Abstract
Spindle assembly checkpoint kinase Mps1 is spatially and temporally regulated during cell cycle progression. Mps1 is predominately localized to the cytosol in interphase cells, whereas it is concentrated on kinetochores in prophase and prometaphase cells. The timing and mechanism of Mps1 redistribution during cell cycle transition is currently poorly understood. Here, we show that Mps1 relocates from the cytosol to the nucleus at the G2/M boundary prior to nuclear envelope breakdown (NEB). This timely translocation depends on two tandem LXXLL motifs in the N terminus of Mps1, and mutations in either motif abolish Mps1 nuclear accumulation. Furthermore, we found that phosphorylation of Mps1 Ser80 (which is located between the two LXXLL motifs) also plays a role in regulating timely nuclear entry of Mps1. Mps1 that is defective in LXXLL motifs has near wild-type kinase activity. Moreover, the kinase activity of Mps1 appears to be dispensable for nuclear translocation, as inhibition of Mps1 by a highly specific small-molecule inhibitor did not perturb its nuclear entry. Remarkably, translocation-deficient Mps1 can mediate activation of spindle assembly checkpoint response; however, it fails to support a sustained mitotic arrest upon prolonged treatment with nocodazole. The mitotic slippage can be attributed to precocious degradation of Mps1 in the arrested cells. Our studies reveal a novel cell cycle-dependent nuclear translocation signal in the N terminus of Mps1 and suggest that timely nuclear entry could be important for sustaining spindle assembly checkpoint responses.
Key words: Mps1, spindle assembly checkpoint, nuclear import, kinase, protein degradation
Introduction
Accurate separation of the duplicated genome is essential for cell survival. Faithful segregation of chromosomes during mitotic cell division is safeguarded by the spindle assembly checkpoint (SAC).1 SAC is an evolutionarily conserved mechanism that operates in prometaphase to prevent the onset of anaphase until every chromosome has successfully attached to the spindles. Defects in spindle assembly checkpoint are implicated in tumorigenesis.2,3 Components of the SAC signaling pathway have been identified from yeast to human.1,2 The SAC signaling converges on Cdc20, a key activator of the ubiquitin E3 ligase anaphase promoting complex/cyclosome (APC/C), which targets Cyclin B and Securin to proteolysis. SAC inactivates Cdc20 by promoting the binding of BubR1, Bub3, Mad2 and Cdc20 to APC/C, thereby precluding substrates recruitment of APC/CCdc20. Mps1, a dual-specificity protein kinase, is a core component of spindle assembly checkpoint and apparently acts upstream in the SAC signaling cascade. The activity of Mps1 is required for activation of SAC in response to unoccupied kinetochores and the lack of tension between the sister chromatids.1 Besides its critical function in SAC, Mps1 is also required for chromosome alignment and cytokinesis during mitosis.4 The abundance and activity of Mps1 is cell cycle regulated,5,6 with both peaking in mitosis. Finally, Mps1 also may play a role in centrosome duplication in mammalian cells, as overexpression or depletion of Mps1 perturbs the normal centrosome duplication cycle.7
Regulated translocation and restricted subcellular localization of signal-transducing proteins are some of the most prevailing mechanisms in controlling of cell signaling pathways.8 Consistent with a myriad of functions of Mps1, the subcellular localization of Mps1 is both spatially and temporally regulated during cell cycle progression.9 In interphase cells, Mps1 primarily resides within the cytosol, centrosomes and nuclear envelope.7,13 Upon mitotic entry, Mps1 relocates to kinetochores, from which it is abruptly disassociated and diffusely redistributed throughout the cytosol as the metaphase plate is formed.13 The binding of Mps1 to kinetochores is a very dynamic process, with retention t1/2 of about 10 sec.10 Timely unloading of Mps1 from kinetochore in metaphase is required for anaphase onset.11 Although spectacular changes in subcellular localization of Mps1 have been observed during cell cycle progression, the biological significance and the underlying mechanism that are responsible for the dynamic behavior of Mps1 remain largely unknown.
We have begun to reveal the dynamic changes of Mps1 and how these changes are coordinated during cell cycle progression. Using a combined approach of live cell imaging, mutagenesis and chemical biology, we characterized the timing and mechanisms that govern Mps1 relocation from cytosol to the nucleus. We found that Mps1 enters the nucleus prior to nuclear envelope breakdown (NEB). Nuclear import of Mps1 requires two LXXLL motifs in the N terminus of Mps1. Disruption of either motif abolishes Mps1 nuclear accumulation without affecting Mps1 kinetochore localization. Perturbation of Mps1 nuclear import has no apparent effect on activation of spindle assembly checkpoint, instead, the timing of nuclear entry may affect the sustainability of spindle assembly checkpoint.
Results
Mps1 is imported to the nucleus prior to nuclear envelope breakdown.
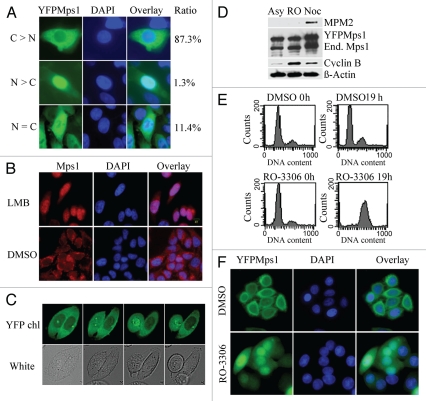
During interphase, Mps1 is found predominately in the cytoplasm, centrosome and nuclear membrane.7,9,12,13 As cells move into mitosis, Mps1 is relocated to kinetochores in pro-phase and remains there until metaphase, at which point Mps1 is abruptly dissociated from kinetochore and redistributed in cytosol. Mps1 is excluded from the nucleus upon reformation of nuclear envelope. The dynamic subcellular localization of Mps1 can be faithfully recapitulated with fluorescence protein-tagged Mps1 (e.g., YFP-Mps1) in live cells.9,13 While subcellular localization of Mps1 in interphase and mitosis are well documented, the timing of Mps1 redistribution during G2/M transition is poorly characterized. Therefore, we choose to investigate this aspect of Mps1 dynamic change using a SW480 cell line that stably expresses YFP-Mps1. As expected, YFP-Mps1 is predominantly localized to the cytoplasm (Fig. 1A). The cytoplasmic localization of Mps1 during interphase could be a result of the rate of export exceeding the rate of import. CRM1-dependent nuclear export is one of major exporting pathways. This pathway can be inactivated with Leptomycin B, a specific and potent inhibitor of CRM1 that perturbs nuclear export by glycosylating a key cysteine residue.14 To test whether CRM1 is involved in Mps1 export from the nucleus, YFP-Mps1 cells were treated with Leptomycin B. As shown in Figure 1B, YFP-Mps1 becomes predominantly localized to within the nucleus upon treatment with Leptomcyin B. This result suggests that Mps1 can shuttle between nucleus, and that cytoplasm and CRM1 is involved in Mps1 export in interphase cells. The rate of Mps1 export exceeds the rate of nuclear import during interphase.
Figure 1.
Mps1 is imported into the nucleus at the G2/M boundary. (A) Subcellular localization of YFP-Mps1 in asynchronized SW480 cells. Distribution of YFP-Mps1 was classified into three categories. Representative images are shown. The percentage of cells in each category was determined from inspecting images of at least 200 cells. (B) Localization of endogenous Mps1 after Leptomycin B (LMB) treatment for 2 h. (C) Live cell imaging of SW480-YFPMps1 cell at the G2/M boundary or in G1/S phase. Cells were incubated in an environmental chamber. Images from YFP channel or bright field were taken every 15 min. (D and E) Treatment of RO-3306 blocks SW480YFP-Mps1 cells at G2/M, with increased Cyclin B expression without phosphorylation of MPM-2, a typical mitotic marker. SW480YFP-Mps1 cells treated with DMSO or RO-3306 for 0 or 19 h were harvested and stained with propidium iodine (PI) prior to FACS analysis using FACScan. RO-3306-treated but not DMOS-treated cells show 4X DNA content, with G2/M arrest. (F) YFP-Mps1 is translocated into the nucleus in cells arrested at G2/M following RO-3306 treatment. Nuclei are identified by DAPI staining.
To precisely measure Mps1 redistribution prior to or after mitotic entry, we performed time-lapse live cell imaging analysis of YFP-Mps1 subcellular localization during G2/M transition. As shown in Figure 1C, we found that Mps1 predominantly resides within the cytoplasm and centrosome during this transition. Mps1 moved into the nucleus at G2/M transition prior to nuclear envelope breakdown (cell on the left), which is characterized by ongoing segregation of the centrosomes (identified by the centrosome-bound YFP-Mps1). Fifteen minutes later, YFP-Mps1 becomes concentrated on kinetochores. In contrast, YFP-Mps1 in the interphase cell remains predominantly in the cytoplasm (cell on the right). Thus, Mps1 is imported into the nucleus prior to nuclear envelope breakdown during unperturbed cell cycle progression.
Mitotic entry is primarily driven by cyclin B-Cdk1. To determine whether Mps1 nuclear import depends on the activity of cyclin B-Cdk1 kinase, we treated YFP-Mps1 cells with a specific Cdk1 inhibitor, RO-3306, which causes reversible cell cycle arrest at G2/M boundary.15 In agreement with our previous observation of wild-type SW480, SW480YFP-Mps1 cells undergo robust growth arrest by RO-3306 at the G2/M boundary, which can be identified by 4 N DNA content and negative staining of mitotic marker MPM-2 (Fig. 1D and E). As seen in the unperturbed G2/M transition, cells arrested by RO-3306 treatment exhibit pronounced nuclear staining of Mps1 (Fig. 1F). This result again suggests that Mps1 is imported into the nucleus prior to mitotic entry and nuclear envelope breakdown. Moreover, nuclear import of Mps1 is independent of the activity of cyclin B-cdk1.
Two LXXLL motifs are required for Mps1 Nuclear localization.
A prevailing mechanism for importing proteins into the nucleus is through the importin pathway. Frequently, importindependent pathways involve the binding of cargo protein to the NLS receptor importin-α, which interacts with the transporter, importin-β, to form a trimeric complex that carries the cargo protein through the nuclear pore.16 Classical NLSs are either monopartite or bipartite and are marked by clusters of basic amino acids.16 These types of NLSs are typically recognized by importin-α. Another class of NLSs directly binds importin β; unfortunately, there is significant diversity of this second type of NLS, making it difficult to predict by sequence alone. A recent study suggests that R/H/KXPY(2–5) may be a consensus NLS for a subtype of importin β (also known as Kapβ2).17 To identify potential NLSs for Mps1 nuclear import, we initially employed a NLS prediction program (NLStradamus or PredictNLS) to search for candidate NLSs in Mps1. The NLS prediction program identified a motif at the extreme C terminus of Mps1(852KKR GKK857) as a putative NLS. We proceeded to construct a mutant YFP-Mps1 without the putative NLS (YFP-Mps1ΔCT), which was tested in SW480 cells. Removal of this putative NLS has little impact on Mps1 nuclear import when cells were blocked at G2/M boundary (data not shown), suggesting that there are either multiple NLSs in Mps1 or other types of NLSs involved in Mps1 nuclear import.
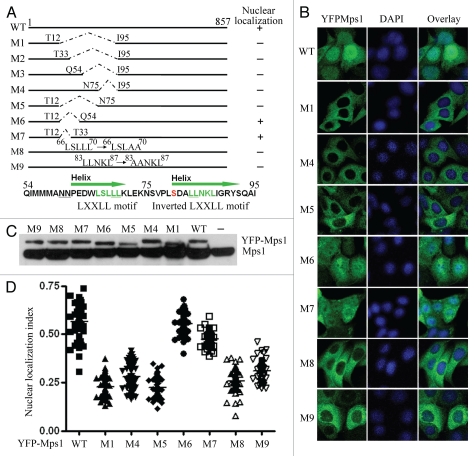
To experimentally define the NLS, we tested an array of Mps1 deletion mutants from our previous studies in reference 6 and 9. Interestingly, we discovered that an N-terminal deletion mutant of Mps1 (M1) abrogates Mps1 nuclear import (Fig. 2A and B). To find the minimal fragment of Mps1 required for its nuclear localization, we generated a set of YFP-tagged Mps1 mutants and stably expressed them in SW480 cells (Fig. 2A and C). The sub-cellular distributions of these mutants were then determined after arrest at the G2/M boundary by the treatment of RO-3306. As shown in Figure 2A and B, the fragment spanning from Q54 to I95 is required for Mps1 nuclear import. After cross-referencing species orthologs, including human, mouse and Xenopus (Fig. S2), we found two LXXLL motifs (L denotes leucine and X represents any amino acid) arranged in tandem that appear to be evolutionarily conserved (Figs. 2A and S2). Replacing leucine residues in either LXXLL motifs (L68L69→AA in M8 or L83 L84→AA in M9) abrogates nuclear import of Mps1 in G2/M (Fig. 2B). In contrast, replacing two tandem Asparagines adjacent to the first LXXLL motif with alanines (N60N61→AA) has no effect on Mps1 nuclear localization (data not shown). The motif LXXLL was initially found in transcriptional cofactors, which mediate the protein-protein interactions of cofactors with nuclear receptors.18–20 The interaction mediated by the LXXLL motif has been seen to influence a variety of processes, including the subcellular localization,21,22 transcriptional activity,23,24 NFκB signaling pathway,25 TGFβ signaling pathway26 and protein degradation.27 The finding that the two LXXLL motifs in Mps1 are critical for nuclear import suggests these motifs likely mediate Mps1 interaction either directly or indirectly, with factors that control the cell cycle-dependent nuclear import pathway.
Figure 2.
Determining the requirement of Mps1 nuclear entry. (A) Schematic map of Mps1 mutants. The sequence elements that are responsible for Mps1 nuclear localization include two LXXLL motifs. (B) Representative images of YFP-tagged wild-type Mps1 and Mps1 mutants after incubation with 9 µM RO-3306 for 19 h. Nuclei were identified by DAPI staining. (C) Immunoblot analysis of the expression of ecotopically expressed YFP-tagged wild-type and mutant Mps1 along with the endogenous Mps1 using an anti-Mps1 (Millipore) raised against the N terminus of Mps1. (D) Quantification of Mps1 nuclear translocation upon RO-3306 treatment. About 50–70 cells for each Mps1 mutant were counted and quantified using the scheme described in Figure S1.
Mps1 kinase activity is dispensable for its nuclear translocation.
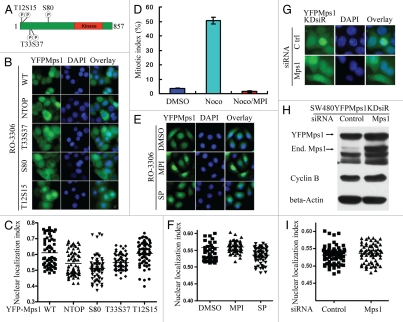
The binding specificity of a coactivator to nuclear receptor can be also determined by the sequence flanking the LXXLL motif.28,29 Phosphorylation of amino acids in the flanking region may influence this specificity.30 Previous results have shown that three serines and two threonines in the N-terminal of Mps1 can be auto-phosphorylated.9,31 Ser 80 is localized between M8 and M9 (Fig. 2A). To test whether phosphorylation modification of this site may potentially regulate Mps1 nuclear import, we analyzed the nuclear entry of YFP-Mps1 with five non-phosphomimicing amino acids (NTOP), single mutant YFP-Mps1S80A, YFP-Mps1T33S37AA and YFP-Mps1T12S15AA. As shown in Figure 3B and C, the mutants YFP fused NTOP, S80A and T33S37AA showed impaired Mps1 nuclear entry, and the effect of YFP-Mps1S80A is most dramatic. These results suggest the phosphorylation of S80, at least in part, affects Mps1 nuclear localization.
Figure 3.
The kinase activity of Mps1 is not required for Mps1 nuclear entry. (A) The relevant phosphorylation sites in the N terminus of Mps1 are shown. (B) Subcellular localization YFP-tagged Mps1 and Mps1 mutants in SW480 cells were imaged after RO-3306 treatment for 19 h. Shown were representative images for each mutant. At least 50–70 cells were inspected. (C) A scatter plot of the distribution of results obtained in (B). The average distribution for each sample is shown as a horizontal bar. Using SNK grouping and the Kruskal-Wallis test, the difference of nuclear localization index between YFPMps1T33S37AA and NTOP is not identified as significant (p > 0.05), while the difference between S80 and NTOP is significant (p < 0.0001). There is no statistical difference between WT and T12S15 (p > 0.05). (D) MPI-0479605(MPI), a specific inhibitor of Mps1 kinase activity abolishes the spindle assembly checkpoint arrest induced by nocodazole (100 ng/ml). (E) Nuclear accumulation of YFP-Mps1 is unaffected by co-treatment with MPI-0479605 (10 µM or SP600125 (10 µM) and RO-3306 for 12 h after released from double thymidine treatment. (F) A scatter plot of the distribution of YFP-Mps1 treated with control, MPI-0479605 or SP600125. About 50–70 cells for each treatment were counted and quantified. (G) Subcellular localization of YFP-Mps1KD upon treatment with RO-3306 in the presence or absence of the endogenous Mps1. SW480 cells stably expressing YFP-Mps1KDsiR were transfected with either control or Mps1 siRNA. (H) Depletion of the endogenous Mps1 is verified by immunoblotting with an anti-Mps1 antibody (Millipore). β-actin was blotted as loading control. Cyclin B expression is indicative of G2/M arrest. (I) Quantitation plot of subcellular localization of YFP-Mps1KDsiR treated with control or Mps1 siRNA. There is no significant statistical difference among DMSO, SP and MPI compound groups or between control and Mps1 siRNA groups (p > 0.05) in (F and I).
Since some of these autophosphorylation sites may also be transphosphorylated by other kinases, we next tested whether Mps1 activity is required for Mps1 nuclear translocation using MPI-0479605 (Myrexis),32 a small-molecule inhibitor of Mps1 similar to reversine33 but with improved selectivity and potency (IC50 < 5 nM). In agreement with the results of siRNA knockdown of Mps112,34,35 or inhibition with other small-molecule inhibitors,33,36–38 cells exposed to MPI-0479605 have no spindle assembly checkpoint (Fig. 3D). However, we failed to observe any nuclear import defect in MPI-0479605-treated cells. A similar result was also obtained using a relatively non-specific Mps1 inhibitor SP600125 (Fig. 3E and F). Further, we examined the localization of YFP-tagged kinase-dead Mps1 (YFPMps1KDsiR) in depletion of the endogenous Mps1 by siRNA at G2/M boundary. As shown in Figure 3G–I, the nuclear accumulation of YFPMps1KDsiR was not affected by Mps1 siRNA treatment. Taken together, these results suggest that, while Mps1 phosphorylation at certain sites may regulate Mps1 nuclear import, the kinase activity of Mps1, per se, is not essential for its translocation into the nucleus during G2/M transition.
Integrity of LXXLL motifs is essential for sustaining spindle assembly checkpoint arrest.
Perturbation of Mps1 nuclear import should only cause a short delay in Mps1's access to components in the nuclear compartment as the nuclear envelope breakdown occurs shortly after Mps1 translocation to the nucleus. To address the potential importance of timely nuclear import of Mps1, we investigated the impact of LXXLL motifs on Mps1 function in mitosis. First, we examined whether inactivation of LXXLL motif impedes kinetochore localization of Mps1 upon triggering of spindle assembly checkpoint by treatment with nocodazole. As shown in Figure 4A, kinetochore localization of YFP-tagged M8 and M9 is indistinguishable from wild-type or N60N61 mutant, suggesting that either mutations alone are unlikely to affect kinetochore recruitment of Mps1. Next, we assessed whether LXXLL motif affects spindle assembly checkpoint activation by Mps1. In order to compare wild-type and mutant Mps1 in the absence of endogenous Mps1, we created SW480 cell lines stably expressing untagged wild-type and mutant Mps1 that are siRNA-insensitive (Mps1R) through scrambling the coding sequence. Upon transfection of siRNA, the endogenous Mps1 will be depleted, leaving cells with ectopically expressed Mps1 or Mps1 mutants to regulate mitosis. Immunofluorescence studies confirm that the sub-cellular localization profiles of wild-type and mutant Mps1 are identical to their YFP counterparts, indicating that the effect of LXXLL mutation on Mps1 nuclear import is independent of the endogenous Mps1 (Fig. S3).
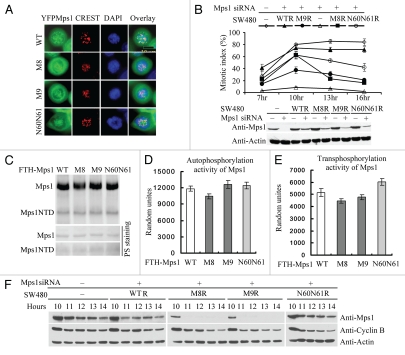
Figure 4.
The two LXXLL motifs in the N terminus of Mps1 were essential for Mps1 stability and sustained spindle assembly checkpoint arrest. (A) The LXXLL motifs in the Mps1 N terminus were dispensable for Mps1 kinetochore localization. YFP-tagged wild-type and Mps1 mutants were treated with nocodazole (100 ng/ml) for 12 h after released from a double thymidine treatment protocol. Representative images of prometaphase arrest cells are shown. (B) The LXXLL motifs are critical for sustained spindle assembly checkpoint maintenance. Codon scrambled wild-type and mutant Mps1 immune to siRNA targeting were stably expressed in SW480 cells. The spindle assembly checkpoint responses were measured after the endogenous Mps1 was depleted by treatment with the Mps1 siRNA at indicated time points. The efficiency of siRNA knockdown was determined by western blotting. (C–E) Mutation of the LXXLL motifs does not significantly affect the kinase activity of Mps1. Flag-tagged wild-type and mutant Mps1 were expressed in 293T cells via transient transfection. Forty-eight h after transfection, cells are harvested and lyzed. Flag-tagged wild-type and mutant Mps1 proteins were immunoprecipitated from cell lysates and subjected to in vitro kinase assay using Mps1NTD as a substrate as described previously in reference 6. (F) Precocious degradation of Mps1 mutants defective in the LXXLL motifs in the checkpoint arrested cells. Stable cell lines expressing wild-type and mutant Mps1 were synchronized by double thymidine treatment and then released into medium in the presence of nocodazole (100 ng/ml) for 12 h. The levels of Mps1 at indicated time points were measured by immunoblotting.
Wild-type or mutant Mps1 cells treated with or without Mps1 siRNA were first synchronized with double thymidine treatment followed by incubation with nocodazole and were harvested at the indicated time after release into nocodazole-containing growth medium. The mitotic index was then determined. As shown in Figure 4B, SW480 cells treated with the control siRNA undergo mitotic arrest for more than 15 h. Mitotic arrest is abolished in parental SW480 cells when the endogenous Mps1 is depleted by Mps1 siRNA. As expected, ectopic expression of siRNA-resistant Mps1 in SW480 cells rescues the defect caused by siRNA depletion. In contrast, SW480 cells expressing siRNA-resistant M8 (M8R) and M9 (M9R) fail to sustain robust mitotic arrest upon treatment with nocodazole, while the control mutant N60N61R is similar to the wild-type Mps1 (WTR). In addition, Both M8R and M9R show significant spindle assembly checkpoint slippage phenotype. Furthermore, the M9R mutant seems to have additional defects in activation of spindle assembly checkpoint. Defects of M8 and M9 in spindle checkpoint function could be a result of compromised Mps1 kinase activity or abundance. The former possibility is unlikely to be the case as the kinase activity of M8 and M9 is very similar to wild type in IP-kinase assay (Fig. 4C–E). To measure the abundance of wild-type and mutant Mps1 at different time points of nocodazole treatment, we blotted cell extracts for Mps1, Cyclin B and Actin. Remarkably, the steady-state levels of wild-type and mutant Mps1 are similar at 10 h post nocodazole treatment. However, M8R and M9R levels decline drastically to undetectable levels after 10 h compared with the level of cyclin B (Fig. 4F). This observation suggests that defects in Mps1 mutants in sustaining spindle assembly checkpoint can probably be attributed to decreased protein levels due to precocious degradation of the mutant proteins. Thus, the LXXLL motifs of Mps1 appear to be important for both nuclear import and sustaining spindle assembly checkpoint arrest.
Discussion
The activity and subcellular localization of Mps1 is dynamically regulated during cell cycle progression. The precise timing of Mps1 redistribution during the G2/M transition is unknown. We show that Mps1 enters the nucleus at the G2/M boundary prior to nuclear envelope breakdown (NEB) and nuclear import of Mps1 requires two tandem LXXLL motifs in the N terminus of Mps1. The kinase activity of Mps1 appears to be dispensable for its nuclear translocation; however, phosphorylation of Mps1 may regulate the efficiency of Mps1 nuclear entry. Furthermore, we demonstrated that Mps1 mutants that are deficient in nuclear import at the G2/M transition cannot sustain spindle assembly checkpoint responses, suggesting that timing of Mps1 accumulation in the nucleus could be important for maintaining robust spindle assembly checkpoint responses.
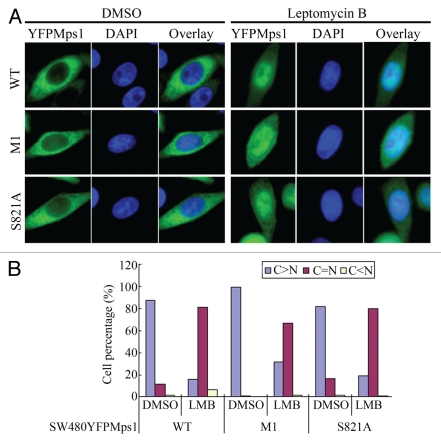
Nuclear accumulation of Mps1 at the G2/M boundary, in theory, could also be a result of a decreasing rate of Mps1 nuclear export. Indeed, the two LXXLL motifs appear to bear more resemblance to the classical nuclear export signal than nuclear import signal. However, this notion is unlikely to be true, as the removal of the two LXXLL motifs does not lead to Mps1 nuclear accumulation (Fig. 5A and B). Furthermore, nuclear accumulation of Mps1 upon treatment with Leptomycin B is unaffected by the removal of the two LXXLL motifs (Fig. 5A and B). Thus, these data are inconsistent with notion that these motifs are nuclear export signals. Future studies are necessary to identify the bona fide CRM1-dependent nuclear export signal in Mps1.
Figure 5.
The region including two LXXLL motifs is not involved in CRM1-dependent nuclear export of Mps1. (A) YFP-tagged wild-type and mutant Mps1 loss two LXXLL motifs (M1) or phosphorylation at S821 are treated with DMSO or Leptomycin B (200 ng/ml) for 2 h prior to imaging. (B) Quantitation of subcellular localization of wild-type, LXXLL deletion and S821 Mps1 upon treatment with Leptomycin B.
Regulated subcellular localization of master mitotic regulators such as cyclin B and Cdk1 have been shown to play an important role in controlling the timing and robustness of mitotic events.39 Orderly progression through mitosis relies not only on oscillating activity of master regulators, but also on a series of peripheral oscillators controlling individual events entrained by the master regulator.40,41 Perhaps the spindle assembly checkpoint timer is entrained with the master regulator, such as the right protein is degraded at the right time.42 Delayed nuclear entry of Mps1 caused by disruption of the LXXLL motif may very well perturb the correct coupling of the checkpoint timer with the master regulator.
The mechanism underlying destabilization of Mps1 in checkpoint-arrested cells is unclear. We can only speculate that disruption of the LXXLL motifs may disengage a normal binding partner of Mps1 that mediates Mps1 nuclear import and protects Mps1 from being recognized by the proteolytic degradation machinery, although we cannot rule out that the LXXLL motifs are recognized by different binding partners in G2/M vs. prometaphase-arrested cells. A likely candidate responsible for precocious degradation of mutant Mps1 is APC/C. In budding yeast, Mps1 is degraded by APC/CCdc20 at the onset of anaphase.43 Ubiquitination of Mps1 by APC/CCdc20 and APC/CCdh1 has been demonstrated in human cells and proposed as a mechanism for clearance of Mps1 in late mitosis and interphase.44 In this vein, Mps1 interaction with its partners via the LXXLL motifs may obstruct recognition of the D-box by Cdc20 or Cdh1 in a cell cycle-dependent manner. Regardless of the exact mechanism of destabilization of Mps1 as a result of disruption of LXXLL motif, dynamic changes in Mps1 subcellular localization and abundance are important for its cellular function.
Materials and Methods
DnA constructs and stable cell lines generation.
Retroviral expression vector YFP-Mps1 and YFP-Mps1R in the pRex background have been described previously in reference 9. The deletion mutants of YFP-Mps1 were generated using the Quikchange mutagenesis kit (Agilent). All mutations were confirmed by DNA sequencing. The procedure for generation of SW480 stable cell lines expressing various Mps1 mutants is described in reference 9.
Cell synchronization.
Synchronization of cells at G1/S phase was accomplished by double thymidine treatment as described by Stucke et al.12 To synchronized cells at the G2/M boundary, cells were incubated with 9 µM RO-3306 (Alexis Chemicals) for 19 h. In some experiments, the double thymidine synchronization protocol was used to obtain S phase-arrested cells, followed by release into growth medium containing 9 µM RO-3306 for 12 h. The latter method was found to be more efficient in synchronizing cells at the G2/M boundary with fewer apoptotic cells.
siRNA transfection.
Control and Mps1 siRNA were purchased from Dharmacon. The sequences of control and Mps1 siRNA were described previously in reference 9. Briefly, cells were split into a 12-well plate and transfected at 20% confluence using Transfectin (Bio-Rad).
Immunoblotting analysis.
For protein gel blot analysis, cells were washed with cold D-PBS twice and lyzed in buffer [20 mM Tris-Cl (pH 8.0), 0.2 M NaCl, 0.5% NP40, 1 mM EDTA, 1 mM PMSF, 20 mM NaF, 0.1 mM Na3VO4, 1x protease inhibitors(Roche)] for 5 min prior to scraping. Cell extracts were clarified by spinning at 13,000 rpm for 10 min. The protein concentrations were determined by BSA assay. Equal amounts of total proteins were resolved in 8% or 10% SDS-PAGE gel and transferred to nitrocellulose membrane. Primary and secondary antibodies were applied sequentially. The blots were developed in Super Signal WesternDura (Pierce) according to manufacture instruction.
Immunofluorescence.
Cells for immunofluorescence were grown on cover glasses. Prior to staining, cells were washed three times with D-PBS and fixed for 10 min in D-PBS plus 1% para-formaldehyde. Anti-Mps1 and Anti-Mad2 were prepared at a 1:100 dilution. Kinetochores were identified by staining with the CREST antisera (Antibody Inc.). Secondary antibodies conjugated with Alexa Fluor 488-conjugated goat anti-mice secondary antibodies or Alexa Fluor 596-conjugated goat anti-human secondary antibodies (Invitrogen, Eugene, OR) were used depending on the source of primary antibodies. After staining, the coverglasses were mounted onto precleaned microscope slides with D-PBS plus 1.5 µg/ml DAPI and 50% glycerol and sealed with nail oil. Images were acquired on a Nikon TE2000 microscope equipped with a 63x oil objective lens and Metamorph software (Molecular Device).
Time-lapse fluorescence imaging.
SW480 cells stably expressing YFP-Mps1 were seeded in a DT 0.15 mm dish (Bioptechs) in DMEM at 50% confluence. Twelve hours prior to imaging, the fresh medium was added, and the DT dish was transferred to an environmental chamber mounted on the microscope.
Acknowledgments
We thank Drs. Daniel Wettstein, Ian McAlexander and Brandi Williams at Myrexis for a generous supply of MPI-0479605. We thank Edward Kee and Douglas Chapnick for critical readings of the manuscript and members of Liu laboratory for discussions. We also thank members of Zhong laboratory and Cao laboratory for providing reagents. This work was supported by grants from National Natural Science Foundation of China to Q.X. (30971444) and the National Institutes of Health (CA107098-01) and (GM083172) to Xuedong Liu.
Supplementary Material
References
- 1.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 2.Wassmann K, Benezra R. Mitotic checkpoints: from yeast to cancer. Curr Opin Genet Dev. 2001;11:83–90. doi: 10.1016/S0959-437X(00)00161-1. [DOI] [PubMed] [Google Scholar]
- 3.Kops GJ, Weaver BA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer. 2005;5:773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- 4.Fisk HA, Mattison CP, Winey M. A field guide to the Mps1 family of protein kinases. Cell Cycle. 2004;3:439–442. doi: 10.4161/cc.3.4.784. [DOI] [PubMed] [Google Scholar]
- 5.Stucke VM, Sillje HHW, Arnaud L, Nigg EA. Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication. EMBO J. 2002;21:1723–1732. doi: 10.1093/emboj/21.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun T, Yang X, Wang W, Zhang X, Xu Q, Zhu S, et al. Cellular abundance of Mps1 and the role of its carboxyl terminal tail in substrate recruitment. J Biol Chem. 2010;285:38730–38739. doi: 10.1074/jbc.M110.177642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisk HA, Winey M. The mouse Mps1p-like kinase regulates centrosome duplication. Cell. 2001;106:95–104. doi: 10.1016/S0092-8674(01)00411-1. [DOI] [PubMed] [Google Scholar]
- 8.Ferrell J., Jr How regulated protein translocation can produce switch-like responses. Trends Biochem Sci. 1998;23:461–465. doi: 10.1016/S0968-0004(98)01316-4. [DOI] [PubMed] [Google Scholar]
- 9.Xu Q, Zhu S, Wang W, Zhang X, Old W, Ahn N, et al. Regulation of kinetochore recruitment of two essential mitotic spindle checkpoint proteins by mps1 phosphorylation. Mol Biol Cell. 2009;20:10–20. doi: 10.1091/mbc.E08-03-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howell BJ, Moree B, Farrar EM, Stewart S, Fang G, Salmon ED. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr Biol. 2004;14:953–964. doi: 10.1016/j.cub.2004.05.053. [DOI] [PubMed] [Google Scholar]
- 11.Jelluma N, Dansen TB, Sliedrecht T, Kwiatkowski NP, Kops GJ. Release of Mps1 from kinetochores is crucial for timely anaphase onset. J Cell Biol. 2010;191:281–290. doi: 10.1083/jcb.201003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stucke VM, Sillje HH, Arnaud L, Nigg EA. Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication. EMBO J. 2002;21:1723–1732. doi: 10.1093/emboj/21.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu ST, Chan GK, Hittle JC, Fujii G, Lees E, Yen TJ. Human MPS1 kinase is required for mitotic arrest induced by the loss of CENP-E from kinetochores. Mol Biol Cell. 2003;14:1638–1651. doi: 10.1091/mbc.02-05-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, et al. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, et al. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc Natl Acad Sci USA. 2006;103:10660–10665. doi: 10.1073/pnas.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mattaj IW, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 17.Lee BJ, Cansizoglu AE, Suel KE, Louis TH, Zhang Z, Chook YM. Rules for nuclear localization sequence recognition by karyopherin beta2. Cell. 2006;126:543–558. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torchia J, Rose DW, Inostroza J, Kamei Y, Westin S, Glass CK, et al. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 19.McInerney EM, Rose DW, Flynn SE, Westin S, Mullen TM, Krones A, et al. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 21.Kawajiri K, Ikuta T, Suzuki T, Kusaka M, Muramatsu M, Fujieda K, et al. Role of the LXXLL-motif and activation function 2 domain in subcellular localization of Dax-1 (dosage-sensitive sex reversal-adrenal hypoplasia congenita critical region on the X chromosome, gene 1) Mol Endocrinol. 2003;17:994–1004. doi: 10.1210/me.2002-0360. [DOI] [PubMed] [Google Scholar]
- 22.Ikuta T, Watanabe J, Kawajiri K. Characterization of the LxxLL motif in the aryl hydrocarbon receptor: effects on subcellular localization and transcriptional activity. J Biochem. 2002;131:79–85. doi: 10.1093/oxfordjournals.jbchem.a003080. [DOI] [PubMed] [Google Scholar]
- 23.Su T, Deguchi A, Yao Y, Luo J, Weinstein IB. Dip1 inhibits growth and gene transcription in MCF-7 breast cancer cells. J Exp Ther Oncol. 2007;6:117–127. [PubMed] [Google Scholar]
- 24.Nakae J, Cao Y, Daitoku H, Fukamizu A, Ogawa W, Yano Y, et al. The LXXLL motif of murine forkhead transcription factor FoxO1 mediates Sirt1-dependent transcriptional activity. J Clin Invest. 2006;116:2473–2483. doi: 10.1172/JCI25518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jang HD, Yoon K, Shin YJ, Kim J, Lee SY. PIAS3 suppresses NFkappaB-mediated transcription by interacting with the p65/RelA subunit. J Biol Chem. 2004;279:24873–24880. doi: 10.1074/jbc.M313018200. [DOI] [PubMed] [Google Scholar]
- 26.Antonson P, Jakobsson T, Almlof T, Guldevall K, Steffensen KR, Gustafsson JA. RAP250 is a coactivator in the transforming growth factor beta signaling pathway that interacts with Smad2 and Smad3. J Biol Chem. 2008;283:8995–9001. doi: 10.1074/jbc.M707203200. [DOI] [PubMed] [Google Scholar]
- 27.Tintignac LA, Lagirand J, Batonnet S, Sirri V, Leibovitch MP, Leibovitch SA. Degradation of MyoD mediated by the SCF (MAFbx) ubiquitin ligase. J Biol Chem. 2005;280:2847–2856. doi: 10.1074/jbc.M411346200. [DOI] [PubMed] [Google Scholar]
- 28.Coulthard VH, Matsuda S, Heery DM. An extended LXXLL motif sequence determines the nuclear receptor binding specificity of TRAP220. J Biol Chem. 2003;278:10942–10951. doi: 10.1074/jbc.M212950200. [DOI] [PubMed] [Google Scholar]
- 29.McInerney EM, Rose DW, Flynn SE, Westin S, Mullen TM, Krones A, et al. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ko L, Cardona GR, Iwasaki T, Bramlett KS, Burris TP, Chin WW. Ser-884 adjacent to the LXXLL motif of coactivator TRBP defines selectivity for ERs and TRs. Mol Endocrinol. 2002;16:128–140. doi: 10.1210/me.16.1.128. [DOI] [PubMed] [Google Scholar]
- 31.Jelluma N, Brenkman AB, McLeod I, Yates JR, 3rd, Cleveland DW, Medema RH, et al. Chromosomal instability by inefficient Mps1 auto-activation due to a weakened mitotic checkpoint and lagging chromosomes. PLoS ONE. 2008;3:2415. doi: 10.1371/journal.pone.0002415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams B, Cimbora D, Bichwal V, Papac D, McAlexander I, Willardsen J, et al. MPI-0479605: A novel small-molecule inhibitor of the mitotic kinase TTK with anti-tumor activity in pre-clinical models; 100th Annual Meeting of the American Association for Cancer Research; 2009; Denver CO. [Google Scholar]
- 33.Santaguida S, Tighe A, D'Alise AM, Taylor SS, Musacchio A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small-molecule inhibitor reversine. J Cell Biol. 2010;190:73–87. doi: 10.1083/jcb.201001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fisk HA, Mattison CP, Winey M. Human Mps1 protein kinase is required for centrosome duplication and normal mitotic progression. Proc Natl Acad Sci USA. 2003;100:14875–14880. doi: 10.1073/pnas.2434156100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang J, Chen Y, Zhao Y, Yu H. Autophosphorylation-dependent activation of human Mps1 is required for the spindle checkpoint. Proc Natl Acad Sci USA. 2007;104:20232–20237. doi: 10.1073/pnas.0710519105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt M, Budirahardja Y, Klompmaker R, Medema RH. Ablation of the spindle assembly checkpoint by a compound targeting Mps1. EMBO Rep. 2005;6:866–872. doi: 10.1038/sj.embor.7400483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hewitt L, Tighe A, Santaguida S, White AM, Jones CD, Musacchio A, et al. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-C-Mad2 core complex. J Cell Biol. 2010;190:25–34. doi: 10.1083/jcb.201002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwiatkowski N, Jelluma N, Filippakopoulos P, Soundararajan M, Manak MS, Kwon M, et al. Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function. Nat Chem Biol. 2010;6:359–368. doi: 10.1038/nchembio.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takizawa CG, Morgan DO. Control of mitosis by changes in the subcellular location of cyclin-B1-Cdk1 and Cdc25C. Curr Opin Cell Biol. 2000;12:658–665. doi: 10.1016/S0955-0674(00)00149-6. [DOI] [PubMed] [Google Scholar]
- 40.Morgan DO. The hidden rhythms of the dividing cell. Cell. 2010;141:224–226. doi: 10.1016/j.cell.2010.03.042. [DOI] [PubMed] [Google Scholar]
- 41.Lu Y, Cross FR. Periodic cyclin-Cdk activity entrains an autonomous Cdc14 release oscillator. Cell. 2010;141:268–279. doi: 10.1016/j.cell.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pines J. Mitosis: a matter of getting rid of the right protein at the right time. Trends Cell Biol. 2006;16:55–63. doi: 10.1016/j.tcb.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 43.Palframan WJ, Meehl JB, Jaspersen SL, Winey M, Murray AW. Anaphase inactivation of the spindle checkpoint. Science. 2006;313:680–684. doi: 10.1126/science.1127205. [DOI] [PubMed] [Google Scholar]
- 44.Cui Y, Cheng X, Zhang C, Zhang Y, Li S, Wang C, et al. Degradation of the human mitotic checkpoint kinase Mps1 is cell cycle-regulated by APC-cCdc20 and APC-cCdh1 ubiquitin ligases. J Biol Chem. 2010;285:32988–32998. doi: 10.1074/jbc. M110.140905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.