Abstract
The nucleotide sequence of the DNA fragment containing the beta-glucosidase gene of Candida pelliculosa was determined. Analysis of the sequence revealed three open reading frames which could encode 65,825, and 412 amino acid residues. The presence of the second frame was found to be sufficient for the expression of the beta-glucosidase gene in a heterologous host Saccharomyces cerevisiae. Putative protein encoded by this gene had hydrophobic amino acids, resembling a signal peptide, at its N-terminal region and 19 potential glycosylation sites. Codon usage of Candida genes had the similar pattern shown in S.cerevisiae. Codon bias of the beta-glucosidase gene of Candida was relatively low, compared with that of the highly expressed genes of S. cerevisiae.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armentrout R. W., Brown R. D. Molecular cloning of genes for cellobiose utilization and their expression in Escherichia coli. Appl Environ Microbiol. 1981 Jun;41(6):1355–1362. doi: 10.1128/aem.41.6.1355-1362.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
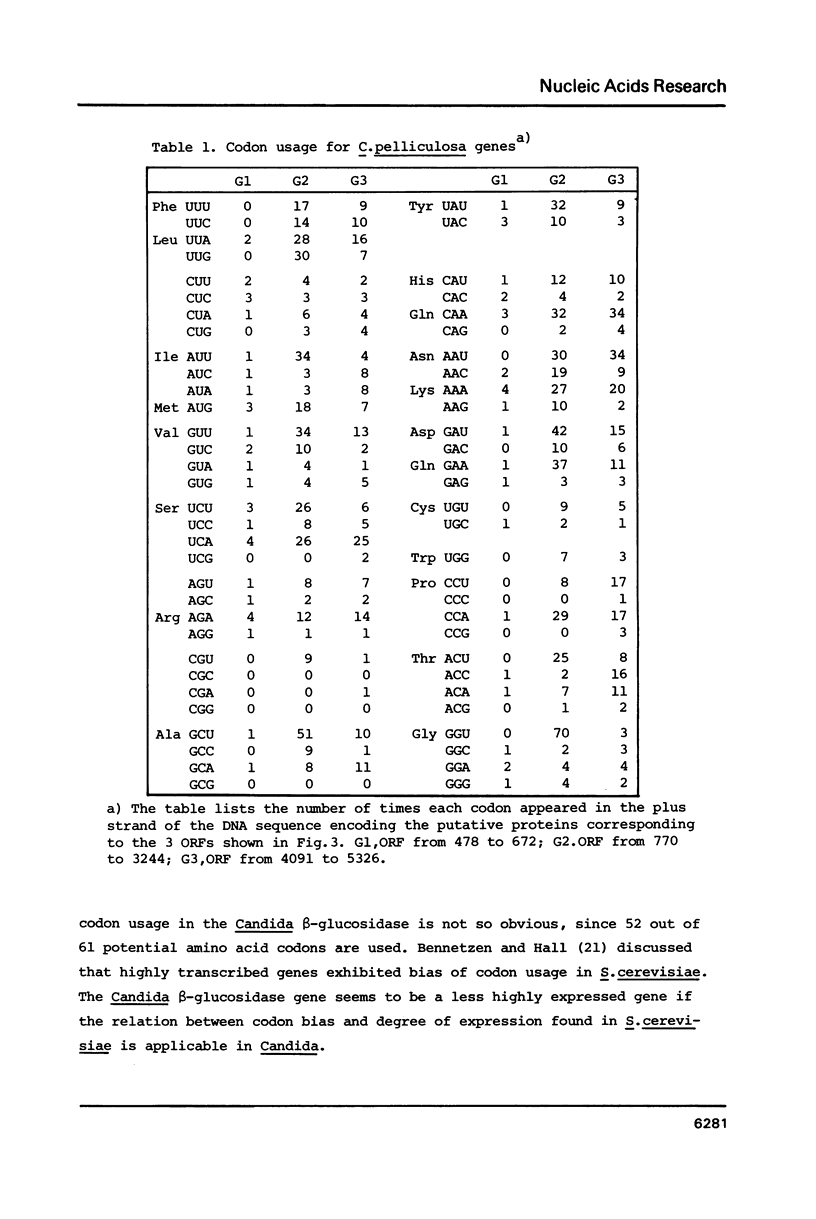
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Clarke L., Carbon J. Functional expression of cloned yeast DNA in Escherichia coli: specific complementation of argininosuccinate lyase (argH) mutations. J Mol Biol. 1978 Apr 25;120(4):517–532. doi: 10.1016/0022-2836(78)90351-0. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Kelly J. D., Cohen E. H. Transcription terminates in yeast distal to a control sequence. Cell. 1983 Jun;33(2):607–614. doi: 10.1016/0092-8674(83)90441-5. [DOI] [PubMed] [Google Scholar]
- Hubbard S. C., Ivatt R. J. Synthesis and processing of asparagine-linked oligosaccharides. Annu Rev Biochem. 1981;50:555–583. doi: 10.1146/annurev.bi.50.070181.003011. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J Mol Biol. 1982 Jul 15;158(4):573–597. doi: 10.1016/0022-2836(82)90250-9. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langford C. J., Klinz F. J., Donath C., Gallwitz D. Point mutations identify the conserved, intron-contained TACTAAC box as an essential splicing signal sequence in yeast. Cell. 1984 Mar;36(3):645–653. doi: 10.1016/0092-8674(84)90344-1. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Taussig R., Carlson M. Nucleotide sequence of the yeast SUC2 gene for invertase. Nucleic Acids Res. 1983 Mar 25;11(6):1943–1954. doi: 10.1093/nar/11.6.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh-e A., Tada S., Oshima Y. 2-micrometers DNA-like plasmids in the osmophilic haploid yeast Saccharomyces rouxii. J Bacteriol. 1982 Sep;151(3):1380–1390. doi: 10.1128/jb.151.3.1380-1390.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M. E. Compilation of published signal sequences. Nucleic Acids Res. 1984 Jul 11;12(13):5145–5164. doi: 10.1093/nar/12.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]



