Abstract
Protein synthesis across kingdoms involves the assembly of 70S (prokaryotes) or 80S (eukaryotes) ribosomes on the mRNAs to be translated. 70S ribosomes are protected from degradation in bacteria during stationary growth or stress conditions by forming dimers that migrate in polysome profiles as 100S complexes. Formation of ribosome dimers in Escherichia coli is mediated by proteins, namely the ribosome modulation factor (RMF), which is induced in the stationary phase of cell growth. It is reported here a similar ribosomal complex of 110S in eukaryotic cells, which forms during nutrient starvation. The dynamic nature of the 110S ribosomal complex (mammalian equivalent of the bacterial 100S) was supported by the rapid conversion into polysomes upon nutrient-refeeding via a mechanism sensitive to inhibitors of translation initiation. Several experiments were used to show that the 110S complex is a dimer of nontranslating ribosomes. Cryo-electron microscopy visualization of the 110S complex revealed that two 80S ribosomes are connected by a flexible, albeit localized, interaction. We conclude that, similarly to bacteria, rat cells contain stress-induced ribosomal dimers. The identification of ribosomal dimers in rat cells will bring new insights in our thinking of the ribosome structure and its function during the cellular response to stress conditions.
Key words: ribosome, translation, stress, starvation, polysome
Introduction
The translation of mRNAs into polypeptides in all living species is conducted by highly specialized RNA protein particles called ribosomes. Eukaryotic 80S ribosomes are composed of two subunits. The small subunit has a sedimentation value of 40S, whereas that of the large subunit is 60S. Similarly, prokaryotic 70S ribosomes are composed of 30S and 50S subunits. The structures of the prokaryotic ribosomal subunits and the 70S ribosome have been studied at atomic resolution using crystallographic approaches.1 Recently, X-ray structures of 40S from Tetrahymena thermophila and 80S ribosome from Saccharomyces cerevisiae have emerged.2,3 In addition, cryo-electron microscopy (cryo-EM) studies of both the 70S and 80S ribosomes have provided snapshots of the ribosomes bound with initiation, elongation, recycling and termination factors.4–7 Together, those structural studies have contributed immensely to the understanding of protein synthesis in bacteria and eukaryotes. It is now well accepted that the ribosome structure is conserved more than any other macromolecule or organelle among living organisms.8
The mechanism of protein synthesis can be described as having four distinct phases: (1) initiation, the small ribosomal subunit binds to the mRNA and upon selection of the initiator AUG codon, the large ribosomal subunit is recruited to form the translation competent 80S (70S in prokaryotes) ribosome; (2) elongation, decoding of protein sequence, delivery of amino acids by aminoacyl-tRNAs and incorporation into the growing polypeptide chain by formation of peptide bonds (3) termination, release of the polypeptide upon recognition of a stop codon on the mRNA and (4) recycling of the ribosomes into free subunits that continue translation of other mRNAs.8
An experimental approach, which is widely used to study structure/function of ribosomes and also monitor the efficiency of mRNA translation, is the use of sucrose gradients to separate free ribosomal subunits from translating ribosomes by velocity sedimentation.9 Because ribosomal subunits are larger than free mRNA protein complexes, this methodology can also separate the two populations from cellular extracts. In addition, translating mRNAs, which are bound to several ribosomes, forming polyribosomes, migrate with the much larger fractions in sucrose gradients. The sedimentation velocity of polyribosomes will depend on their size, which relates to the efficiency of translation of the corresponding mRNAs. The larger the corresponding mRNA and the number of bound ribosomes, the higher the sedimentation velocity. Sedimentation of particles is monitored by measuring the absorbance of the fractions at 254 nm, which provides the so-called polysome profile. A typical polysome profile shows distinct peaks for the free ribosomal subunits (small 40S in eukaryotes/30S in prokaryotes) and large (60S/50S), followed by the 80S (70S in prokaryotes) ribosomes and heavier polyribosomes. The sedimentation of polyribosomes for a single mRNA depends on the number of ribosomes bound, such as disomes, trisomes, etc. The development of the polysome profile techniques, along with other molecular biology approaches, allowed studies on the factors involved in ribosomal subunit joining and dissociation.10
Certain conditions are known to inhibit translational initiation (stress, environmental factors, exposure to drugs, etc.). The result is an increased accumulation of free ribosomal subunits and nontranslating monosomes and a decrease in the polyribosome pool.11–13 Sustained association of mRNAs with polyribosomes under these conditions is associated with efficient translation of the corresponding mRNAs.12,14 The molecular mechanisms that control the levels of free ribosomes under these conditions are not well known. However, it has been suggested that ribophagy (degradation of ribosomes) is induced in yeast under conditions of nutrient starvation.15
In Escherichia coli, it was shown that during the transition from exponential to stationary growth phase, 70S ribosome dimers are formed, which were detected on sucrose gradient fractionations as an 100S peak.16 This resting state of the ribosomes was termed “ribosomal hibernation.”17 Ribosome dimerization was promoted by a small basic protein RMF (ribosome modulation factor), which increased in levels when cells entered the stationary growth phase.18,19 RMF was shown to bind close to the peptidyl transferase center, rendering the ribosome translationally inactive.16,20 70S ribosomal-dimer formation was also supported by protein HPF (hibernation promoting factor, also known as YhbH), which was induced during the stationary growth phase.21 Ribosomes from the stationary phase had lower affinity to initiation factor 3 (IF3), promoting dissociation of ribosomes into translationally competent subunits.22 In contrast to the RMF and HPF proteins, which were found to bind exclusively the 100S ribosomes, protein YfiA, an inhibitor of translational activity of ribosomes, was also induced during stationary growth phase23 and was detected with both 70S and 100S ribosomes.24 YfiA was shown to bind to the subunit interface of the 70S, thus stabilizing the 30S and 50S interaction.25,26
The dynamic nature of ribosomal dimer formation in bacteria was further supported by the observation of the quick dissociation of the 100S dimers and their conversion into translationally competent ribosomal particles upon return to exponential growth conditions.24,27 Experimental approaches using crosslinking and electron microscopy techniques demonstrated that the interaction between the 70S ribosomes in the dimers was via the small ribosomal subunits.17 Recently, these dimers were studied by cryo-electron microscopy (cryo-EM) via the use of “in gradient” chemical crossliniking (GraFix), which increased the stability of ribosomal dimers. It was claimed that some degree of flexibility was present between the small ribosomal subunits within the 100S dimers.28 Moreover, in situ cryo-EM studies of intact E. coli cells proved that the 100S ribosomes do exist in vivo.29
Dimerization of 70S ribosomes has been suggested as a universal survival mechanism in bacteria during the stationary growth phase.18,30 RMF and HPF homologs were found in the proteobacteria γ group (Gram-negative), while other bacteria contain proteins homologous to HPF but not RMF.31 HPF was sufficient to promote formation of ribosomal dimers in Staphylococcus aureus cells during the transition to stationary growth phase.32
Hibernating/resting ribosomes in eukaryotic cells have not been described. In our work with rat C6 glioma cells, we observed the formation of a peak similar to 100S when cells were challenged with amino acid depletion. Similar to the bacterial model, this complex was unstable, and its formation was reversed after restoration of optimal growth conditions. This led us to investigate the possibility of the existence of hibernating ribosomes in mammalian cells. We found that the rat 110S ribosomal population consists of 80S dimers and 60S:80S heterodimers and was induced both by, different stresses that cause translation initiation inhibition and independently of stress in cell-free extracts after 80S ribosome run-off. Here, we provide a detailed analysis of the mammalian 80S dimers. As opposed to the stalled 70S dimers, our work suggests that the 110S ribosomal dimers formed under stress conditions are bound together via interactions mediated by the 60S subunits. Cryo-EM analysis of the 110S pool indicates 80S-80S dimers via a flexible connection.
The presence of dimers similar to those in this report, were found earlier in polysome profiles from rat and hamster cells.33–35 To address the question of the species specificity of ribosomal dimer formation, we tested several commonly used rat, mouse and human cell lines. After amino acid starvation in the same experimental conditions, only rat cell lines were able to form ribosomal dimers. Our data point to fundamental differences between mammalian ribosomes and raise awareness for the use of rat cells for studies on translational control mechanisms related to human diseases.
Results
Stress induces a novel ribonucleoprotein complex in C6 cells.
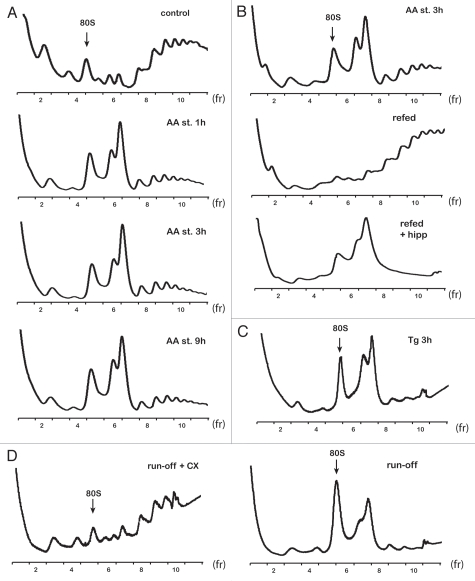
Amino acid starvation activates the protein kinase GCN2, which phosphorylates the α subunit of the translation initiation factor eIF2 (eIF2α), with a subsequent decrease of global protein synthesis.36 Phosphorylation of eIF2α initiates translational and transcriptional reprogramming of the stressed cells, thus preserving cellular resources and allowing cells to adapt.36 Polysome profile analysis of cytoplasmic extracts from amino acid-starved C6 cells revealed the presence of a novel double peak at 110S preceeding the polysomes (Fig. 1A). The population of the 110S peaks will be referred to throughout as a 110S peak. Formation of the novel 110S peak occurred as early as 15 min (data not shown) with maximal induction at 1 h of starvation. The ratio of the 110S/80S fractions did not change between 1 and 9 h in cells treated with amino acid starvation (Fig. 1A). At the same time, polysomes decreased (Fig. 1A). These data are consistent with decreased protein synthesis rates caused by eIF2α phosphorylation, as previously reported in reference 11. Restoration of optimal growth conditions by transferring cells to amino acid-containing medium resulted in disassembly of the 110S peak and an increase in heavy polysomes (Fig. 1B). The ratio of 110S/80S fraction area was similar between control and cells refed for 1 h after 3 h treatment with amino acid starvation. The dynamic nature of the 110S peak formation upon starvation and refeeding provides strong evidence that the peak is likely composed of non-translating ribosomes, which associate with mRNAs and shifts to polyribosomes during relief from amino acid starvation. In order to further support that the 110S peak is formed under conditions of inhibition of translation initiation, we hypothesized that refeeding of starved cells in the presence of such inhibitors will not cause disassembly of the 110S complex. It is shown that the 110S complex was maintained in cells refed in the presence of hippuristanol (Fig. 1B). Hippuristanol is an inhibitor of the helicase activity of the translation initiation factor eIF4A,37 and refeeding in its presence probably prevented 110S peak-associated ribosomes to be recruited to mRNAs and the resumption of mRNA translation.
Figure 1.
110S peak formation in polysome profiles of rat C6 glioma cells. (A) Polysome profiles from control and amino acid-starved (AA st) cells at the indicated times. Ten OD260 units of cell extracts were loaded on 10–50% sucrose gradients and analyzed as described in Materials and Methods. The position of 80S is indicated. (B) Polysome profiles from amino acid-starved and amino acid-refed cells for 1 h after 3 h of starvation, in the absence (refed) or presence of hippuristanol (refed + hipp). (C) Polysome profiles from cells treated with Tg for 3 h. Cell extracts were loaded on 10–35% sucrose gradients. (D) Polysome profiles from cell extracts that were subjected to translational runoff in the absence (run-off) or presence of CX (run-off + CX). Cell extracts were loaded on 10–35% sucrose gradients. Data are shown from represantive experiments.
In order to further verify the hypothesis that the 110S peak formation is the result of stress-induced inhibition of protein synthesis, we tested whether other types of cellular stress, which are known to inhibit translation initiation in mammalian cells, would induce 110S formation. Thapsigargin (Tg) is a well-characterized chemical that induces the unfolded protein response by calcium release from the endoplasmic reticulum (ER). Treatment of cells with Tg activates the eIF2α kinase PERK, which phosphorylates eIF2α and thereby inhibits mRNA translation initiation.38 Polysome profile analysis of cell lysates from 3 h Tg-treated C6 cells showed formation of the 110S peak (Fig. 1C). This lead us to the conclusion that in C6 cells, the presence of the 110S peak in the polysome profiles is tightly associated with the global translation inhibition, which prevents free ribosomes from being recruited to mRNAs. The characteristics of the 110S peak in C6 cells are similar to the bacterial 100S ribosomes. In both cases, the presence of an additional peak in the polysome profile is induced in conditions with limited availability of nutrients, and such a complex disappears shortly after restoration of optimal growth conditions.27 Our data support the hypothesis that the 110S peak forms by association of free ribosomes generated by stress-induced inhibition of protein biosynthesis.
Formation of the 110S peak in non-stressed cell-free extracts following ribosome run-off.
In order to test whether the 110S peak consists of resting ribosomes, we used a cell-free system and C6 cytoplasmic extracts and performed a ribosome run-off experiment. Cell lysates from control C6 cells were incubated in vitro under conditions that promote translation elongation of existing polysome-associated polypeptides and release of ribosomes from the mRNAs. In order to ensure that the release of the ribosomes is the result of completion of translation of mRNAs bound with polyribosomes, we used the inhibitor of translation elongation CX as a control for this analysis. Following cell-free translation of the cell extracts, the reaction mixtures were analyzed on the sucrose gradients. Polysome profiles from CX-treated extracts showed the presence of polysomes and absence of the 110S peak (Fig. 1D). In contrast, polysome profiles from extracts not-treated with CX, showed the 110S peak and absence of polysomes (Fig. 1D). These data again suggested that 110S peak formation is promoted by all cellular conditions that cause release of the ribosomes from the mRNAs without subsequent recycling to new rounds of translation initiation.
The 110S peak is composed of ribosomes.
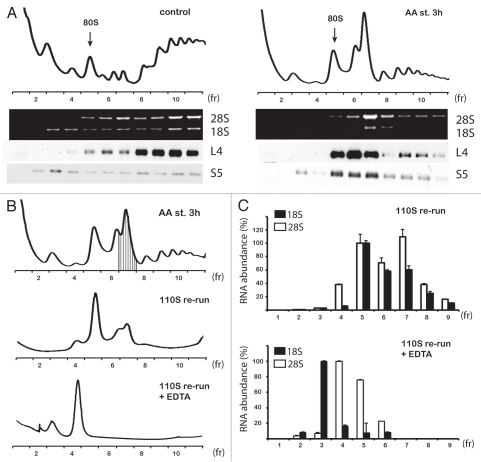
The hypothesis that the 110S peak consists of ribosomes is supported by two pieces of data: (1) the quick disassembly of the peak and build-up of polysomes upon transfer of cells from starvation to optimal growth media (Fig. 1B) and (2) the assembly of the 110S peak in vitro following ribosome runoff (Fig. 1D). To further support this hypothesis, we analyzed the ribosomal protein and RNA content of the fractions obtained from polysome profile gradients of amino acid-starved and control cells. We tested the distribution of the large ribosomal subunit protein L4 and the small ribosomal subunit protein S5 across the gradients. From the same fractions, RNA was isolated and analyzed by agarose gel electrophoresis for the presence of the rRNAs: 18S, small ribosomal subunit and 28S, large ribosomal subunit. Both proteins showed the expected distribution across the polysome profiles from control cells; L4 was present in fractions corresponding to 60S, 80S and polysomes when S5 was present in fractions corresponding to 40S, 80S and polysomes (Fig. 2A). Distribution of those ribosomal proteins in polysome profiles from amino acid-starved cells showed that both proteins were present in the 110S peak fractions (Fig. 2A). A similar pattern of distribution of the ribosomal subunits was confirmed by the presence of 18S and 28S rRNAs (Fig. 2A). These data directly demonstrate that the 110S fraction contains both ribosomal subunits. As an additional test, we analyzed fraction 7, which contained the 110S peak (Fig. 2B) on a new set of gradients. We performed the analysis in the presence or absence of EDTA, which induces dissociation of ribosomal particles into large and small subunits. Fractionation of the untreated 110S complex revealed its unstable nature: free 60S, 80S and 110S particles were present in the gradient (Fig. 2B). Subsequent quantitative qPCR analysis of the 18S and 28S rRNA across the gradient brought the interesting observation that the ratio of 18S to 28S in the 110S peak fraction was lower than the ratio in the fractions corresponding to 80S (Fig. 2C). These data suggested that the 110S peak is likely composed of two sub-populations of complexes, light heterodimers of 60S with 80S and heavier 80S homodimers. In agreement with this conclusion, dissociation of the peak with EDTA resulted into free subunits containing 18S rRNA in the 40S and 28S in the 60S fractions (Fig. 2C). The unstable nature of the 110S ribosomal peak was further confirmed by treatment of the peak fractions with a high salt concentration. We noted that KCl in concentrations higher than 200 mM in the cell extract and in the gradient buffer dissociated the 110S peak (data not shown). These findings provide an additional similarity between the bacterial hibernating 100S dimers20 and the ribosomal 110S complex in stressed C6 cells.
Figure 2.
110S peak in rat C6 glioma cells contains ribosomal subunits. (A) Detection of ribosomal proteins (L4 and S5) and rRNAs (18S and 28S) in the fractions of polysome profiles of control and amino acid-starved (AA st) cells. (B) Re-run of peak fraction (no. 7) on sucrose gradients in the absence (110S re-run) or presence of EDTA (110S re-run + EDTA). (C) qPCRanalysis of rRNAs (18S and 28S) in fractions of the gradients described in (B). Values are expressed as percentage of the signal obtained in fraction 5 (100%) corresponding to 80S (top part) and fractions 3 (100%, for 18S) corresponding to 40S and 4 (100%, for 28S) corresponding to 60S (bottom part). Data are average from three independent experiments.
Cryo-Em visualization of ribosomal dimmers.
Bacterial 100S ribosomal dimers were previously visualized by electron microscopy after negative staining17 and three-dimensional processing of cryo-EM images.28 These studies revealed that bacterial 100S ribosomes are interacting via the 30S subunits. We used a similar approach to demonstrate that the 110S peak in C6 cells is a ribosomal dimer.
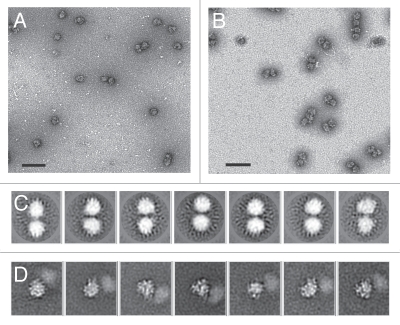
We first used negative staining combined with electron microscopy to examine the nature of the 110S fractions from the sucrose gradient. The micrographs revealed that there were several ribosome dimers, but about half the total population was monomeric (Fig. 3A). We reasoned that since the interaction between the dimers may be relatively unstable, many of the dimers were dissociating into monomers during the grid preparation. Therefore, to stabilize the dimer interactions, we isolated ribosomes using in-gradient chemical crosslinking39 of the starved cell lysates. As expected, presence of glutaraldehyde significantly improved stability of the ribosomal dimers. The negative-stained micrographs revealed that the vast majority of particles were dimers after chemical fixation (Fig. 3B). In addition to the faster sedimenting dimer population, the in-gradient cross-linking revealed peaks for monomeric 40S, 60S and 80S fractions. This indicated that cross-linking stabilized the pre-formed dimers but did not artificially contribute to dimer formation from free 80S.
Figure 3.
Electron microscopy images of ribosomal dimers present in the 110S peak from rat C6 glioma cells. Electron micrographs showing negatively stained complexes from the 110S peak in the absence (A) or presence (B) of in-gradient crosslinkin. Scale bar size is 1 µm. (C) Cryo-EM 2D averages of crossilnked dimers. Dimer 80S particles were picked, centered and normalized. After orienting all particles, such that the two ribosomes were at 12 o'clock and 6 o'clock positions, dimer projections were subjected to multiple rounds of reference-free alignment and K-means classification procedures. (D) Reference-based alignment of an individual 80S ribosome within the dimer. Ribosomes within the dimer particle were aligned to an 80S reference using projection-matching techniques. The pre-aligned projections within each aligned class were then subjected to multivariate statistical analysis to identify the relative location of the second ribosome within each dimer. Representative averages are shown here, with a more complete data set presented in Supplemental Figure 1. The window dimensions for (C and D) are 740 Å2.
To further analyze the dimeric population, we employed cryo-electron microscopy (cryo-EM). Cryo-EM is used to flash freeze the specimen in its native state and provides superior resolution over negative staining. Fifty-seven micrographs were collected, and a total of 3,495 ribosome dimers were manually selected and subjected to two-dimensional alignment and classification procedures. As expected, the 2D averages revealed dumbbell shaped dimers (Fig. 3C). The large size and globular shape of the individual ribosomes within the 2D averages indicates that the dimers are composed of 80S or potentially 60S subunits. There was no evidence of a more elongated subunit (indicative of 40S) in any of the 2D averages.
The inability to distinguish features within the 2D averages of ribosomal dimers suggests that the connection between the two particles is highly flexible. It is difficult to conceive that the connection would be random, because if that were true, we would expect to see larger oligomers as opposed to distinct dimers. To obtain a better understanding of how the two ribosomes are interacting, we focused on aligning only one of the two ribosomes within each dimer. Dimeric particles selected from the cryo-EM micrographs were shifted, such that one of the dimers was centered in the boxed projections. The centered particles were first subjected to reference-based alignment via projection matching algorithms40 using a vacant 80S ribosome from HeLa cells as the reference.41 Pre-aligned classes were further classified using multivariate statistical analysis.42 These results yielded individual particles with visible features in the 2D averages of the centered particle (Fig. 3D). Many of the 2D averages showed a smudge where the second particle resided (Fig. 3D and Fig. S1). While the smudge is indicative of the relative location of the second particle with respect to the aligned 80S ribosome, the connection between the two ribosomes appears dynamic or flexible, as evidenced by the lack of detail in the second ribosome within the 2D averages. These images support a distinct connection between the two ribosome particles.
What is linking 60S subunits?
The diameter of individual particles within the 2D averages was calculated to be approximately 300 Å. The size and shape of the particles as determined by cryo-EM and image classification supports a connection between the 60S subunits of the stressed ribosomes with potential subpopulations of 80S-80S and 80S-60S. Although we cannot exclude the presence of 60S-60S subpopulations, it is unlikely that they exist under the 110S peak that we analyzed (Fig. 2B; fraction 7, shaded). We identified no potential 40S-40S or 40S-80S populations in our data. This analysis further supports the biochemical data localizing the intersubunit dimer connection to reside within a component of the 60S subunit. Provided that our 2D averages show the second particle in the dimers as a smudge (Fig. 3D), we reasoned that the connection is through a highly flexible linker within the large ribosomal subunit. The most obvious choice for a flexible linker in the 60S subunit would be one of the stalks: either the L1 stalk, which is localized near the exit-site (E-site) of the 60S subunit43 or the P proteins that comprise the L7/L12 stalk in bacteria and reside near the aminoacyl-site (A-site) of the 60S subunit.44
To identify probable candidates responsible for dimer interaction, we used two-dimensional gel electrophoresis and proteomics. While we were unable to identify novel proteins in the 110S fraction from starved cells, we noted changes in the phosphorylation status of stalk protein P2, with the 110S starved peak containing a less phosphorylated P2 protein (data not shown). The ribosomal stalk is a hetero-pentameric assembly with a highly flexible structure that associates with the 60S ribosome. It is believed to be the landing pad for translation factors and factor-mediated GTP hydrolysis.45 Rat ribosomal stalk P proteins can also form oligomers in vitro.46 It is therefore possible that the interaction of the rat 80S dimers is mediated via modulation of the phosphorylation status of stalk proteins. Bacterial and eukaryotic stalk proteins (except rRNA binding domain of L10 and P0), although they do not share sequence similarity, they function in a similar manner.44 It was shown that bacterial stalk proteins are involved in recruitment of initiation factor 2 (IF2), elongation factors Tu (EF-Tu) and G (EF-G) and release factor 3 (RF3), which catalyze major steps of mRNA translation in a GTP-dependent fashion.47 A working hypothesis for future studies might be that stalk protein-mediated ribosomal dimer formation during stress blocks translation factor recruitment and ribosome recycling, thus limiting continuation of translation initiation of mRNAs.
Formation of the 110S dimers does not require synthesis of stress-induced factors.
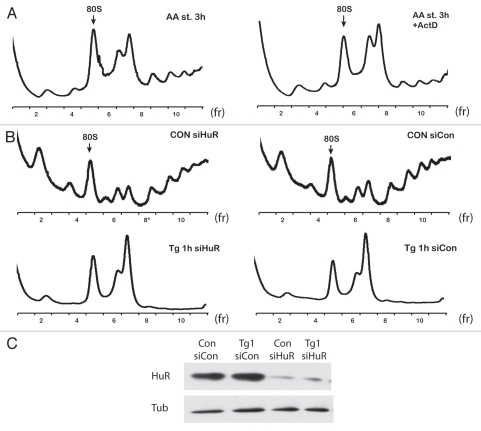
In bacteria, transition to the stationary growth phase (induced by the limited supply of nutrients) requires increased accumulation of RMF and/or HPF proteins, which promote the 100S ribosomal dimer formation.18,31 Here we tested whether formation of ribosomal dimers in C6 starved cells requires execution of the stress-induced transcription program. To answer this question we used Actinomycin D, a well-studied inhibitor of RNA transcription. Addition of actinomycin D during amino acid starvation did not prevent dimer formation, which suggested that complex formation is independent of the synthesis of stress-induced factors (Fig. 4A). Moreover, actinomycin D induced formation of the dimers in amino acid-fed cells during 3 h of treatment (data not shown). The latter was likely the result of inhibition of protein synthesis due to induction of phosphorylation of eIF2α, as previously reported in reference 48. These data and the unsuccessful attempt to identify novel proteins bound to the 110S dimers via proteomic analysis suggested that there is a fundamental difference between bacteria and mammalian cells with regard to the mechanism that controls ribosomal dimer formation during poor nutritional conditions.
Figure 4.
Formation of dimers in C6 cells does not require synthesis of new factors and is not dependent on stress granule formation. (A) Polysome profiles from cells treated with amino acid starvation in the absence (AA st 3 h) or presence of actinomycin D (AA st 3 h + ActD). (B) Polysome profiles of cells from control and amino acid-starved cells treated with siHuR or con siRNAs. (C) Western blot analysis of extracts from cells treated with siRNAs and blotted for HuR and tubulin.
An additional test was performed to determine whether ribosomal dimer formation depends on stress granule assembly. In eukaryotic cells under stress conditions, translation initiation is impaired, and stress granules (SGs) are formed.49 SGs are complexes composed of mRNA, translation initiation factors, 40S ribosomal subunits most likely assembled into 48S preinitiation complexes and several other proteins, including RNA helicases, translation and mRNA stability regulators as well as factors involved in cell signaling.49 Because of the common elements between SGs and ribosomal dimers (under stress conditions, both have dynamic nature and contain ribosomal subunits), we tested whether ribosomal dimers in C6 cells under stress conditions can be a form of SG-related complex. One of the marker proteins involved in SG assembly is HuR.50 We tested whether siRNA-mediated silencing of HuR would affect ribosomal dimer formation during stress induced by Tg-treatment. The efficiency of HuR silencing was monitored by western blotting (Fig. 4C). Depletion of HuR levels by 80% did not affect dimer formation (Fig. 4B). Moreover, other well-characterized components of SGs, like TIA-1 and TIAR,51,52 also had no effect on ribosomal dimer formation (data not shown).
The physiological significance of dimer formation in bacteria during poor nutritional conditions is a contribution to inhibition of protein synthesis and preservation of cellular resources.53 The levels of RMF and HPF proteins are induced under these conditions, with a primary role to decrease translational activity of the cells.16,31 In mammalian cells, protein synthesis is regulated during nutrient limitation or other stress conditions by sophisticated signaling mechanisms.54,55 For example, diverse stress conditions decrease global protein synthesis via increased phosphorylation of eIF2α, which leads to decreased ternary complex availability.56 In addition, inhibition of the mTOR pathway causes decreased phosphorylation of p70-S6 kinase 1 (S6K1), a positive regulator of translation,57–59 and decreased 4E-BP1 phosphorylation, which negatively regulates mRNA translation.58,60 It is clear that in contrast to bacteria, mammalian cells use signaling networks to control protein synthesis during stress. The physiological significance of ribosomal dimer formation in mammalian cells during stress is not clear. Because in both bacteria and C6 cells, formation of ribosomal dimers is the result of translational inhibition, it is possible that ribosomal dimer formation during stress is an evolutionary remnant of translational control.
Presence of ribosomal dimers in stressed mammalian cells is species-specific.
110S ribosome dimers were previously reported in sucrose gradients from rat and hamster cell extracts.33–35 These peaks were suggested to contain 40S and 60S subunits.61 It was also suggested that in liver extracts from rats treated with the hepatotoxic agent carbon tetrachloride, a similar peak to the 110S was formed and contained ribosomes. These ribosomes were suggested to be targets of ribosomal degradation.62 These ribosomal peaks were also observed in the rat adrenals after adrenocorticotropin (ACTH) injection.63 Of particular interest was a report demonstrating that dimer formation in the liver of fasted rats could be reversed by amino acid reperfusion in vivo.64 Our studies demonstrate a direct relationship between formation of the ribosomal peak and inhibition of protein synthesis. This conclusion can also explain all the earlier reports on 110S peak formation in rat cells.
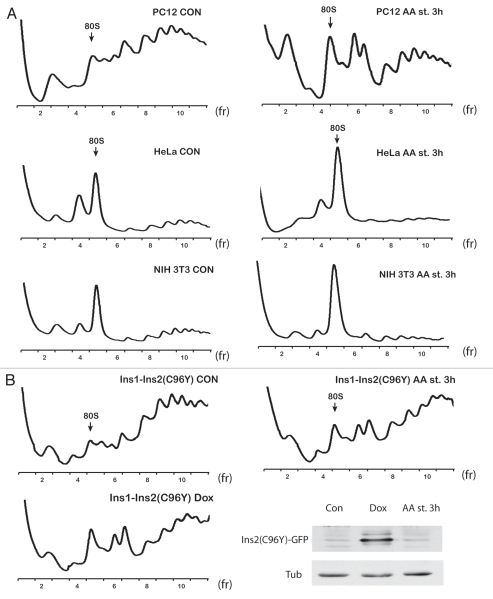
We next determined whether human and mouse cells can also form dimers upon amino acid starvation. Polysome profiles from amino acid-starved human HeLa or mouse NIH 3T3 cell extracts showed a decrease in polysomes and an increase in the 80S peak area. However, we did not observe any 110S dimer formation (Fig. 5A). We tested several human, mouse and rat cell lines and searched for dimer formation upon stress conditions. We observed a 110S peak formation only in rat cells, such as C6 glioma (Fig. 1A), PC12 derived from a phenochromocytoma of the rat adrenal medulla (Fig. 5A) and NRK kidney cells (data not shown). We also tested rat cells, which are being used as an experimental model system for studies on stress-induced diabetes. Diabetes has been associated with increased endoplasmic reticulum stress in pancreatic β cells, leading to apoptosis and decreased plasma insulin.65 Ins1 insulinoma cells expressing an inducible mutant insulin 2 gene (C96Y-so called Akita mutation) undergo ER stress-mediated apoptosis and are being used as a model to study the stress-induced factors that are involved in pancreatic β-cell apoptosis.66 Induction of the mutant insulin protein in Ins1-Ins2 (C96Y) cells via the use of doxycycline (Dox) mimics the induction of ER stress in vivo in pancreatic β cells.66 Treatment with Dox increased eIF2α phosphorylation, decreased protein synthesis (data not shown) and induced 110S peak formation in Ins1-Ins2 (C96Y) cells (Fig. 5B). Similar results were obtained by treating Ins1-Ins2 (C96Y) cells with amino acid deficient media. We observed decreased protein biosynthesis by 50% (data not shown), which could explain the relatively high level of remaining polysomes (Fig. 5B). These data suggested that rat cells form ribosomal dimers during stress conditions, which cause inhibition of protein synthesis. The presence of ribosomal dimers in rat cell lines may have implications for studies that use such cells to study the molecular mechanisms of human diseases. Almost all studies that use polysome profile analysis use the position of the 40S, 60S and 80S ribosomes as markers to evaluate data of distribution of proteins or mRNAs across the gradients. Our data show that the 110S peak is a resting state of ribosomal dimers and should not be confused with either 80S monosomes or polysomal disomes.
Figure 5.
Ribosomal dimers are formed only in rat cells. (A) Polysome profiles of control and amino acid-starved rat PC12, human HeLa and mouse NIH 3T3 cells. (B) Polysome profiles of rat insulinoma cells expressing an inducible fusion gene of Ins2 (C96Y)-GFP. Control, amino acid-starved and doxycycline-treated cells were analyzed. The expression of the fusion protein was determined by western blotting using GFP antibodies. Tubulin was used as a loading control.
Discussion
Hibernating or aggregating ribosomes?
In bacteria, expression of RMF is induced during the transition to the stationary phase of growth as well as under stress conditions of hyperosmolarity67 or acidic media.68 Additionally, in E. coli deficient in the protein FtsY, which mediates delivery of proteins to the bacterial outer-membrane, RMF is induced to attenuate global protein synthesis due to the accumulation of outer membrane proteins in cytoplasmic inclusion bodies.69 All these data support a role for RMF as an important regulator of mRNA translation in bacteria. Mutants deficient in RMF have low survival rates in the stationary growth phase.18,30 In addition, they are more sensitive to heat and acidic conditions during stationary growth.68,70 Because ribosomal stability in an RMF mutant strain of E. coli decreased dramatically under stress conditions, it was speculated that dimerization is a mechanism to protect ribosomes from degradation.53 In contrast to this suggestion, application of heat shock to bacteria in stationary phase revealed complete dissociation of the 100S complexes without any apparent degradation of rRNA.68 Despite all the progress in understanding formation of ribosomal dimers in bacteria, the involvement of structural changes of ribosomes in this process are still not clear.29
Formation of ribosomal dimers in rat cells has common elements with bacteria: (1) all cellular conditions that cause inhibition of translation resulted in formation of the 110S peak in polysome profiles and (2) assembly and disassembly of ribosomal dimers was dynamic in response to nutrient availability. However, we also found several differences: (1) mammalian dimer formation does not appear to require a stress-induced factor; (2) mammalian ribosomal dimers have a mixed population of 80S:80S and 60S:80S dimers, with the interaction being most likely via the 60S subunit, while in bacteria, it is via the small 30S subunit and (3) Unlike bacterial dimers, mammalian ribosomal dimers probably do not have a well-defined interaction site or interaction occurs via a highly flexible element, although this conclusion needs additional experimental evidence.
Our studies show that rat but not human or mouse cells formed ribosomal dimers in polysome profiles when analyzed in parallel using identical experimental conditions. These data do not exclude that human or mouse cells may form ribosomal dimers with lower stability. It is also possible, that monomers could be in a hibernating state in species other than rat. Our data raise questions about possible ribosome differences between species or ribosome-associated proteins that have species-specific modifications. Future studies will address these possibilities. It will also be a challenge to determine the physiological significance of mammalian ribosomal dimers. Ribosomal dimerization in bacteria is considered a protective mechanism to prevent ribosomal degradation.53 In eukaryotic cells, ribosomal autophagy (ribophagy) has been described under stress conditions.15 It will be interesting to determine, in future studies, whether ribosomal dimerization regulates recruitment of ribosomal particles to autophagosomes. If the latter is the case, perhaps rat cells have decreased activities for ribophagy, leading to accumulation of ribosomal dimers. Although the present work describes the formation of resting ribosomes in rat cells during stress conditions, determining the significance of ribosomal dimerization in different species will require additional studies. We have presented here some ideas and directions for future work, which may bring to light an unrecognized aspect in translational control in mammalian cells.
Materials and Methods
Cells and media.
C6 rat glioma, human HeLa and mouse NIH 3T3 cells were grown in DMEM medium supplemented with 10% fetal bovine serum (FBS) and 2 mM L-glutamine with antibiotics (penicillin and streptomycin). PC12 cells were maintained in DMEM medium supplemented with 5% calf serum and 5% horse donor serum, 2 mM L-glutamine, 10 mM HEPES and antibiotics (penicillin and streptomycin). Rat insulinoma cells (Ins1) cells stably expressing the Ins2 (C96Y)-GFP fusion protein were cultured in RPMI 1640 supplemented with 10% FBS, 2 mM L-glutamine, 1 mM sodium pyruvate, 10 mM HEPES, 55 µM β-mercaptoethanol, and antibiotics (penicillin and streptomycin) and selection drugs (200 µg/ml G418 and 50 µg/ml hygromycin). Expression of Ins2 (C96Y)-EGFP was induced by treating cells with 5 µg/ml doxycycline for 72 h and was confirmed by western blot analysis using a GFP monoclonal antibody (BD PharMingen). Amino acid-starved cells, were incubated in Krebs-Ringer Bicarbonate Buffer (KRB-Sigma Aldrich) supplemented with 10% dialyzed FBS. Control cells were treated at the same time and transferred to fresh medium supplemented with 10% dialyzed FBS. For drug treatments, thapsigargin (Tg) (Sigma-Aldrich) was added to a final concentration of 400 nM, actinomycin D (Sigma-Aldrich) to a final concentration of 10 µg/ml (8 µM) and hippuristanol to a final concentration of 1 µM.
Polysome profile.
100 µg/ml cycloheximide (CX) was added to cells for 5 min at 37°C. Cells were then washed twice with PBS containing CX at 100 µg/ml, scraped and pelleted at 3,500 rpm for 10 min. The cell pellets were suspended in 500 µl of lysis buffer (10 mM HEPES-KOH, pH 7.4; 2.5 mM MgCl2; 100 mM KCl; 0.25% Nonidet NP-40, 200 units/ml) RNase inhibitor (RNaseOUT, Invitrogen) and EDTA-free protease inhibitor mixture (Roche Applied Science), kept on ice for 10 min then passed 15 times through a 23-gauge needle. Lysates were centrifuged at 10,000 γ for 10 min and supernatants (cytosolic cell extracts) were collected; absorbance was measured at 260 nm. About 10 ODs were layered over 10–50% or 10–35% sucrose gradients in buffer (10 mM HEPES-KOH, pH 7.4; 2.5 mM MgCl2; 100 mM KCl). Gradients were centrifuged at 17,000 rpm in a Beckman SW28 rotor for 15 h at 4°C. After centrifugation, 14 fractions (1.2 ml/each) were collected, and the first 11 were analyzed further. Because CX was used during polysome preparation to freeze the ribosomes on the translating mRNAs, we tested whether the presence of CX had any effect in our findings. Starved cells without CX treatment also formed a 110S peak, suggesting that CX did not induce peak formation (Data not shown). For chemical in-gradient cross-linking, gradients were prepared with 0–0.1% glutaraldehyde gradient. The crosslinking reactions were quenched by adding in the fractions TRIS-HCl pH: 7.5 to a final concentration of 20 mM.
Analysis of 110S fraction on sucrose gradients.
Fractions corresponding to the 110S complex were combined and concentrated using Amicon Ultra-4 (10K cut off). Treatment with EDTA (15 mM) for 30 min on ice was applied as indicated. Resulting concentrated samples were loaded on 10–50% sucrose gradients, and the polysome profile analysis was performed as described.
Run-off experiments.
Cells were lysed in polysome profile lysis buffer according to the previously described protocol followed by addition of an equal volume of translation mix described before in reference 71 (2x concentration: 1.6 mM MgCl2; 160 mM KAc; 80 mM amino acids; 188 µg/ml creatine phosphate kinase; 0.8 U/µl Rnasin; 1.6 mM ATP; 0.2 mM GTP; 0.2 mM Spermidine; 32 mM Hepes, pH: 7.6 and 40 mM creatine phosphate) and samples were incubated at 30°C for 30 min. This procedure allows completion and release of started polypeptides. As a control, the same experiment was performed in the presence of CX at a final concentration of 100 µg/ml. The latter inhibits runoff of ribosomes and completion of peptide synthesis.
Electron microscopy.
Negative staining was performed using 2% uranyl acetate and carbon coated copper grids. Cryo-EM was performed using Quantifoil grids plunge-frozen in liquid ethane. All images were acquired on a FEI Tecnai electron microscope operating at 200 kV. Cryo-EM imaging was performed at a set magnification of 40,510x under low-dose conditions and an underfocus range of 1.7–5.5 microns. Images were collected on a Gatan 4Kx4K CCD camera with 15 micron pixel size. The calibrated pixel size was 3.70 Å/pixel. Images were processed using SPIDER.72 For ribosomal dimers, projections were first normalized then subjected to multiple iterations of reference-free alignment and K-means classification. For reference-based alignment and classification procedures, an individual 80S ribosome within the dimeric structure was aligned to an 80S ribosome from HeLa cells.41 Reference-based alignment was performed by back-projecting the reference at 15° intervals, resulting in 83 reference projections. Each aligned class was then subjected to multivariate data analysis and classification to produce class averages as previously described in reference 42.
Other methods.
Total RNAs and proteins from each fraction were prepared by using Trizol LS reagent (Invitrogen) and the TCA precipitation method respectively. The distributions of 18S-28S rRNAs and ribosomal proteins were monitored by agarose gel electrophoresis and western blotting, respectively.73,74 qPCR analysis for rRNAs was performed using the following primer sets: (1) 18S: (forward: TTG ACG GAA GGG CAC CAC CAG; reverse: GCA CCA CCA CCC ACG GAA TCG) (2) 28S: (forward: GGC CGA AAC GAT CTC AAC CT; reverse: GCC ACC GTC CTG CTG TCT AT). siRNA against HuR was obtained from Dharmacon RNAi Technologies; control siRNA was from Ambion. Transfection was performed by using Lipofectamine (Invitrogen) according to the manufacturer instructions. Antibodies used were directed against tubulin (Sigma-Aldrich), ribosomal protein L4 kind gift from Vincent P. Mauro (The Scripps Research Institute, CA), ribosomal protein S573 and HuR (Santa Cruz).
Acknowledgments
We would like to thank Ibrahim Yaman for making the initial observation of the 110S peak in C6 cells while a graduate student in the laboratory (M.H.).
Financial Support
This work was supported by the National Institute of Health R01 (grant number DK060596 and DK053307 to M.H.); and the Canadian Institutes of Health Research (grant number MOP-12182 to B.P.).
Supplementary Material
References
- 1.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 2.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 4.Taylor DJ, Devkota B, Huang AD, Topf M, Narayanan E, Sali A, et al. Comprehensive molecular structure of the eukaryotic ribosome. Structure. 2009;17:1591–1604. doi: 10.1016/j.str.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korostelev A, Ermolenko DN, Noller HF. Structural dynamics of the ribosome. Curr Opin Chem Biol. 2008;12:674–683. doi: 10.1016/j.cbpa.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, et al. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao H, Zhou Z, Rawat U, Huang C, Bouakaz L, Wang C, et al. RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors. Cell. 2007;129:929–941. doi: 10.1016/j.cell.2007.03.050. [DOI] [PubMed] [Google Scholar]
- 8.Ramakrishnan V. The ribosome: some hard facts about its structure and hot air about its evolution. Cold Spring Harb Symp Quant Biol. 2009;74:25–33. doi: 10.1101/sqb.2009.74.032. [DOI] [PubMed] [Google Scholar]
- 9.Lecocq RE, Cantraine F, Keyhani E, Claude A, Delcroix C, Dumont JE. Quantitative evaluation of polysomes and ribosomes by density gradient centrifugation and electron microscopy. Anal Biochem. 1971;43:71–79. doi: 10.1016/0003-2697(71)90109-6. [DOI] [PubMed] [Google Scholar]
- 10.Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, et al. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaccioliy F, Huang CC, Wang C, Bevilacqua E, Franchi-Gazzola R, Gazzola GC, et al. Amino acid starvation induces the SNAT2 neutral amino acid transporter by a mechanism that involves eukaryotic initiation factor 2alpha phosphorylation and cap-independent translation. J Biol Chem. 2006;281:17929–17940. doi: 10.1074/jbc.M600341200. [DOI] [PubMed] [Google Scholar]
- 12.Bevilacqua E, Wang X, Majumder M, Gaccioli F, Yuan CL, Wang C, et al. eIF2alpha phosphorylation tips the balance to apoptosis during osmotic stress. J Biol Chem. 2010;285:17098–17111. doi: 10.1074/jbc.M110.109439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sells BH, Ennis HL. Polysome stability in relaxed and stringent strain of Escherichia coli during amino acid starvation. J Bacteriol. 1970;102:666–671. doi: 10.1128/jb.102.3.666-671.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cataldo L, Mastrangelo MA, Kleene KC. A quantitative sucrose gradient analysis of the translational activity of 18 mRNA species in testes from adult mice. Mol Hum Reprod. 1999;5:206–213. doi: 10.1093/molehr/5.3.206. [DOI] [PubMed] [Google Scholar]
- 15.Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 16.Wada A, Igarashi K, Yoshimura S, Aimoto S, Ishihama A. Ribosome modulation factor: stationary growth phase-specific inhibitor of ribosome functions from Escherichia coli. Biochem Biophys Res Commun. 1995;214:410–417. doi: 10.1006/bbrc.1995.2302. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida H, Maki Y, Kato H, Fujisawa H, Izutsu K, Wada C, et al. The ribosome modulation factor (RMF) binding site on the 100S ribosome of Escherichia coli. J Biochem. 2002;132:983–989. doi: 10.1093/oxfordjournals.jbchem.a003313. [DOI] [PubMed] [Google Scholar]
- 18.Yamagishi M, Matsushima H, Wada A, Sakagami M, Fujita N, Ishihama A. Regulation of the Escherichia coli rmf gene encoding the ribosome modulation factor: growth phase- and growth rate-dependent control. EMBO J. 1993;12:625–630. doi: 10.1002/j.1460-2075.1993.tb05695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Izutsu K, Wada A, Wada C. Expression of ribosome modulation factor (RMF) in Escherichia coli requires ppGpp. Genes Cells. 2001;6:665–676. doi: 10.1046/j.1365-2443.2001.00457.x. [DOI] [PubMed] [Google Scholar]
- 20.Yoshida H, Yamamoto H, Uchiumi T, Wada A. RMF inactivates ribosomes by covering the peptidyl transferase centre and entrance of peptide exit tunnel. Genes Cells. 2004;9:271–278. doi: 10.1111/j.1356-9597.2004.00723.x. [DOI] [PubMed] [Google Scholar]
- 21.Ueta M, Yoshida H, Wada C, Baba T, Mori H, Wada A. Ribosome binding proteins YhbH and YfiA have opposite functions during 100S formation in the stationary phase of Escherichia coli. Genes Cells. 2005;10:1103–1112. doi: 10.1111/j.1365-2443.2005.00903.x. [DOI] [PubMed] [Google Scholar]
- 22.Yoshida H, Ueta M, Maki Y, Sakai A, Wada A. Activities of Escherichia coli ribosomes in IF3 and RMF change to prepare 100S ribosome formation on entering the stationary growth phase. Genes Cells. 2009;14:271–280. doi: 10.1111/j.1365-2443.2008.01272.x. [DOI] [PubMed] [Google Scholar]
- 23.Agafonov DE, Kolb VA, Spirin AS. Ribosome-associated protein that inhibits translation at the aminoacyl-tRNA binding stage. EMBO Rep. 2001;2:399–402. doi: 10.1093/embo-reports/kve091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maki Y, Yoshida H, Wada A. Two proteins, YfiA and YhbH, associated with resting ribosomes in stationary phase Escherichia coli. Genes Cells. 2000;5:965–974. doi: 10.1046/j.1365-2443.2000.00389.x. [DOI] [PubMed] [Google Scholar]
- 25.Agafonov DE, Kolb VA, Nazimov IV, Spirin AS. A protein residing at the subunit interface of the bacterial ribosome. Proc Natl Acad Sci USA. 1999;96:12345–12349. doi: 10.1073/pnas.96.22.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vila-Sanjurjo A, Schuwirth BS, Hau CW, Cate JH. Structural basis for the control of translation initiation during stress. Nat Struct Mol Biol. 2004;11:1054–1059. doi: 10.1038/nsmb850. [DOI] [PubMed] [Google Scholar]
- 27.Wada A. Growth phase coupled modulation of Escherichia coli ribosomes. Genes Cells. 1998;3:203–208. doi: 10.1046/j.1365-2443.1998.00187.x. [DOI] [PubMed] [Google Scholar]
- 28.Kato T, Yoshida H, Miyata T, Maki Y, Wada A, Namba K. Structure of the 100S ribosome in the hibernation stage revealed by electron cryomicroscopy. Structure. 2010;18:719–724. doi: 10.1016/j.str.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 29.Ortiz JO, Brandt F, Matias VR, Sennels L, Rappsilber J, Scheres SH, et al. Structure of hibernating ribosomes studied by cryoelectron tomography in vitro and in situ. J Cell Biol. 2010;190:613–621. doi: 10.1083/jcb.201005007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Apirakaramwong A, Fukuchi J, Kashiwagi K, Kakinuma Y, Ito E, Ishihama A, et al. Enhancement of cell death due to decrease in Mg2+ uptake by OmpC (cation-selective porin) deficiency in ribosome modulation factor-deficient mutant. Biochem Biophys Res Commun. 1998;251:482–487. doi: 10.1006/bbrc.1998.9494. [DOI] [PubMed] [Google Scholar]
- 31.Ueta M, Ohniwa RL, Yoshida H, Maki Y, Wada C, Wada A. Role of HPF (hibernation promoting factor) in translational activity in Escherichia coli. J Biochem. 2008;143:425–433. doi: 10.1093/jb/mvm243. [DOI] [PubMed] [Google Scholar]
- 32.Ueta M, Wada C, Wada A. Formation of 100S ribosomes in Staphylococcus aureus by the hibernation promoting factor homolog SaHPF. Genes Cells. 2010;15:43–58. doi: 10.1111/j.13652443.2009.01364.x. [DOI] [PubMed] [Google Scholar]
- 33.Martin TE, Rolleston FS, Low RB, Wool IG. Dissociation and reassociation of skeletal muscle ribosomes. J Mol Biol. 1969;43:135–149. doi: 10.1016/0022-2836(69)90084-9. [DOI] [PubMed] [Google Scholar]
- 34.Reader RW, Stanners CP. On the significance of ribosome dimers in extracts of animal cells. J Mol Biol. 1967;28:211–223. doi: 10.1016/S0022-2836(67)80004-4. [DOI] [PubMed] [Google Scholar]
- 35.Eliceiri GL. The RNA of a dimerized ribosomal structure from hamster cells. Biochim Biophys Acta. 1972;269:450–452. doi: 10.1016/0005-2787(72)90132-3. [DOI] [PubMed] [Google Scholar]
- 36.Zhang P, McGrath BC, Reinert J, Olsen DS, Lei L, Gill S, et al. The GCN2 eIF2alpha kinase is required for adaptation to amino acid deprivation in mice. Mol Cell Biol. 2002;22:6681–6688. doi: 10.1128/MCB.22.19.6681-8.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bordeleau ME, Mori A, Oberer M, Lindqvist L, Chard LS, Higa T, et al. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat Chem Biol. 2006;2:213–220. doi: 10.1038/nchembio776. [DOI] [PubMed] [Google Scholar]
- 38.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 39.Stark H. GraFix: stabilization of fragile macromolecular complexes for single particle cryo-EM. Methods Enzymol. 2010;481:109–126. doi: 10.1016/S0076-6879(10)81005-5. [DOI] [PubMed] [Google Scholar]
- 40.Penczek PA, Grassucci RA, Frank J. The ribosome at improved resolution: new techniques for merging and orientation refinement in 3D cryo-electron microscopy of biological particles. Ultramicroscopy. 1994;53:251–270. doi: 10.1016/03043991(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 41.Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell. 2004;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 42.Shaikh TR, Trujillo R, LeBarron JS, Baxter WT, Frank J. Particle-verification for single-particle, reference-based reconstruction using multivariate data analysis and classification. J Struct Biol. 2008;164:41–48. doi: 10.1016/j.jsb.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cornish PV, Ermolenko DN, Staple DW, Hoang L, Hickerson RP, Noller HF, et al. Following movement of the L1 stalk between three functional states in single ribosomes. Proc Natl Acad Sci USA. 2009;106:2571–2576. doi: 10.1073/pnas.0813180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grela P, Bernado P, Svergun D, Kwiatowski J, Abramczyk D, Grankowski N, et al. Structural relationships among the ribosomal stalk proteins from the three domains of life. J Mol Evol. 2008;67:154–167. doi: 10.1007/s00239-008-9132-2. [DOI] [PubMed] [Google Scholar]
- 45.Tchórzewski M. The acidic ribosomal P proteins. Int J Biochem Cell Biol. 2002;34:911–915. doi: 10.1016/S1357-2725(02)00012-2. [DOI] [PubMed] [Google Scholar]
- 46.Gonzalo P, Lavergne JP, Reboud JP. Pivotal role of the P1 N-terminal domain in the assembly of the mammalian ribosomal stalk and in the proteosynthetic activity. J Biol Chem. 2001;276:19762–19769. doi: 10.1074/jbc.M101398200. [DOI] [PubMed] [Google Scholar]
- 47.Helgstrand M, Mandava CS, Mulder FA, Liljas A, Sanyal S, Akke M. The ribosomal stalk binds to translation factors IF2, EF-Tu, EF-G and RF3 via a conserved region of the L12 C-terminal domain. J Mol Biol. 2007;365:468–479. doi: 10.1016/j.jmb.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 48.Jiang HY, Wek SA, McGrath BC, Scheuner D, Kaufman RJ, Cavener DR, et al. Phosphorylation of the alpha subunit of eukaryotic initiation factor 2 is required for activation of NFkappaB in response to diverse cellular stresses. Mol Cell Biol. 2003;23:5651–5663. doi: 10.1128/MCB.23.16.5651-63.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Masuda K, Marasa B, Martindale JL, Halushka MK, Gorospe M. Tissue- and age-dependent expression of RNA-binding proteins that influence mRNA turnover and translation. Aging (Albany NY) 2009;1:681–698. doi: 10.18632/aging.100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gallouzi IE. Could stress granules be involved in age-related diseases? Aging (Albany NY) 2009;1:753–757. doi: 10.18632/aging.100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.El-Sharoud WM. Ribosome inactivation for preservation: concepts and reservations. Sci Prog. 2004;87:137–152. doi: 10.3184/003685004783238517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Komar AA, Hatzoglou M. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle. 2011;10:229–240. doi: 10.4161/cc.10.2.14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34:7–11. doi: 10.1042/BST0340007. [DOI] [PubMed] [Google Scholar]
- 57.Holz MK, Blenis J. Identification of S6 kinase 1 as a novel mammalian target of rapamycin (mTOR)-phosphorylating kinase. J Biol Chem. 2005;280:26089–26093. doi: 10.1074/jbc.M504045200. [DOI] [PubMed] [Google Scholar]
- 58.Panieri E, Toietta G, Mele M, Labate V, Ranieri SC, Fusco S, et al. Nutrient withdrawal rescues growth factor-deprived cells from mTOR-dependent damage. Aging (Albany NY) 2010;2:487–503. doi: 10.18632/aging.100183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hands SL, Proud CG, Wyttenbach A. mTOR's role in ageing: protein synthesis or autophagy? Aging (Albany NY) 2009;1:586–597. doi: 10.18632/aging.100070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence J, Jr, et al. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 61.Eliceiri GL. The ribosomal RNA of hamster-mouse hybrid cells. J Cell Biol. 1972;53:177–184. doi: 10.1083/jcb.53.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lilja HS, Longnecker DS, Curphey TJ, Daniel DS, Adams WE. Studies of DNA damage in rat pancreas and liver by 6-diazo-5-oxo-L-norleucine, ethyl diazoacetate and azaserine. Cancer Lett. 1981;12:139–146. doi: 10.1016/0304-3835(81)90049-5. [DOI] [PubMed] [Google Scholar]
- 63.Genot A, Dazord A. Transitory increase of run-off single ribosomes in rat adrenocortical cells, following ACTH administration, in vivo. Biochem Biophys Res Commun. 1982;109:1172–1179. doi: 10.1016/0006-291X(82)91900-3. [DOI] [PubMed] [Google Scholar]
- 64.McGown E, Richardson AG, Henderson LM, Swan PB. Effect of amino acids on ribosome aggregation and protein synthesis in perfused rat liver. J Nutr. 1973;103:109–116. doi: 10.1093/jn/103.1.109. [DOI] [PubMed] [Google Scholar]
- 65.Volchuk A, Ron D. The endoplasmic reticulum stress response in the pancreatic beta-cell. Diabetes Obes Metab. 2010;12:48–57. doi: 10.1111/j.1463-326.2010.01271.x. [DOI] [PubMed] [Google Scholar]
- 66.Hartley T, Siva M, Lai E, Teodoro T, Zhang L, Volchuk A. Endoplasmic reticulum stress response in an INS-1 pancreatic beta-cell line with inducible expression of a folding-deficient proinsulin. BMC Cell Biol. 2010;11:59. doi: 10.1186/1471-2121-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garay-Arroyo A, Colmenero-Flores JM, Garciarrubio A, Covarrubias AA. Highly hydrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit. J Biol Chem. 2000;275:5668–5674. doi: 10.1074/jbc.275.8.5668. [DOI] [PubMed] [Google Scholar]
- 68.Niven GW. Ribosome modulation factor protects Escherichia coli during heat stress, but this may not be dependent on ribosome dimerisation. Arch Microbiol. 2004;182:60–66. doi: 10.1007/s00203-004-0698-9. [DOI] [PubMed] [Google Scholar]
- 69.Bürk J, Weiche B, Wenk M, Boy D, Nestel S, Heimrich B, et al. Depletion of the signal recognition particle receptor inactivates ribosomes in Escherichia coli. J Bacteriol. 2009;191:7017–7026. doi: 10.1128/JB.00208-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.El-Sharoud WM, Niven GW. The influence of ribosome modulation factor on the survival of stationary-phase Escherichia coli during acid stress. Microbiology. 2007;153:247–253. doi: 10.1099/mic.0.2006/001552-0. [DOI] [PubMed] [Google Scholar]
- 71.Zeenko VV, Wang C, Majumder M, Komar AA, Snider MD, Merrick WC, et al. An efficient in vitro translation system from mammalian cells lacking the translational inhibition caused by eIF2 phosphorylation. RNA. 2008;14:593–602. doi: 10.1261/rna.825008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 73.Majumder M, Yaman I, Gaccioli F, Zeenko VV, Wang C, Caprara MG, et al. The hnRNA-binding proteins hnRNP L and PTB are required for efficient translation of the Cat-1 arginine/lysine transporter mRNA during amino acid starvation. Mol Cell Biol. 2009;29:2899–2912. doi: 10.1128/MCB.01774-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li Y, Bevilacqua E, Chiribau CB, Majumder M, Wang C, Croniger CM, et al. Differential control of the CCAAT/enhancer-binding protein beta (C/EBPbeta) products liver-enriched transcriptional activating protein (LAP) and liver-enriched transcriptional inhibitory protein (LIP) and the regulation of gene expression during the response to endoplasmic reticulum stress. J Biol Chem. 2008;283:22443–22456. doi: 10.1074/jbc.M801046200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.