]
- 12.Kansal RG, McGreer A, Low DE, Norrby- Teglund A, Kotb M. Inverse relation between disease severity and expression of the streptococcal cysteine protease, SpeB, among clonal M1T1 isolates recovered from invasive group A streptococcal infection cases. Infect Immun. 2000;68:6362–6369. doi: 10.1128/iai.68.11.6362-6369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-wide analysis of group A streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2006;2:e5. doi: 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walker MJ, Hollands A, Sanderson-Smith M, Cole JN, Kirk JK, Henningham A, McArthur JD, Dinkla K, Aziz RK, Kansal RG, Simpson AJ, Buchanan JT, Chhatwal GS, Kotb M, Nizet V. DNase Sda1 provides selection pressure for a genetic and phenotypic switch promoting invasive group A streptococcal infection. Nat Med. 2007;13:981–985. doi: 10.1038/nm1612. [DOI] [PubMed] [Google Scholar]
- 15.Levin JC, Wessels MR. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A Streptococcus. Mol Microbiol. 1998;30:209–219. doi: 10.1046/j.1365-2958.1998.01057.x. [DOI] [PubMed] [Google Scholar]
- 16.Sumby P, Porcella SF, Madrigal AG, Barbian KD, Virtaneva K, Ricklefs SM, Sturdevant DE, Graham MR, Vuopio-Varkila J, Hoe NP, Musser JM. Evolutionary origin and emergence of a highly successful clone of serotype M1 group A Streptococcus involved multiple horizontal gene transfer events. J Infect Dis. 2005;192:771–782. doi: 10.1086/432514. [DOI] [PubMed] [Google Scholar]
- 17.Aziz RK, Edwards RA, Taylor WW, Low DE, McGreer A, Kotb M. Mosaic prophages with horizontally acquired genes account for the emergence and diversification of the globally disseminated M1T1 clone of Streptococcus pyogenes. J Bacteriol. 2005;187:3311–3318. doi: 10.1128/JB.187.10.3311-3318.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sumby P, Barbian KD, Gardner DJ, Whitney AR, Welty DM, Long RD, Bailey JR, Parnell MJ, Hoe NP, Adams GG, Deleo FR, Musser JM. Extracellular deoxyribonuclease made by group A Streptococcus assists pathogenesis by enhancing evasion of the innate immune response. Proc Natl Acad Sci USA. 2005;102:1679–1684. doi: 10.1073/pnas.0406641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buchanan JT, Simpson A, Aziz R, Liu G, Kristian S, Kotb M, Feramisco J, Nizet V. DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr Biol. 2006;16:396–400. doi: 10.1016/j.cub.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 20.Aziz RK, Pabst MJ, Jeng A, Kansal R, Low DE, Nizet V, Kotb M. Invasive M1T1 group A Streptococcus undergoes a phase-shift in vivo to prevent proteolytic degradation of multiple virulence factors by SpeB. Mol Microbiol. 2004;51:123–134. doi: 10.1046/j.1365-2958.2003.03797.x. [DOI] [PubMed] [Google Scholar]
- 21.Ikebe T, Ato M, Matsumura T, Hasegawa H, Sata T, Kobayashi K, Watanabe H. Highly frequent mutations in negative regulators of multiple virulence genes in group A streptococcal toxic shock syndrome isolates. PLoS Pathog. 2010;6:e1000832. doi: 10.1371/journal.ppat.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hytönen J, Haataja S, Gerlach D, Podbielski A, Finne J. The SpeB virulence factor of Streptococcuspyogenes, a multifunctional secreted and cell surface molecule with strepadhesin, laminin-binding and cysteine protease activity. Mol Microbiol. 2001;39:512–519. doi: 10.1046/j.1365-2958.2001.02269.x. [DOI] [PubMed] [Google Scholar]
- 23.Ashbaugh CD, Warren HB, Carey VJ, Wessels MR. Molecular analysis of the role of the group A streptococcal cysteine protease, hyaluronic acid capsule, and M protein in a murine model of human invasive soft-tissue infection. J Clin Invest. 1998;102:550–560. doi: 10.1172/JCI3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashbaugh CD, Wessels MR. Absence of a cysteine protease effect on bacterial virulence in two murine models of human invasive group A streptococcal infection. Infect Immun. 2001;69:6683–6688. doi: 10.1128/IAI.69.11.6683-6688.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hollands A, Aziz RK, Kansal RG, Kotb M, Nizet V, Walker MJ. A naturally occuring mutation in ropB suppresses SpeB expression and reduces M1T1 group A streptococcal systemic virulence. PLoS One. 2008;3:e4102. doi: 10.1371/journal.pone.0004102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun H, Ringdahl U, Homeister JW, Fay WP, Engleberg NC, Yang AY, Rozek LS, Wang X, Sjöbring U, Ginsburg D. Plasminogen is a critical host pathogenicity factor for group A streptococcal infection. Science. 2004;305:1283–1286. doi: 10.1126/science.1101245. [DOI] [PubMed] [Google Scholar]
- 27.Sanderson-Smith ML, Walker MJ, Ranson M. The maintenance of high affinity plasminogen binding by group A streptococcal plasminogen-binding M-like protein is mediated by arginine and histidine residues within the a1 and a2 repeat domains. J Biol Chem. 2006;281:25965–25971. doi: 10.1074/jbc.M603846200. [DOI] [PubMed] [Google Scholar]
- 28.Sanderson-Smith ML, Dinkla K, Cole JN, Cork AJ, Maamary PG, McArthur JD, Chhatwal GS, Walker MJ. M protein-mediated plasminogen binding is essential for the virulence of an invasive Streptococcus pyogenes isolate. FASEB J. 2008;22:2715–2722. doi: 10.1096/fj.07-105643. [DOI] [PubMed] [Google Scholar]
- 29.Lyon WR, Gibson CM, Caparon MG. A role for Trigger Factor and an Rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 1998;17:6263–6275. doi: 10.1093/emboj/17.21.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neely MN, Lyon WR, Runft DL, Caparon M. Role of RopB in growth phase expression of the SpeB cysteine protease of Streptococcus pyogenes. J Bacteriol. 2003;185:5166–5174. doi: 10.1128/JB.185.17.5166-5174.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nizet V, Beall B, Bast DJ, Datta V, Kilburn L, Low DE, De Azavedo JCS. Genetic locus for streptolysin S production by group A Streptococcus. Infect Immun. 2000;68:4245–4254. doi: 10.1128/iai.68.7.4245-4254.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Unnikrishnan M, Cohen J, Sriskandan S. Growth-phase-dependent expression of virulence factors in an M1T1 clinical isolate of Streptococcus pyogenes. Infect Immun. 1999;67:5495–5499. doi: 10.1128/iai.67.10.5495-5499.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Falkow S. Molecular Koch's postulates applied to microbial pathogenicity. Rev Infect Dis. 1988;10:S274–S276. doi: 10.1093/cid/10.supplement_2.s274. [DOI] [PubMed] [Google Scholar]
- 34.Cunningham MW. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kotb M, Norrby-Teglund A, McGeer A, El-Sherbini H, Dorak MT, Khurshid A, Green K, Peeples J, Wade J, Thomson G, Schwartz B, Low DE. An immunogenetic and molecular basis for differences in outcomes of invasive group A streptococcal infections. Nat Med. 2002;8:1398–1404. doi: 10.1038/nm1202-800. [DOI] [PubMed] [Google Scholar]
- 36.Walker MJ, McArthur JD, McKay F, Ranson M. Is plasminogen deployed as a Streptococcus pyogenes virulence factor? Trends Microbiol. 2005;13:308–313. doi: 10.1016/j.tim.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 37.McKay FC, McArthur JD, Sanderson-Smith ML, Gardam S, Currie BJ, Sriprakash KS, Fagan PK, Towers RJ, Batzloff MR, Chhatwal GS, Ranson M, Walker MJ. Plasminogen binding by group A streptococcal isolates from a region of hyperendemicity for streptococcal skin infection and a high incidence of invasive infection. Infect Immun. 2004;72:364–370. doi: 10.1128/IAI.72.1.364-370.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fiorentino TR, Beall B, Mshar P, Bessen DE. A genetic-based evaluation of the principal tissue reservoir for group A streptococci isolated from normally sterile sites. J Infect Dis. 1997;176:177–182. doi: 10.1086/514020. [DOI] [PubMed] [Google Scholar]
- 39.Miyoshi-Akiyama T, Ikebe T, Watanabe H, Uchiyama T, Kirikae T, Kawamura Y. Use of DNA arrays to identify a mutation in the negative regulator, csrR, responsible for the high virulence of a naturally occurring type M3 group A Streptococcus clinical isolate. J Infect Dis. 2006;193:1677–1684. doi: 10.1086/504263. [DOI] [PubMed] [Google Scholar]
- 40.Turner CE, Kurupati P, Jones MD, Edwards RJ, Sriskandan S. Emerging role of the interleukin-8 cleaving enzyme SpyCEP in clinical Streptococcus pyogenes infection. J Infect Dis. 2009;200:555–563. doi: 10.1086/603541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okamoto H, Shoin S, Minami M, Koshimur S, Shimizu R. Experimental anticancer studies 30. Factors influencing streptolysin S-forming ability of streptococci having anticancer activity. Jpn J Exp Med. 1966;36:161. [PubMed] [Google Scholar]
- 42.Svensson MD, Sjöbring U, Luo F, Bessen DE. Roles of the plasminogen activator streptokinase and the plasminogen-associated M protein in an experimental model for streptococcal impetigo. Microbiol. 2002;148:3933–3945. doi: 10.1099/00221287-148-12-3933. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.