Abstract
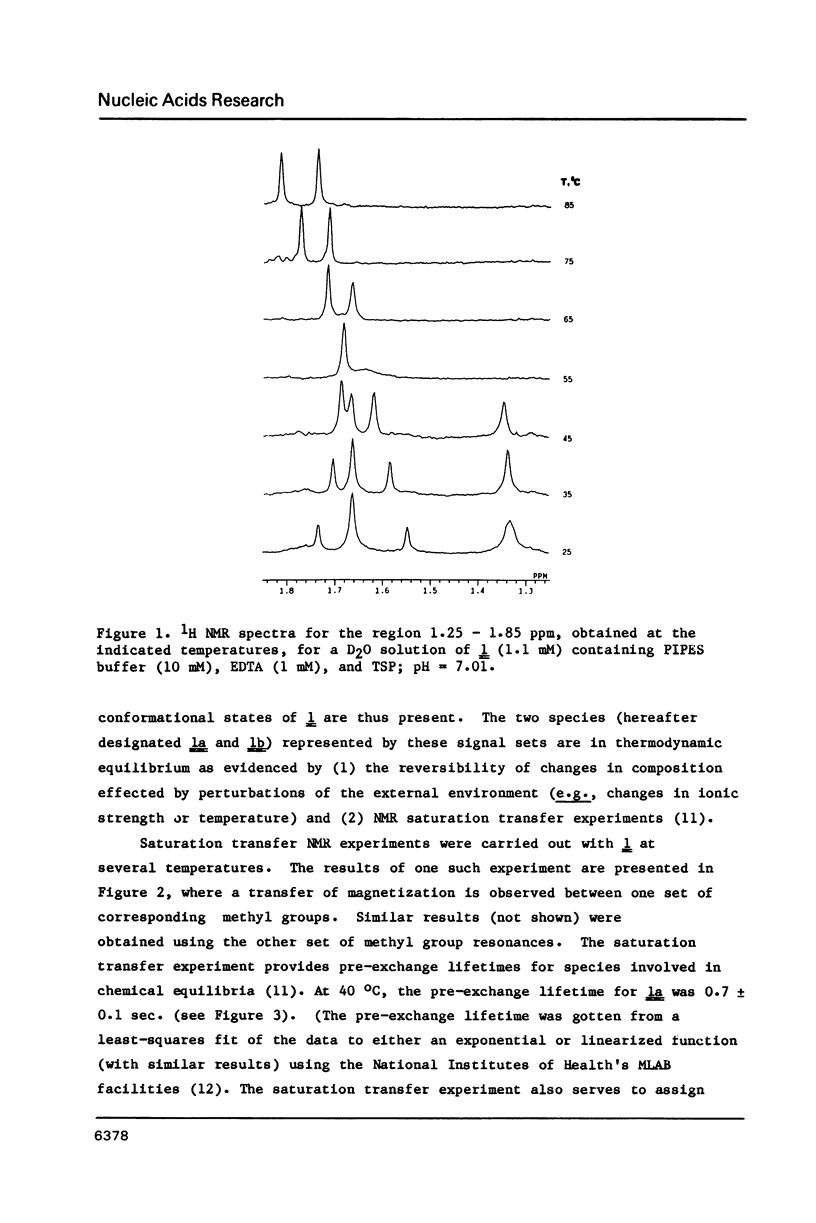
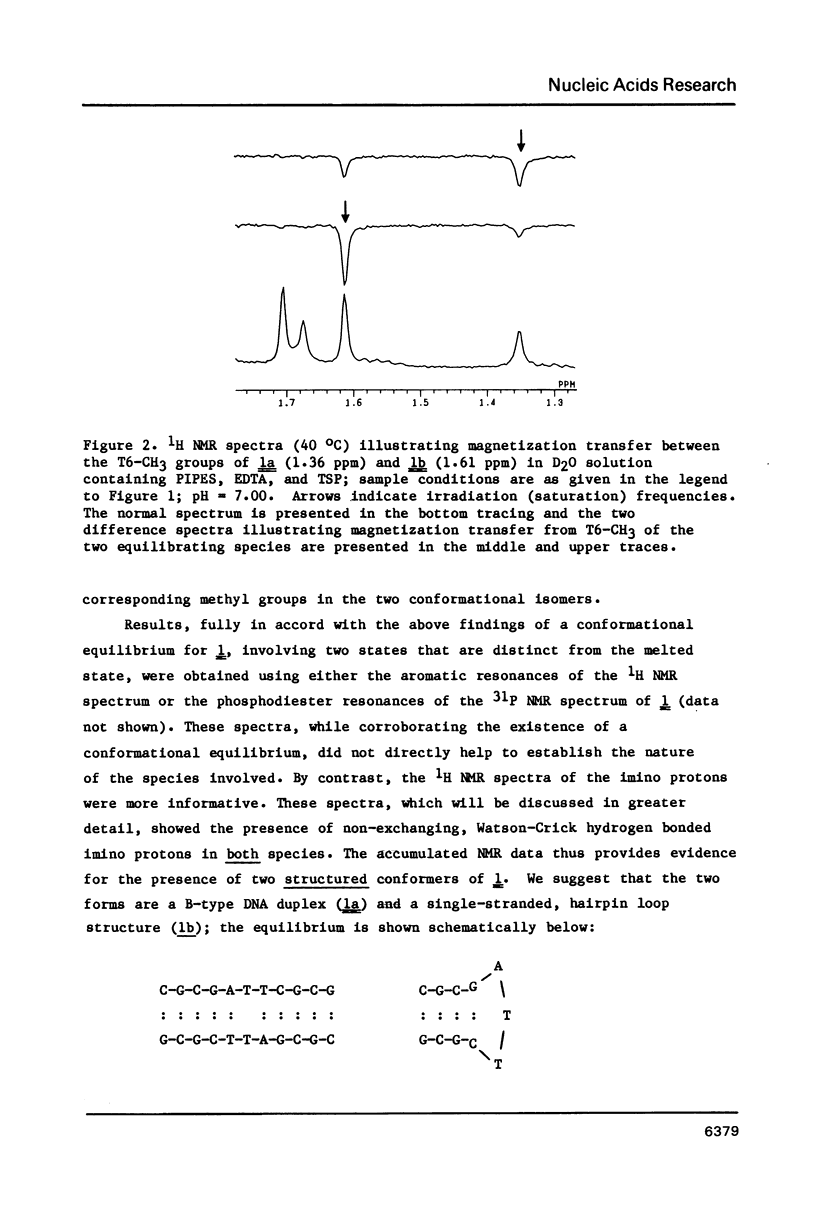
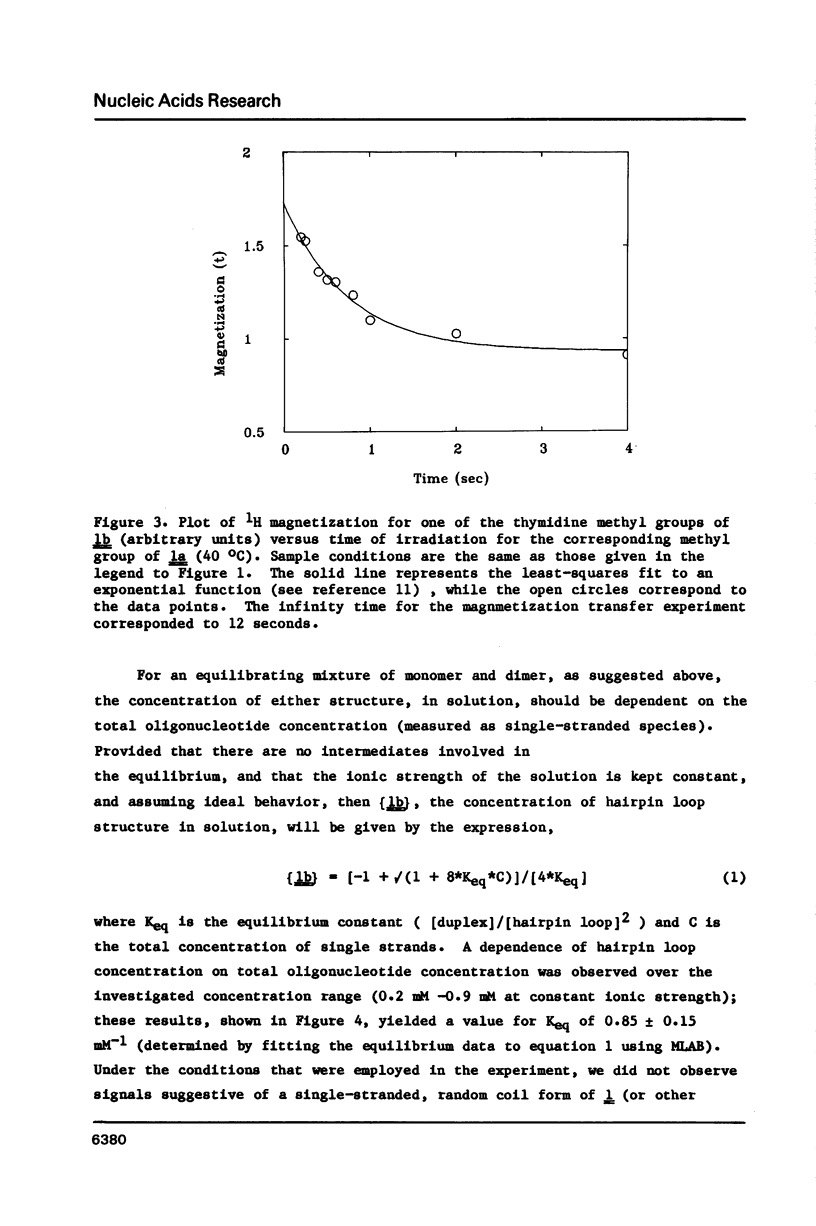
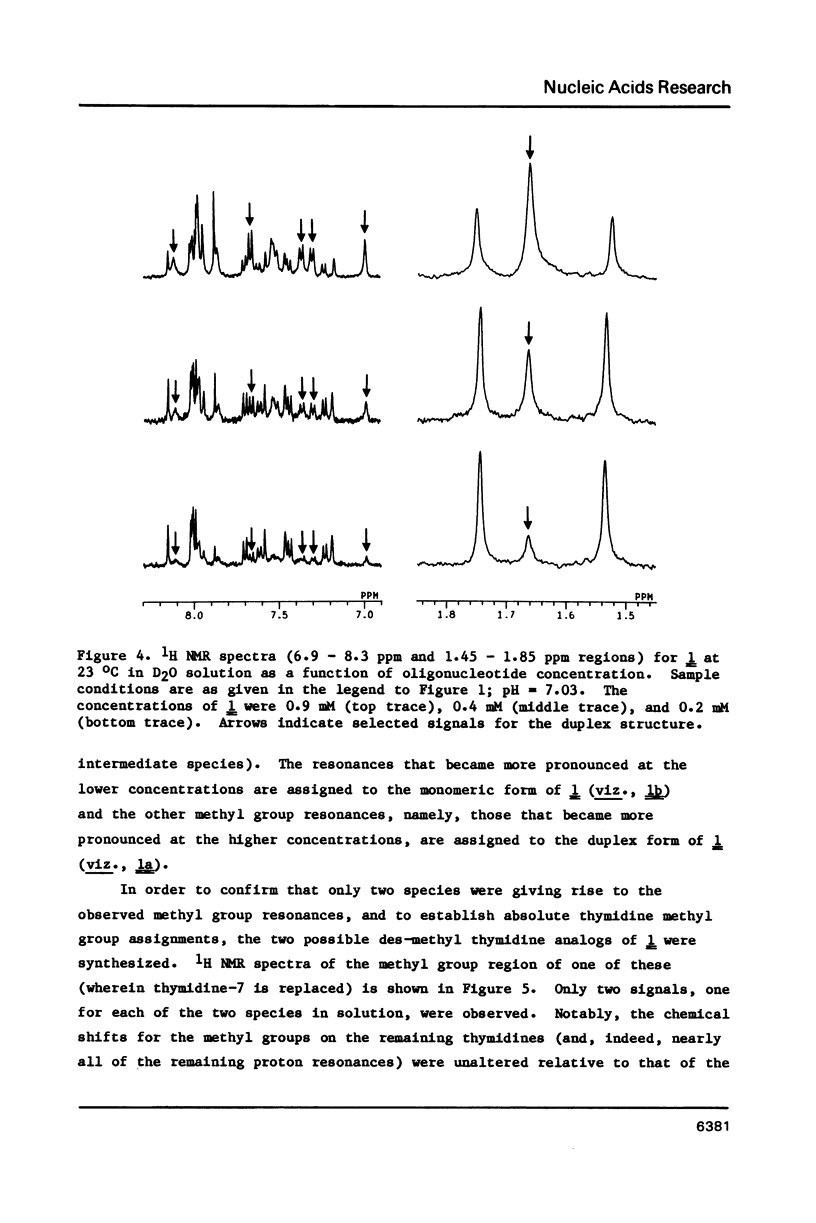
Nuclear magnetic resonance (NMR) and circular dichroism (CD) studies have been carried out with the oligodeoxyribonucleotide mismatch sequence, d(CGCGATTCGCG), 1. It has been found that 1 exists, in solution, as an equilibrium mixture of slowly interconverting, structured conformational isomers, 1a and 1b. On the basis of the concentration dependence of the 1a-1b equilibrium, the 1H NMR spectrum of the imino protons of the nucleotide bases, and the individual CD spectra of 1a and 1b, it is suggested that the two species correspond to a B-type DNA duplex and a single-stranded, hairpin-loop structure; the portion of the single-stranded species not involved in the loop appears to have a B-type DNA structure (on the basis of the CD measurements). To facilitate 1H NMR resonance assignments, the two possible des-methyl thymidine derivatives of 1 were synthesized; the effect of this substitution on the physical chemical properties of 1 was explored. The 1H NMR spectra of 1, as a function of temperature, showed that, under conditions wherein both species were present to a significant extent, the duplex form melted at a lower temperature than the single-stranded, hairpin loop structure.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cornelis A. G., Haasnoot J. H., den Hartog J. F., de Rooij M., van Boom J. H., Cornelis A. Local destabilisation of a DNA double helix by a T--T wobble pair. Nature. 1979 Sep 20;281(5728):235–236. doi: 10.1038/281235a0. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Dickerson R. E. Structure of a B-DNA dodecamer. III. Geometry of hydration. J Mol Biol. 1981 Sep 25;151(3):535–556. doi: 10.1016/0022-2836(81)90009-7. [DOI] [PubMed] [Google Scholar]
- Haasnoot C. A., de Bruin S. H., Berendsen R. G., Janssen H. G., Binnendijk T. J., Hilbers C. W., van der Marel G. A., van Boom J. H. Structure, kinetics and thermodynamics of DNA hairpin fragments in solution. J Biomol Struct Dyn. 1983 Oct;1(1):115–129. doi: 10.1080/07391102.1983.10507429. [DOI] [PubMed] [Google Scholar]
- Haasnoot C. A., den Hartog J. H., de Rooij J. F., van Boom J. H., Altona C. Loopstructures in synthetic oligodeoxynucleotides. Nucleic Acids Res. 1980 Jan 11;8(1):169–181. doi: 10.1093/nar/8.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keepers J. W., Schmidt P., James T. L., Kollman P. A. Molecular-mechanical studies of the mismatched base analogs of d(CGCGAATTCGCG)2:d(CGTGAATTCGCG)2, d(CGAGAATTCGCG)2, d(CGCGAATTCACG)2, d(CGCGAATTCTCG)2, and d(CGCAGAATTCGCG).d(CGCGAATTCGCG). Biopolymers. 1984 Dec;23(12):2901–2929. doi: 10.1002/bip.360231214. [DOI] [PubMed] [Google Scholar]
- Knott G. D. Mlab--a mathematical modeling tool. Comput Programs Biomed. 1979 Dec;10(3):271–280. doi: 10.1016/0010-468x(79)90075-8. [DOI] [PubMed] [Google Scholar]
- Marky L. A., Blumenfeld K. S., Kozlowski S., Breslauer K. J. Salt-dependent conformational transitions in the self-complementary deoxydodecanucleotide d(CGCAATTCGCG): evidence for hairpin formation. Biopolymers. 1983 Apr;22(4):1247–1257. doi: 10.1002/bip.360220416. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Ikuta S., Itakura K., Bhatt R., Hare D. R. NMR studies of DNA conformation and dynamics in solution. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):197–206. doi: 10.1101/sqb.1983.047.01.025. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Ikuta S., Itakura K. Deoxyadenosine-deoxycytidine pairing in the d(C-G-C-G-A-A-T-T-C-A-C-G) duplex: conformation and dynamics at and adjacent to the dA X dC mismatch site. Biochemistry. 1984 Jul 3;23(14):3218–3226. doi: 10.1021/bi00309a016. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Marky L. A., Broka C., Rice J. A., Itakura K., Breslauer K. J. Premelting and melting transitions in the d(CGCGAATTCGCG) self-complementary duplex in solution. Biochemistry. 1982 Feb 2;21(3):428–436. doi: 10.1021/bi00532a002. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Marky L. A., Rice J. A., Broka C., Itakura K., Breslauer K. J. Extra adenosine stacks into the self-complementary d(CGCAGAATTCGCG) duplex in solution. Biochemistry. 1982 Feb 2;21(3):445–451. doi: 10.1021/bi00532a004. [DOI] [PubMed] [Google Scholar]
- Tibanyenda N., De Bruin S. H., Haasnoot C. A., van der Marel G. A., van Boom J. H., Hilbers C. W. The effect of single base-pair mismatches on the duplex stability of d(T-A-T-T-A-A-T-A-T-C-A-A-G-T-T-G) . d(C-A-A-C-T-T-G-A-T-A-T-T-A-A-T-A). Eur J Biochem. 1984 Feb 15;139(1):19–27. doi: 10.1111/j.1432-1033.1984.tb07970.x. [DOI] [PubMed] [Google Scholar]


