Abstract
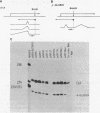
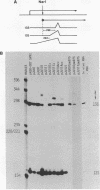
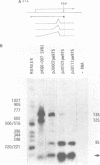
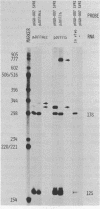
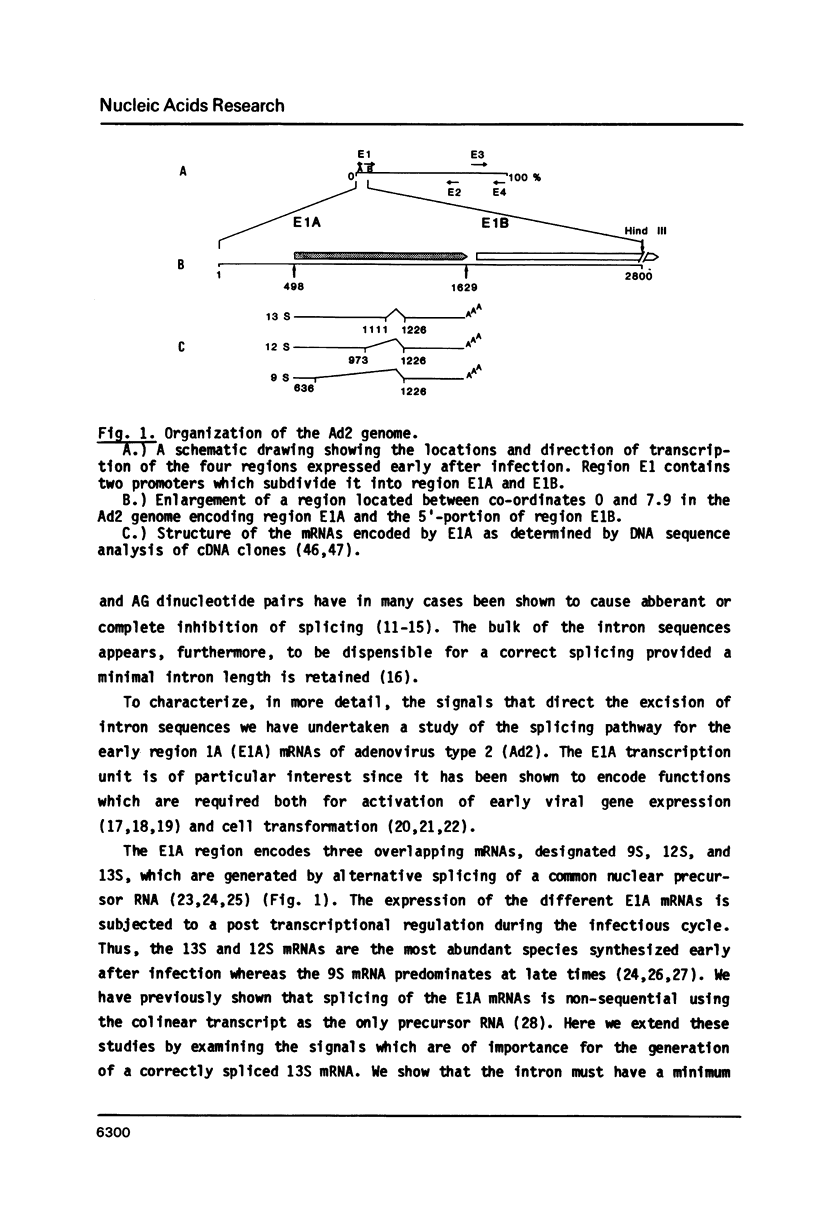
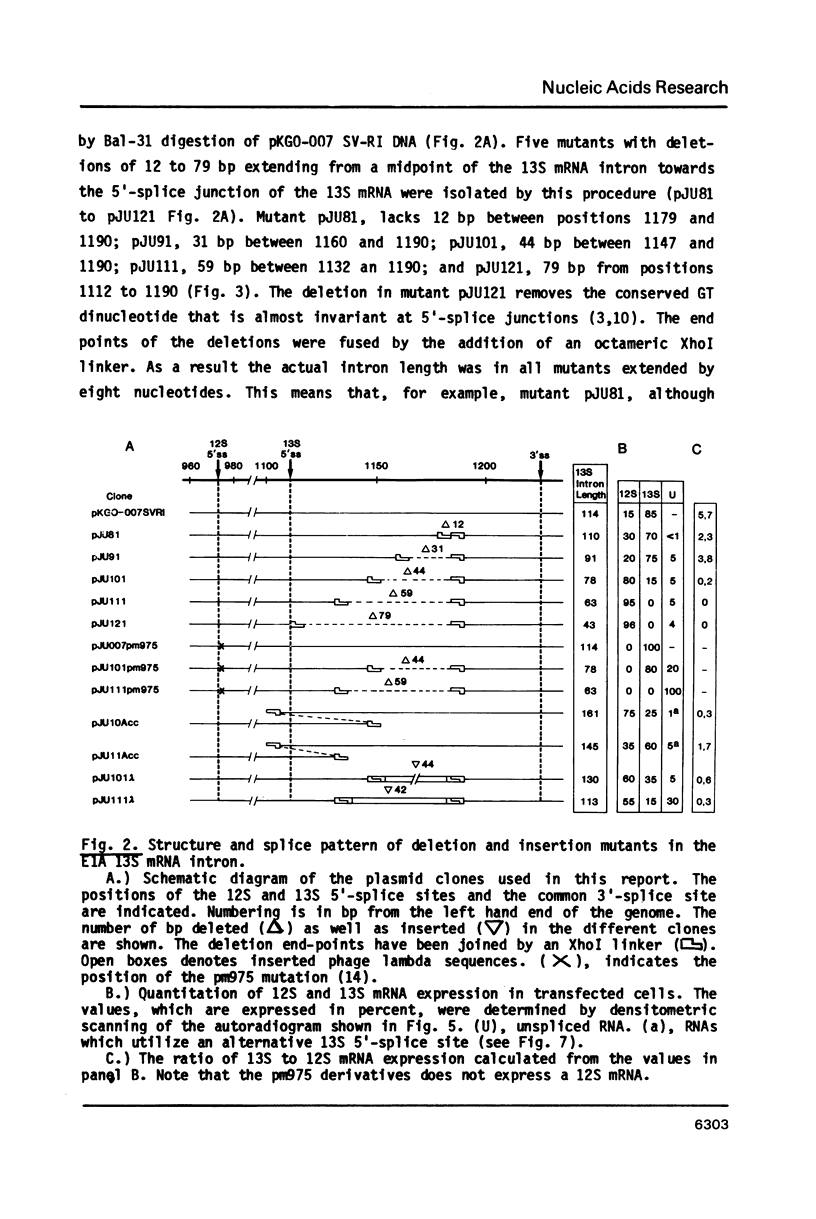
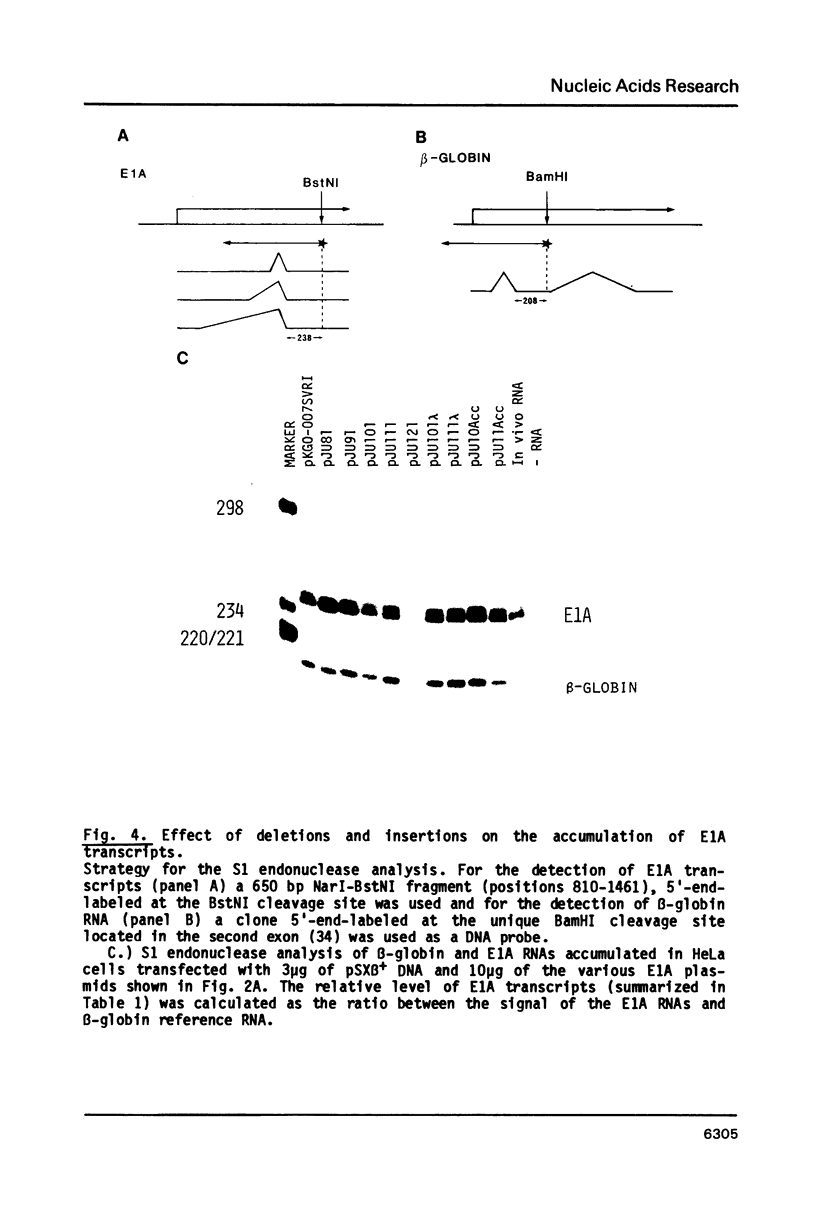
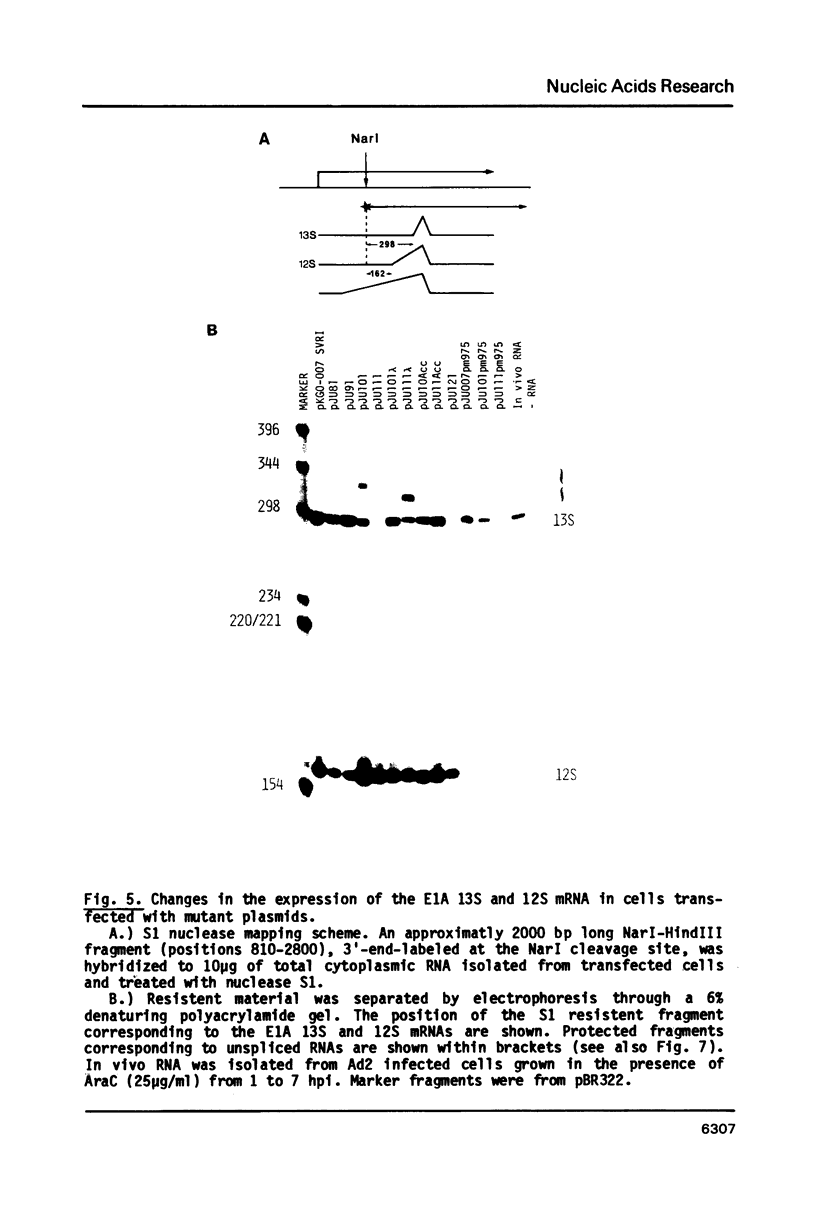
The adenovirus E1A region encodes three overlapping mRNAs, designated 9S, 12S and 13S. They differ from each other with regard to the length of the intron which is removed by RNA splicing. We have constructed E1A genes with deletions and insertions in the intervening sequence that is common to all three E1A mRNAs, in a search for signals which influence splicing of the 13S mRNA. Mutant plasmids were transfected into HeLa cells and the transiently expressed E1A mRNAs characterized by the S1 protection assay. The results show that five upstream and 20 downstream nucleotides are sufficient to allow for a correct utilization of the 5'-splice junction for the E1A 13S mRNA. Moreover, we show that a minimal intron length of 78 nucleotides is required for efficient 13S mRNA splicing. The ability of mutants with large intron deletions to maturate a 13S mRNA could partially be restored by expanding the intron length with phage lambda sequences. However, in no case was the normal splicing efficiency obtained with these mutants. In contrast, one mutant in which sequences from the authentic 13S mRNA intron were used to expand the intron expressed almost normal levels of 13S mRNA, thus suggesting that signals which specifically promote 13S mRNA splicing exist.
Full text
PDF




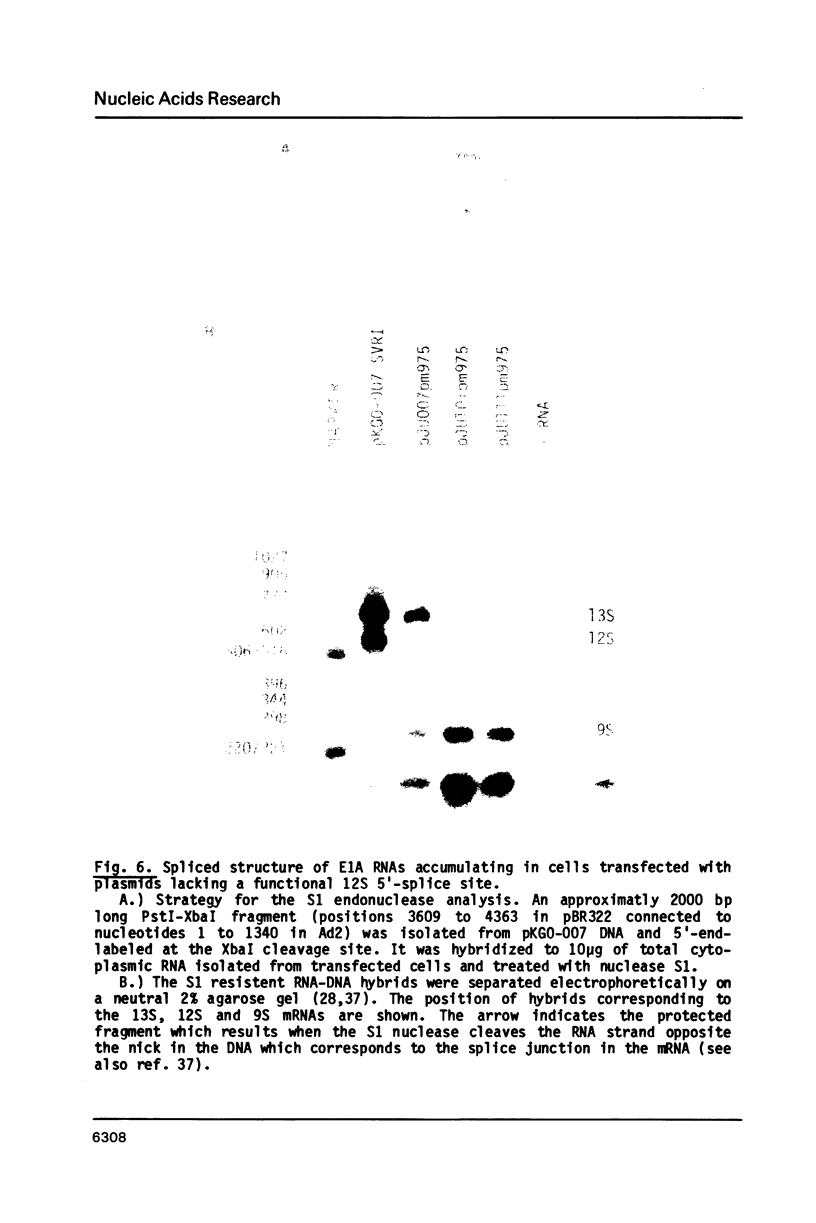


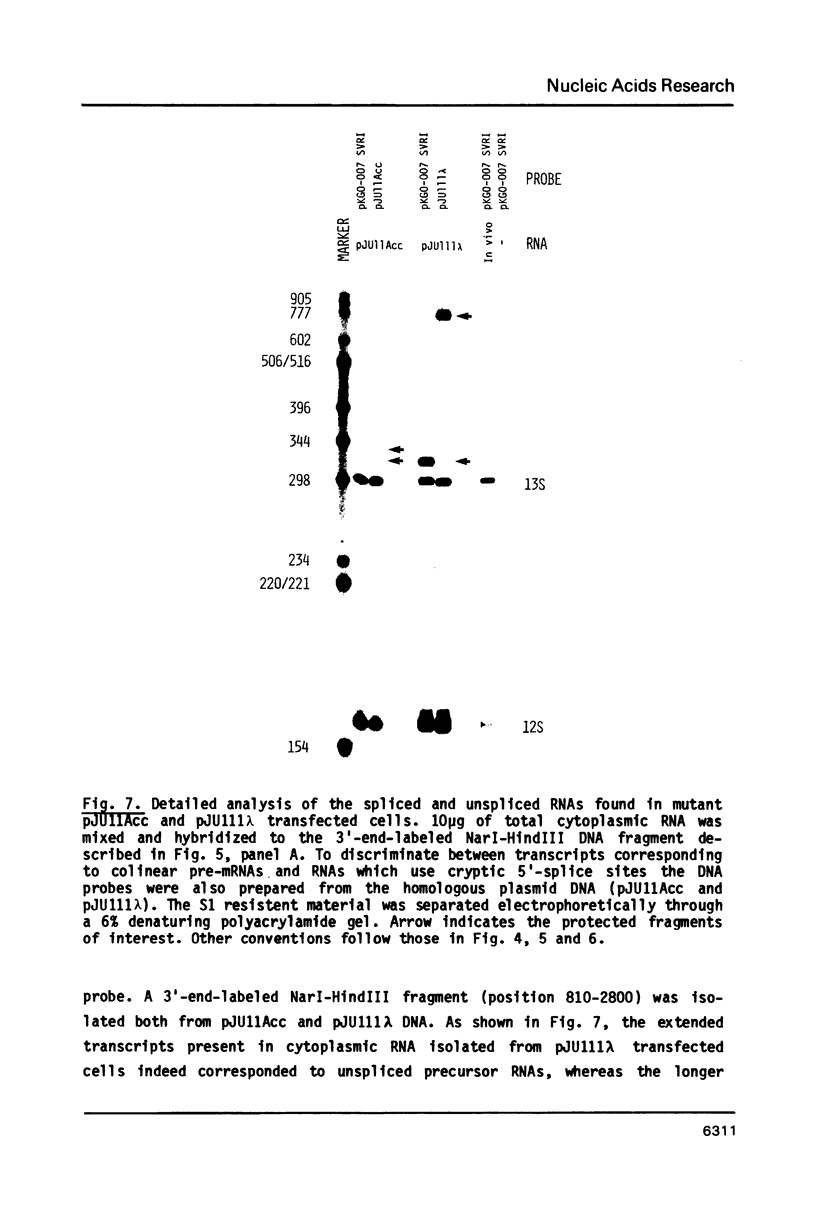




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banerji J., Rusconi S., Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981 Dec;27(2 Pt 1):299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- Berget S. M., Moore C., Sharp P. A. Spliced segments at the 5' terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3171–3175. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Lee F., Harrison T., Williams J., Sharp P. A. Pre-early adenovirus 5 gene product regulates synthesis of early viral messenger RNAs. Cell. 1979 Aug;17(4):935–944. doi: 10.1016/0092-8674(79)90333-7. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Structure of the adenovirus 2 early mRNAs. Cell. 1978 Jul;14(3):695–711. doi: 10.1016/0092-8674(78)90252-0. [DOI] [PubMed] [Google Scholar]
- Borrelli E., Hen R., Chambon P. Adenovirus-2 E1A products repress enhancer-induced stimulation of transcription. Nature. 1984 Dec 13;312(5995):608–612. doi: 10.1038/312608a0. [DOI] [PubMed] [Google Scholar]
- Brawerman G., Mendecki J., Lee S. Y. A procedure for the isolation of mammalian messenger ribonucleic acid. Biochemistry. 1972 Feb 15;11(4):637–641. doi: 10.1021/bi00754a027. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Gallwitz D. Construction of a yeast actin gene intron deletion mutant that is defective in splicing and leads to the accumulation of precursor RNA in transformed yeast cells. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3493–3497. doi: 10.1073/pnas.79.11.3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. K., Reddy V. B., Swinscoe J., Lebowitz P., Weissman S. M. Heterogeneity and 5'-terminal structures of the late RNAs of simian virus 40. J Mol Biol. 1978 Dec 25;126(4):813–846. doi: 10.1016/0022-2836(78)90022-0. [DOI] [PubMed] [Google Scholar]
- Graham F. L., Harrison T., Williams J. Defective transforming capacity of adenovirus type 5 host-range mutants. Virology. 1978 May 1;86(1):10–21. doi: 10.1016/0042-6822(78)90003-x. [DOI] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Hardy S. F., Grabowski P. J., Padgett R. A., Sharp P. A. Cofactor requirements of splicing of purified messenger RNA precursors. Nature. 1984 Mar 22;308(5957):375–377. doi: 10.1038/308375a0. [DOI] [PubMed] [Google Scholar]
- Hernandez N., Keller W. Splicing of in vitro synthesized messenger RNA precursors in HeLa cell extracts. Cell. 1983 Nov;35(1):89–99. doi: 10.1016/0092-8674(83)90211-8. [DOI] [PubMed] [Google Scholar]
- Houweling A., van den Elsen P. J., van der Eb A. J. Partial transformation of primary rat cells by the leftmost 4.5% fragment of adenovirus 5 DNA. Virology. 1980 Sep;105(2):537–550. doi: 10.1016/0042-6822(80)90054-9. [DOI] [PubMed] [Google Scholar]
- Jones N., Shenk T. An adenovirus type 5 early gene function regulates expression of other early viral genes. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3665–3669. doi: 10.1073/pnas.76.8.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones N., Shenk T. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell. 1979 Jul;17(3):683–689. doi: 10.1016/0092-8674(79)90275-7. [DOI] [PubMed] [Google Scholar]
- Kitchingman G. R., Westphal H. The structure of adenovirus 2 early nuclear and cytoplasmic RNAs. J Mol Biol. 1980 Feb 15;137(1):23–48. doi: 10.1016/0022-2836(80)90155-2. [DOI] [PubMed] [Google Scholar]
- Konarska M. M., Grabowski P. J., Padgett R. A., Sharp P. A. Characterization of the branch site in lariat RNAs produced by splicing of mRNA precursors. Nature. 1985 Feb 14;313(6003):552–557. doi: 10.1038/313552a0. [DOI] [PubMed] [Google Scholar]
- Krainer A. R., Maniatis T., Ruskin B., Green M. R. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984 Apr;36(4):993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- Montell C., Berk A. J. Elimination of mRNA splicing by a point mutation outside the conserved GU at 5' splice sites. Nucleic Acids Res. 1984 May 11;12(9):3821–3827. doi: 10.1093/nar/12.9.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins J. R. Mechanism of activation of early viral transcription by the adenovirus E1A gene product. Cell. 1981 Oct;26(2 Pt 2):213–220. doi: 10.1016/0092-8674(81)90304-4. [DOI] [PubMed] [Google Scholar]
- Ofverstedt L. G., Hammarström K., Balgobin N., Hjertén S., Pettersson U., Chattopadhyaya J. Rapid and quantitative recovery of DNA fragments from gels by displacement electrophoresis (isotachophoresis). Biochim Biophys Acta. 1984 Jun 16;782(2):120–126. doi: 10.1016/0167-4781(84)90014-9. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Konarska M. M., Grabowski P. J., Hardy S. F., Sharp P. A. Lariat RNA's as intermediates and products in the splicing of messenger RNA precursors. Science. 1984 Aug 31;225(4665):898–903. doi: 10.1126/science.6206566. [DOI] [PubMed] [Google Scholar]
- Perricaudet M., Akusjärvi G., Virtanen A., Pettersson U. Structure of two spliced mRNAs from the transforming region of human subgroup C adenoviruses. Nature. 1979 Oct 25;281(5733):694–696. doi: 10.1038/281694a0. [DOI] [PubMed] [Google Scholar]
- Rautmann G., Matthes H. W., Gait M. J., Breathnach R. Synthetic donor and acceptor splice sites function in an RNA polymerase B (II) transcription unit. EMBO J. 1984 Sep;3(9):2021–2028. doi: 10.1002/j.1460-2075.1984.tb02085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruskin B., Krainer A. R., Maniatis T., Green M. R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984 Aug;38(1):317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Hong G. F., Hill D. F., Petersen G. B. Nucleotide sequence of bacteriophage lambda DNA. J Mol Biol. 1982 Dec 25;162(4):729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- Spector D. J., McGrogan M., Raskas H. J. Regulation of the appearance of cytoplasmic RNAs from region 1 of the adenovirus 2 genome. J Mol Biol. 1978 Dec 15;126(3):395–414. doi: 10.1016/0022-2836(78)90048-7. [DOI] [PubMed] [Google Scholar]
- Svensson C., Akusjärvi G. Adenovirus 2 early region 1A stimulates expression of both viral and cellular genes. EMBO J. 1984 Apr;3(4):789–794. doi: 10.1002/j.1460-2075.1984.tb01886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson C., Pettersson U., Akusjärvi G. Splicing of adenovirus 2 early region 1A mRNAs is non-sequential. J Mol Biol. 1983 Apr 15;165(3):475–495. doi: 10.1016/s0022-2836(83)80214-9. [DOI] [PubMed] [Google Scholar]
- Treisman R., Novak U., Favaloro J., Kamen R. Transformation of rat cells by an altered polyoma virus genome expressing only the middle-T protein. Nature. 1981 Aug 13;292(5824):595–600. doi: 10.1038/292595a0. [DOI] [PubMed] [Google Scholar]
- Treisman R., Orkin S. H., Maniatis T. Specific transcription and RNA splicing defects in five cloned beta-thalassaemia genes. Nature. 1983 Apr 14;302(5909):591–596. doi: 10.1038/302591a0. [DOI] [PubMed] [Google Scholar]
- Velcich A., Ziff E. Adenovirus E1a proteins repress transcription from the SV40 early promoter. Cell. 1985 Mar;40(3):705–716. doi: 10.1016/0092-8674(85)90219-3. [DOI] [PubMed] [Google Scholar]
- Virtanen A., Pettersson U. The molecular structure of the 9S mRNA from early region 1A of adenovirus serotype 2. J Mol Biol. 1983 Apr 15;165(3):496–499. doi: 10.1016/s0022-2836(83)80215-0. [DOI] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieringa B., Hofer E., Weissmann C. A minimal intron length but no specific internal sequence is required for splicing the large rabbit beta-globin intron. Cell. 1984 Jul;37(3):915–925. doi: 10.1016/0092-8674(84)90426-4. [DOI] [PubMed] [Google Scholar]
- Wieringa B., Meyer F., Reiser J., Weissmann C. Unusual splice sites revealed by mutagenic inactivation of an authentic splice site of the rabbit beta-globin gene. Nature. 1983 Jan 6;301(5895):38–43. doi: 10.1038/301038a0. [DOI] [PubMed] [Google Scholar]
- Wigler M., Pellicer A., Silverstein S., Axel R. Biochemical transfer of single-copy eucaryotic genes using total cellular DNA as donor. Cell. 1978 Jul;14(3):725–731. doi: 10.1016/0092-8674(78)90254-4. [DOI] [PubMed] [Google Scholar]
- Wilson M. C., Fraser N. W., Darnell J. E., Jr Mapping of RNA initiation sites by high doses of uv irradiation: evidence for three independent promoters within the left 11% of the Ad-2 genome. Virology. 1979 Apr 15;94(1):175–184. doi: 10.1016/0042-6822(79)90447-1. [DOI] [PubMed] [Google Scholar]