Abstract
Purpose
The impact of epidermal growth factor receptor (EGFR) and KRAS genotypes on outcomes with erlotinib or gefitinib therapy continues to be debated. This study combines patient data from five trials in predominantly Western populations to assess the impact of EGFR and KRAS mutations on first-line therapy with an EGFR–tyrosine kinase inhibitor (TKI) and compare clinical versus molecular predictors of sensitivity.
Experimental Design
Chemotherapy-naïve patients with advanced non–small cell lung cancer and known EGFR mutation status treated with erlotinib or gefitinib monotherapy as part of a clinical trial were eligible for inclusion. Patients received daily erlotinib (150 mg) or gefitinib (250 mg) until disease progression or unacceptable toxicity. Data were collected in a password-protected web database. Clinical outcomes were analyzed to look for differences based on EGFR and KRAS genotypes, as well as clinical characteristics.
Results
Patients (223) from five clinical trials were included. Sensitizing EGFR mutations were associated with a 67% response rate, time to progression (TTP) of 11.8 months, and overall survival of 23.9 months. Exon 19 deletions were associated with longer median TTP and overall survival compared with L858R mutations. Wild-type EGFR was associated with poorer outcomes (response rate, 3%; TTP, 3.2 months) irrespective of KRAS status. No difference in outcome was seen between patients harboring KRAS transition versus transversion mutations. EGFR genotype was more effective than clinical characteristics at selecting appropriate patients for consideration of first-line therapy with an EGFR-TKI.
Conclusion
EGFR mutation status is associated with sensitivity to treatment with an EGFR-TKI in patients with advanced non–small cell lung cancer. Patients harboring sensitizing EGFR mutations should be considered for first-line erlotinib or gefitinib.
Tyrosine kinase inhibitors (TKI) of the epidermal growth factor receptor (EGFR) have become an important therapeutic option for patients with advanced non–small cell lung cancer worldwide. Considerable effort has been directed toward identification of clinical and molecular markers predictive of response, prolonged time to progression (TTP), and longer overall survival (OS) for patients treated with erlotinib and gefitinib. To date, the clinical variables identified include female sex, Asian ethnicity, adenocarcinoma histology, and never-smoking status (1–4). Efforts to identify a predictive biomarker have focused on the EGFR and have included detection of the receptor by immunohistochemical testing, assessment of DNA copy number, and detection of mutations in EGFR and KRAS (4–11). In clinical trials of first-line therapy with erlotinib or gefitinib across the globe, the most commonly studied and reported biomarker has been the presence or absence of EGFR mutations. Our study has focused on this biomarker to allow for pooling of multiple clinical trials and to enable future comparison of outcomes between Asian patients and those from the United States and Europe.
Mutations in EGFR and KRAS have emerged as promising biomarkers for response to EGFR-TKI therapy. Although two randomized trials comparing an EGFR-TKI with placebo failed to show a clear relationship between EGFR mutations and benefit to EGFR-TKI therapy in patients who had progressed after one or two prior regimens (12, 13), preliminary results of the more recent Iressa Pan-Asia Study show improved progression-free survival with first-line gefitinib rather than platinum-based chemotherapy in Asian patients harboring a sensitizing EGFR mutation (14). Given the debate, it is important to try to gain information from existing and ongoing trials, particularly in patients from the United States and Europe, to determine the clinical significance of EGFR genotype in first-line therapy decisions and to explore any ethnic variation in response to EGFR-TKI therapy. There are currently no published randomized trials of EGFR-TKIs versus combination chemotherapy in previously untreated patients from the United States and Europe. To explore the potential impact of EGFR and KRAS genotypes on clinical outcomes of chemotherapy-naïve Western patients with non–small cell lung cancer treated with an EGFR-TKI, we pooled data from smaller phase II trials to achieve a more powerful analysis. The study will provide potential insights into the applicability of the findings from the Iressa Pan-Asia Study for previously untreated patients with sensitizing mutations of the EGFR to our Western populations.
We established a web-based registry of clinical trials that use EGFR-TKIs in chemotherapy-naïve patients whose tumors were screened for mutations in EGFR and KRAS. By focusing on trials of first-line EGFR-TKIs, we aimed to eliminate potential modulating effects of prior chemotherapy. This collection of data from several trials enabled us to compare clinical outcomes associated with specific genetic changes (EGFR exon 19 deletions versus L858R; KRAS transition versus transversion mutations), as well as to assemble more information about less frequently described mutations or combinations of mutations. The study is also intended to compare the impact of clinical versus genomic characteristics in patients treated with an EGFR-TKI. Because only two of the clinical trials of first-line EGFR-TKIs included in our study routinely had collected information about EGFR fluorescence in situ hybridization status, the role of fluorescence in situ hybridization is not addressed in this analysis.
Patients and Methods
Trial and patient eligibility
Clinical trials were eligible for inclusion in this study if they involved prospective administration of gefitinib or erlotinib monotherapy in previously untreated patients with advanced non–small cell lung cancer. All trials were required to have routinely analyzed tumor specimens for EGFR mutations. Although KRAS analysis was not an eligibility requirement for this study, any available KRAS mutation information was included. Investigators from eligible prospective trials were contacted to determine their willingness to contribute individual patient and genotype data to this effort.
All patients had histologically or cytologically confirmed non–small cell lung cancer, stage IV or III-B with a malignant pleural effusion, and had provided written informed consent to treatment and data collection as part of their respective clinical trials. Institutional Review Board approval was obtained from the Dana-Farber/Harvard Cancer Center Institutional Review Board for this analysis. All data was de-identified to assure patient anonymity.
Patient treatment and data collection
Information on race, ethnicity, and smoking status was collected as part of each clinical trial. Patients were classified as never smokers if they had smoked <100 cigarettes and as former smokers if they had quit at least 1 y before enrollment. Two trials did not capture information about pack-years smoked nor differentiate between current versus former smokers (15, 16). Baseline performance status was determined by the treating physician at the time of initial patient enrollment.
Patients were treated with recommended doses of either erlotinib 150 mg daily or gefitinib 250 mg daily. Dose reductions and delays were permitted as per each trial protocol (7, 15–18). Patients were treated until development of progression or unacceptable toxicity. Patients underwent planned radiologic staging assessments at baseline and then every 6 to 8 wk while on therapy. In each of the included clinical trials, Response Evaluation Criteria in Solid Tumors criteria were used to determine radiologic response, stability, or progression. There was no central radiologic or pathologic review for the cases included in this study.
EGFR and KRAS mutation testing
Mutation detection was done in each of the five included trials with the use of well-established methods of direct DNA sequencing of exons 18 to 21 of the EGFR, and exons 1 and 2 of KRAS (19–22). In two of the trials (15, 17, 18), samples that were insufficient for direct sequencing or whose results on direct sequencing were indeterminate were then screened for mutations with the use of Surveyor DNA endonuclease combined with the WAVE HS high-performance liquid chromatography system as previously published (Transgenomic, Inc.; ref. 23). This platform was also used to verify the mutation status of patients in one of the trials in which primary testing had not been done at our center (15).
Statistical analysis
The response rate and disease control rate (percentage of response plus stable disease) were calculated. OS and TTP were measured from the start of TKI, and were analyzed by Kaplan-Meier estimates and log-rank testing. The proportional hazards model was used for multivariate analysis to assess the independent effects of different mutations and to obtain their hazard ratio estimates. Statistical calculations were done with the use of the SPSS statistical package (SPCC Inc.). All P-values were two-sided.
Clinical outcomes (response, TTP, and OS) were determined for all patients with known sensitizing mutations in the EGFR. Known sensitizing mutations included deletions in exon 19; point mutations L858R, L861Q, and G719X; and duplications in exon 19. In addition, specific subsets (exon 19 deletion versus L858R mutations, erlotinib-treated versus gefitinib-treated patients) were analyzed to look for potential differences in outcome based on genotype.
In a separate analysis, patients with complete information on potential clinical predictors (gender, race, smoking history, and histology) were divided into two clinical subsets: those with 3 or 4 clinical predictors, and those with ≤2 clinical predictors. Outcomes of therapy (response rate, TTP, OS) were compared between the two groups and by EGFR mutation status. The predictive abilities of EGFR mutation status and clinical predictors were assessed by the c-index, which is a rank correlation measure of concordance between the predicted probabilities or risks, and observed data. The c-index for response rate was computed by the logistic regression procedure in SAS 9.1 (SAS Institute Inc.), whereas the rcorr.cens function in the Hmisc package was used in R 2.6.2 for the analysis of TTP and OS.
Results
Patient characteristics
At the time of analysis, there were six eligible trials of first-line treatment with erlotinib or gefitinib monotherapy in predominantly Western populations in which EGFR mutations were systematically studied; our study includes five of these clinical trials (Table 1; refs. 7, 15, 17, 18, 24). EGFR mutation analysis has not yet been completed in the sixth trial, so that trial has not yet been incorporated into the database (25).
Table 1.
Trials of first-line EGFR-TKI therapy included in this study
| Trial/Author | Patient selection | Drug | 1st-line pts treated, n |
1st-line pts tested for EGFR, n |
1st-line pts tested for KRAS, n |
|---|---|---|---|---|---|
| Giaccone et al. (1) | Unselected | Erlotinib | 53 | 28 | 26 |
| Jackman et al. (2) | Patients ≥ age 70 y | Erlotinib | 80 | 43 | 41 |
| Miller et al. (3) | Adenocarcinoma with BAC features | Erlotinib | 75 | 57 | 56 |
| Sequist et al. (4) | Known EGFR mutations | Gefitinib | 31 | 31 | 0 |
| Jackman et al. (5) | Women, adenocarcinoma, not current smokers | Erlotinib | 78 | 63 | 52 |
Abbreviations: pts, patients; BAC, bronchioloalveolar carcinoma.
Between March 2003 and July 2008, a total of 317 chemotherapy-naïve patients were treated with erlotinib or gefitinib in one of these five trials. Four of the five trials have been published in peer-reviewed journals. The fifth has completed accrual, and has been presented at national and international meetings. Of these, tumor specimens from 223 patients had undergone successful mutation testing for EGFR and/or KRAS. The remaining patients have not undergone mutation testing due to inadequate or insufficient tissue available. Demographic and outcomes data were collected on those patients whose EGFR mutation status has been determined. Baseline patient characteristics are listed in Table 2. The majority of patients were Caucasian (95%) and female (69%); one third of patients had never smoked. Eighty-six percent of patients had tumors with adenocarcinoma histology, with 35% of these patients exhibiting bronchioloalveolar carcinoma features. This histologic distribution reflects, in part, the eligibility criteria for two of the trials: one required adenocarcinoma histology, whereas another required adenocarcinoma with bronchioloalveolar carcinoma features. Eighty-six percent received erlotinib, whereas 14% received gefitinib.
Table 2.
Baseline demographic and molecular characteristics of the entire study
| Total, N | 223 |
|---|---|
| Age, median (range) | 69 (26–91) |
| Sex, n (%) | |
| Male | 70 (31%) |
| Female | 153 (69%) |
| Race | |
| Caucasian | 212 (95%) |
| Asian | 8 (4%) |
| Black | 3 (1%) |
| Smoking status | |
| Never | 73 (33%) |
| Former/Current | 150 (67%) |
| Performance status | |
| 0 | 48 (22%) |
| 1 | 158 (71%) |
| 2 | 16 (7%) |
| Unreported | 1 (<1%) |
| Histology | |
| Adenocarcinoma (non-BAC) | 113 (51%) |
| Adenocarcinoma with predominant BAC features | 78 (35%) |
| Other | 32 (14%) |
NOTE: Percentages may not add up to 100 due to rounding.
The median potential follow-up was 55.9 months. At the time of analysis, 46 patients were known to be still alive, 143 have died, and another 34 patients were censored. Nine patients were known to be progression-free on continuing therapy with an EGFR inhibitor.
EGFR mutations
Within this study, tumors from 84 patients were found to harbor a sensitizing mutation in EGFR in the absence of any concomitant known resistance mutation. The majority of patients were women (81%), had adenocarcinoma histology (89%), and had no history of tobacco use (58%). Ninety percent of the EGFR-mutant patients in this study were Caucasian.
Of the 84 patients harboring a sensitizing EGFR mutation who were treated with either erlotinib or gefitinib, 56 patients (67%) achieved an objective response (1 complete response, 55 partial response). The disease control rate (responses plus stable disease) was 96%, with two patients not evaluable for response and one patient developing rapid disease progression. As previously described (16), the patient with unexpected progressive disease was a 42-year-old man with an exon 19 deletion and de novo MET amplification, which has been associated with resistance to gefitinib and erlotinib (26, 27).
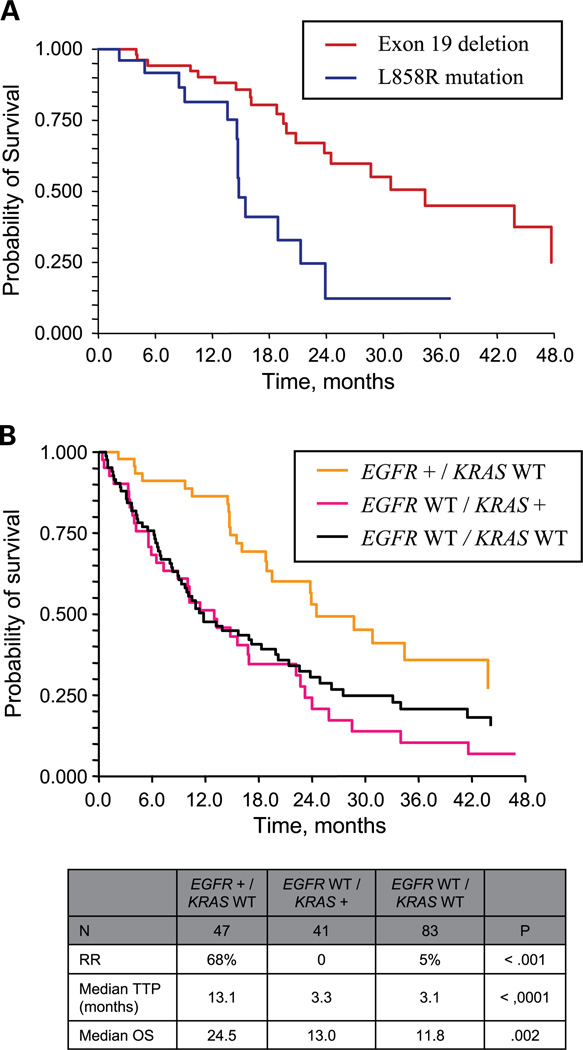
The 84 patients with sensitizing EGFR mutations had a median TTP of 11.8 months (95% confidence interval, 9.3–14.6 months), with a median OS of 23.9 months (95% confidence interval, 19.5–34.4 months). Patients with exon 19 deletions had a longer median TTP (14.6 versus 9.7 months; P = 0.02) and OS (30.8 versus 14.8 mo; P < 0.001) compared with those harboring the L858R mutation (Fig. 1A). Response rates were not significantly different between exon 19 deletions and L858R mutations (63% versus 50%; P = 0.39).
Fig. 1.
A, OS in patients with exon 19 deletions versus L858R point mutations. B, OS based on EGFR and KRAS status.
Outcomes of the 84 patients with known sensitizing mutations were also compared by drug-given erlotinib (n = 56) versus gefitinib (n = 28). There were no statistically significant differences in response rate (erlotinib, 70%; gefitinib, 60%; P = 0.47), median TTP (erlotinib, 13.0 months; gefitinib, 11.4 months; P = 0.49), and median OS (erlotinib, 28.7 months; gefitinib, 20.8 months; P = 0.10).
Role of clinical predictors
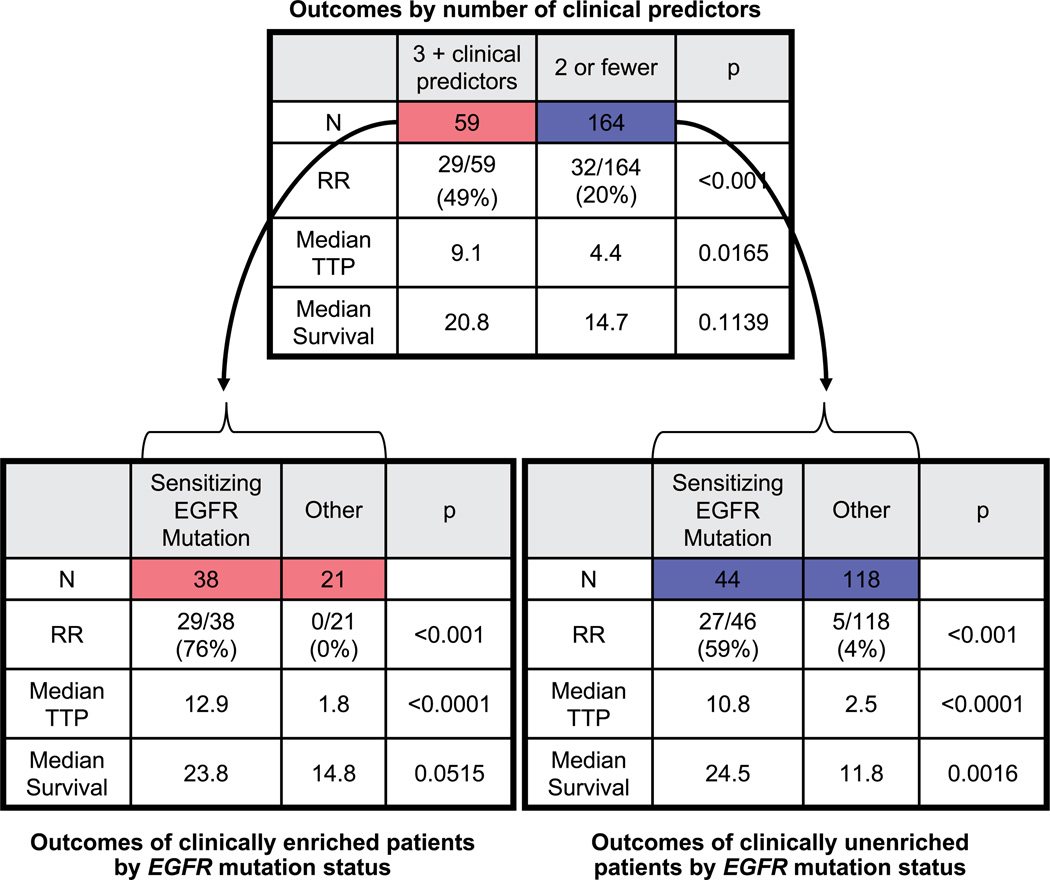
Patients were divided into two groups based on four major clinical predictors (race, gender, smoking status, and tumor histology): those with 3 or 4 clinical predictors versus those with ≤2 (Fig. 2). Such a clinical distinction did indeed help to select a subset of clinically enriched patients with an increased likelihood of response (49% versus 20%; P < 0.001) and prolonged median TTP (9.1 versus 4.4 months; P = 0.0165).
Fig. 2.
Comparison of outcomes by clinical enrichment and EGFR mutation analysis.
Although clinical predictors showed some value in selecting patients for first-line EGFR-TKI therapy, further analyses show EGFR mutation status was a better predictor of outcome. Within the high clinical prediction group (3 or 4 clinical predictors), EGFR mutation status was able to divide these 59 patients into two clear subsets: the 38 patients with sensitizing mutations (response rate, 76%; TTP, 12.9 months) and the 21 patients without a sensitizing mutation (response rate, 0; TTP, 1.8 months). Within the low clinical prediction group (0–2 clinical predictors), patients had a wide range of outcomes, and EGFR mutation status was again able to determine more clearly who would benefit: those with an EGFR mutation had improved response rate (59% versus 4%; P < 0.001), prolonged median TTP (10.8 versus 2.5 months; P < 0.0001), and longer median OS (24.5 versus 11.8 months; P = 0.002) compared with those with wild-type EGFR. When similar analyses were done with the use of stricter (patients with all four clinical predictors) or looser (patients with ≥2 clinical predictors) criteria, the impact of EGFR genotype was similar.
Logistic regression models of response resulted in a c-index of 0.87 for EGFR mutation status and 0.65 for clinical predictors. The predictive ability was not as strong for TTP in general, with a c-index of 0.68 for EGFR mutation status and 0.55 for clinical predictors.
For the OS outcome, the c-index was 0.61 for EGFR mutation status and 0.54 for clinical predictors. Overall, we conclude that EGFR mutation status yields consistently a higher predictive ability than the clinical predictors.
Impact of EGFR and KRAS mutations on clinical outcomes
There were 175 patients with complete information on both EGFR and KRAS genotypes. Four of these patients were excluded from analysis due to the presence of a known non-KRAS resistance mutation (one exon 20 insertion, one T790M), or due to concomitant EGFR and KRAS mutations (two patients). Outcomes for these patients are reported separately. The remaining 171 patients were categorized into three groups: (a) sensitizing EGFR mutation but KRAS wild-type, (b) EGFR wild-type but KRAS mutant, or (c) wild-type for both EGFR and KRAS (Fig. 1B).
Sensitizing EGFR mutations were associated with higher response rates, and longer TTP and OS when compared with patients without EGFR mutations. The two other patient groups—patients with KRAS mutations, and patients who were wild-type for both EGFR and KRAS—fared similarly: response rate was low (0%–5%), and median TTP was <4 months in each group. Median OS was ~1 year, similar to the estimated survival noted in other trials of patients with metastatic non–small cell lung cancer. These observations were confirmed by multivariate analysis to assess the independent effects of each mutation status on TTP and OS. Compared with patients with sensitizing EGFR mutations, the EGFR wild-type group had almost four times the risk of progression (hazard ratio, 3.8; P < 0.001) and twice the risk of death (hazard ratio, 2.0; P = 0.004). In contrast, KRAS mutation status was clearly not associated with TTP (hazard ratio, 1.0; P = 0.955) or OS (hazard ratio, 1.2; P = 0.406).
Further analyses of 39 patients harboring specific KRAS mutations revealed differences in the incidence of transition mutations (purine->purine or pyrimidine->pyrimidine) versus transversion mutations (purine->pyrimidine or pyrimidine->purine). KRAS mutations in never smokers were more likely to be transitions (7 of 9; 78%), whereas KRAS mutations in former or current smokers were more likely to be transversions (21 of 30; 70%; (Fisher's P = 0.019), as has been shown previously (28). Information about the specific type of KRAS mutation was unavailable in two patients.
Despite differences in patterns of incidence in KRAS mutations by smoking status, clinical outcomes were similar between transition or transversion muations: there were no objective responses in either group, and there were no significant differences in TTP (transition 3.3 months versus transversion 1.9 months; P = 0.24) or OS (transition 10.2 months, transversion 13.3 months; P = 0.30), although the analysis is limited by small sample size.
Patients with rarer or multiple mutations
The genotype and clinical outcomes of patients with rarer mutation or multiple mutations are listed in Table 3. Some of these include mutations known to be associated with sensitivity (L861Q) or resistance (exon 20 insertions) to EGFR-TKI. Other mutations or combinations of mutations, however, occur much less frequently. Of particular note is a patient with both an EGFR exon 19 deletion and a KRAS mutation; this patient did well with erlotinib therapy, achieving stable disease for 25.8 months.
Table 3.
Clinical outcomes for rare mutations or rare combinations of mutations
| Mutation(s) | N | Response | TTP, mo | OS, mo |
|---|---|---|---|---|
| Suspected sensitizing mutation | ||||
| L861Q | 1 | SD | 4 | 8.8 |
| G719A | 1 | SD | 3.9 | 5.2 |
| Exon 19 duplication | 1 | PR | 22.1+ | 22.1+ |
| Suspected resistance mutation | ||||
| Exon 20 insertion | 31 | 1 SD, 2 PD | 1.0 (median) | 1.5 (median) |
| Multiple mutations | ||||
| Exon 19 del + L861Q + G873E | 1 | SD | 20.5 | 34.9+ |
| L858R + T790M | 1 | PD | 1.9 | 17.5 |
| T790M + KRAS mutation | 1 | PD | 0.8 | 0.8 |
| Exon 19 del + KRAS mutation | 1 | SD | 25.8 | 44.7+ |
| V802I + KRAS mutation | 1 | PD | 0.9 | 4.0 |
| G719A + G779C | 1 | PR | 2.9+ | 2.9+ |
Abbreviations: SD, stable disease; PR, partial response; PD, progressive disease.
Discussion
Preliminary results from the Iressa Pan-Asia Study provide evidence that EGFR sensitizing mutations are predictive of longer progression-free survival when treated with gefitinib compared with chemotherapy, potentially sparing a subset of patients from initial therapy with more toxic agents (14). Although our study is not randomized, it does confirm the impressive outcomes with an EGFR-TKI in Western patients with sensitizing EGFR mutations compared with EGFR wild-type patients receiving an EGFR-TKI. Moreover, our data suggest that patients who are EGFR wild-type should be considered for combination chemotherapy rather than an EGFR-TKI in the first-line setting.
Furthermore, in the Western population studied in this trial, specific EGFR genotype has an impact. Similar to earlier observations from smaller datasets, exon 19 deletions are associated with longer TTP and OS when compared with L858R point mutations. A waterfall plot of changes in tumor measurements of indicator lesions from baseline to the time of best response provides a visual depiction of the impact of EGFR and KRAS genotypes on sensitivity to EGFR-TKI therapy (Fig. 3).
Fig. 3.
Maximal reduction for indicator lesions (Response Evaluation Criteria in Solid Tumors classification) by EGFR and KRAS genotype.
There are several observations from KRAS analysis. The genotype of KRAS mutations was different in smokers versus nonsmokers, as has been previously observed (28). However, this difference between transition and transversion mutations did not have an impact on the clinical outcome of EGFR-TKI therapy. Secondly, KRAS genotype in general did not have a significant independent impact on outcomes of treatment with an EGFR-TKI: patients with KRAS mutations were almost uniformly wild-type for EGFR, and the EGFR wild-type group had low response rate and short TTP across the board, irrespective of KRAS mutation status.
This study also shows the potential value of using genomic assessments of EGFR rather than using clinical characteristics to select patients for initial treatment with EGFR-TKIs. Whereas the use of clinical characteristics can be helpful in settings in which EGFR mutation testing is neither feasible nor available, this study provides evidence that genomic testing is more accurate in selecting a group of patients with increased chance of sensitivity to therapy with an EGFR-TKI. Based on the data presented here, it is appropriate to consider patients with known sensitizing mutations in EGFR for first-line therapy with an EGFR-TKI.
Online databases, such as our own, can help to expedite the collection and analysis of patients across multiple centers to allow for more powerful analysis of clinical outcomes associated with specific genotypes. In addition to confirming results that had been seen in smaller series, we look forward to planned collaborations to study differences in the incidence of various genotypic changes and clinical outcomes of therapy with EGFR-TKIs.
Translational Relevance.
The impact of epidermal growth factor receptor (EGFR) and KRAS mutations on clinical outcomes to EGFR–tyrosine kinase inhibitor (TKI) therapy in non–small cell lung cancer remains an area of investigation and debate. Although preliminary data from randomized trials of first-line therapy in Asia have suggested a benefit for gefitinib therapy over cytotoxic chemotherapy in patients with known EGFR mutations, no such randomized data yet exist for predominantly Western populations. We have created a database of five clinical trials from the United States and Europe, in which chemotherapy-naïve patients with advanced non–small cell lung cancer were treated with an EGFR-TKI, and EGFR status was tested. Based on the findings presented here, clinicians can derive a stronger understanding of the importance of EGFR mutation testing rather than clinical characteristics in selecting appropriate patients for first-line EGFR-TKI therapy; moreover, they can get a clearer estimation of the impact of specific changes in EGFR and KRAS on patient outcomes.
Acknowledgments
Grant support: The American Society of Clinical Oncology (ASCO) Cancer Foundation Translational Research Professorship, NIH grant 5RO1 CA 114465-05, the International Association for the Study of Lung Cancer Fellowship, and the Alice and Stephen D. Cutler Investigator Fund in Thoracic Oncology at Dana-Farber Cancer Institute. Any opinions, findings, and conclusions expressed in this material are those of the authors and do not necessarily reflect those of the ASCO or the ASCO Cancer Foundation.
Footnotes
Disclosure of Potential Conflicts of Interest
D.M. Jackman, consultant, Genentech; honoraria, Roche. V.A. Miller, consultant, Genentech. P.A. Janne, consultant, AVEO Pharmaceuticals, Boehringer Ingelheim, Roche; research funding, Pfizer, Genentech; patent holder, Genzyme. G.J. Riely, consultant, Astra Zeneca, Roche. M.I. Gallegos Ruiz, employment, Roche. B.E. Johnson, consultant, patent holder, Genzyme. The other authors report no conflicts of interest.
References
- 1.Fukuoka M, Yano S, Giaccone G, et al. Multiinstitutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer (The IDEAL 1 trial) [corrected] J Clin Oncol. 2003;21:2237–2246. doi: 10.1200/JCO.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 2.Kris MG, Natale RB, Herbst RS, et al. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: a randomized trial. JAMA. 2003;290:2149–2158. doi: 10.1001/jama.290.16.2149. [DOI] [PubMed] [Google Scholar]
- 3.Perez-Soler R, Chachoua A, Hammond LA, et al. Determinants of tumor response and survival with erlotinib in patients with non-small-cell lung cancer. J Clin Oncol. 2004;22:3238–3247. doi: 10.1200/JCO.2004.11.057. [DOI] [PubMed] [Google Scholar]
- 4.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 5.Cappuzzo F, Hirsch FR, Rossi E, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005;97:643–655. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- 6.Hirsch FR, Varella-Garcia M, McCoy J, et al. Increased epidermal growth factor receptor gene copy number detected by fluorescence in situ hybridization associates with increased sensitivity to gefitinib in patients with bronchioloalveolar carcinoma subtypes: a Southwest Oncology Group study. J Clin Oncol. 2005;23:6838–6845. doi: 10.1200/JCO.2005.01.2823. [DOI] [PubMed] [Google Scholar]
- 7.Miller VA, Riely GJ, Zakowski MF, et al. Molecular characteristics of bronchioloalveolar carcinoma and adenocarcinoma, bronchioloalveolar carcinoma subtype, predict response to erlotinib. J Clin Oncol. 2008;26:1472–1478. doi: 10.1200/JCO.2007.13.0062. [DOI] [PubMed] [Google Scholar]
- 8.Mitsudomi T, Kosaka T, Endoh H, et al. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with postoperative recurrence. J Clin Oncol. 2005;23:2513–2520. doi: 10.1200/JCO.2005.00.992. [DOI] [PubMed] [Google Scholar]
- 9.Niho S, Kubota K, Goto K, et al. First-line single agent treatment with gefitinib in patients with advanced non-small-cell lung cancer: a phase II study. J Clin Oncol. 2006;24:64–69. doi: 10.1200/JCO.2005.02.5825. [DOI] [PubMed] [Google Scholar]
- 10.Taron M, Ichinose Y, Rosell R, et al. Activating mutations in the tyrosine kinase domain of the epidermal growth factor receptor are associated with improved survival in gefitinib-treated chemorefractory lung adenocarcinomas. Clin Cancer Res. 2005;11:5878–5885. doi: 10.1158/1078-0432.CCR-04-2618. [DOI] [PubMed] [Google Scholar]
- 11.Inoue A, Suzuki T, Fukuhara T, et al. Prospective phase II study of gefitinib for chemotherapy-naive patients with advanced non-small-cell lung cancer with epidermal growth factor receptor gene mutations. J Clin Oncol. 2006;24:3340–3346. doi: 10.1200/JCO.2005.05.4692. [DOI] [PubMed] [Google Scholar]
- 12.Thatcher N, Chang A, Purvish P, et al. Gefitinib plus best supportive care in previously treated patients with refractory advanced non-small-cell lung cancer: results from a randomised, placebo-controlled, multicentre study (Iressa Survival Evaluation in Lung Cancer) Lancet. 2005;366:1527–1537. doi: 10.1016/S0140-6736(05)67625-8. [DOI] [PubMed] [Google Scholar]
- 13.Tsao MS, Sakurada A, Cutz JC, et al. Erlotinib in lung cancer - molecular and clinical predictors of outcome. N Engl J Med. 2005;353:133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 14.Mok T, Wu YL, Thongprasert S, et al. Phase III, randomised, open-label, first-line study of gefitinib (G) vs carboplatin/paclitaxel (C/P) in clinically selected patients (pts) with advanced non-small cell lung cancer (NSCLC) (IPASS) Stockholm, Sweden: 2008. [Google Scholar]
- 15.Giaccone G, Gallegos Ruiz M, Le Chevalier T, et al. Erlotinib for frontline treatment of advanced non-small cell lung cancer: a phase II study. Clin Cancer Res. 2006;12:6049–6055. doi: 10.1158/1078-0432.CCR-06-0260. [DOI] [PubMed] [Google Scholar]
- 16.Sequist LV, Martins RG, Spigel D, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26:2442–2449. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 17.Jackman DM, Lindeman N, Lucca J, et al. Phase II study of erlotinib in chemo-naive women with advanced pulmonary adenocarcinoma. J Clin Oncol. 2007;25 abstract 7591. [Google Scholar]
- 18.Jackman DM, Yeap BY, Lindeman NI, et al. Phase II clinical trial of chemotherapy-naive patients > or = 70 years of age treated with erlotinib for advanced non-small-cell lung cancer. J Clin Oncol. 2007;25:760–766. doi: 10.1200/JCO.2006.07.5754. [DOI] [PubMed] [Google Scholar]
- 19.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 20.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pao W, Wang TY, Riely GJ, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 23.Janne PA, Borras AM, Kuang Y, et al. A rapid and sensitive enzymatic method for epidermal growth factor receptor mutation screening. Clin Cancer Res. 2006;12:751–758. doi: 10.1158/1078-0432.CCR-05-2047. [DOI] [PubMed] [Google Scholar]
- 24.Sequist LV, Martins RG, Spigel DR, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. JCO. 2008;26:2442–2449. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 25.West HL, Franklin WA, McCoy J, et al. Gefitinib therapy in advanced bronchioloalveolar carcinoma: Southwest Oncology Group study S0126. JCO. 2006;24:1807–1813. doi: 10.1200/JCO.2005.04.9890. [DOI] [PubMed] [Google Scholar]
- 26.Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. PNAS. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 28.Riely GJ, Kris MG, Rosenbaum D, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res. 2008;14:5731–5734. doi: 10.1158/1078-0432.CCR-08-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]