Abstract
Methanogen populations and their domains are poorly understood; however, in recent years, research on this topic has emerged. The relevance of this field has also been enhanced by the growing economic interest in methanogen skills, particularly the production of methane from organic substrates. Management attention turned to anaerobic wastes digestion because the volume and environmental impact reductions. Methanogenesis is the biochemically limiting step of the process and the industrially interesting phase because it connects to the amount of biogas production. For this reason, several studies have evaluated the structure of methanogen communities during this process. Currently, it is clear that the methanogen load and diversity depend on the feeding characteristics and the process conditions, but not much data is available. In this study, we apply a Real-Time Polymerase Chain Reaction (RT-PCR) method based on mcrA target to evaluate, by specific probes, some subgroups of methanogens during the mesophilic anaerobic digestion process fed wastewater sludge and organic fraction of the municipal solid waste with two different pre-treatments. The obtained data showed the prevalence of Methanomicrobiales and significantly positive correlation between Methanosarcina and Methanosaetae and the biogas production rate (0.744 p < 0.01 and 0.641 p < 0.05). Methanosarcina detected levels are different during the process after the two pre-treatment of the input materials (T-test p < 0.05). Moreover, a role as diagnostic tool could be suggested in digestion optimisation.
Keywords: methanogen, anaerobic digestion, biogas production, Methanosarcina, Archaea communities
Introduction
Methanogenesis is a characteristic unique to the Archaea (Woese 2007). Biological methane production involves 25 genes and numerous specific proteins and coenzymes. However, the gene number involved in the different aspects of methane production is much higher (Galagan et al. 2002). Methane can be produced through different pathways, each of which has a different substrate. Among the precursor organic molecules, we find CO2, formate, acetate and methyl groups. The CO2, with H2 as an electron donor, is reduced to methane via the hydrogenotrophic mechanism. Acetate is involved in the aceticlastic pathway, and the methyl group acts as the starting point of the methylotrophic pathway (Ferry 2010a, b). Anaerobic digestors are one typical habitat, especially for the following genera: Methanobacterium, Methanothermobacter, Methanomicrobium, Methanoculleus, Methanofollis, Methanospirillum, Methanocorpusculum, Methanosarcina and Methanosaeta (Liu and Whitman 2008). Two genera of Archaea, Methanosarcina and Methanosaeta, are methane producing from acetate, and this acetoclastic mechanism produces higher proportions of biogenic methane. These two genera are also the most studied in recent years with the advent of the complete genome sequencing of some strains (Barber et al. 2011). Methanogenesis is the final step of the anaerobic digestion process in the reactor. Other microorganisms, such as hydrolytic acidogens and acetogens, are involved in the previous steps. These microorganisms prepare the substrates for methanogenesis, which is considered to be the rate-limiting step (Rozzi and Remigi 2004). Anaerobic digestion technologies vary throughout Europe. For example, Germany has more than 4000 digesters (Dolan et al. 2011) and there are numerous examples of integrated management of waste and biomethane fuel production to provide public transport in Sweden and France (Lantz et al. 2007; Dolan et al. 2011). Recently, other countries have begun promotional projects to encourage anaerobic digestion methodology (Dolan et al. 2011). In Italy, the number of anaerobic digestion reactors is growing rapidly, especially farm-scale digesters (De Baere 2006). The fermentation of other organic waste is also financially appraised (Schievano et al. 2009a; Schievano et al. 2009b) in urban aggregation, where organic waste, such as the organic fraction of municipal solid organic waste (OFMSW) and wastewater sludge, are produced (Tambone et al. 2009; Pognani et al. 2009). To optimize the digestion benefits in terms of biogas production, waste volume reduction and waste impact on the environment, many research projects have begun in the past 10 years (Mata-Alvarez et al. 2011). The main results concern the parameters controlling the anaerobic process in technology configurations (Amani et al. 2010; Boe et al. 2010). Moreover, with recent technological and financial achievements, the microbiological aspects of anaerobic digestion have become relevant topics (Weiss et al. 2008; Cardinali-Rezende et al. 2009). This attention has led to the optimization of this process, which has paid for itself. Among the many microorganisms present in the reactor, methanogens are the most sensitive; however, they are difficult to study in culture-based methods, despite their critical role (Liu and Whitman 2008). In recent years, culture-independent techniques have been developed (Sekiguchi et al. 1998). These techniques are based on phylogenetic markers such as the 16S rRNA or methyl coenzyme M reductase (Mcr) genes (Nunoura et al. 2008; Rastogi et al. 2008). The 16S rRNA gene is the most widely used target for gene surveys (Nayak et al. 2009), whereas the Mcr is exclusive to the methanogens, with the exception of the methane-oxidising Archaea (Knittel and Boetius 2009; Whitman et al. 2006). The primary aim of this work is to study methanogen populations in order to find a bioindicator of a productive digestion process. To achieve this purpose, we determined, during anaerobic co-digestions, the abundance of methanogen subgroups utilising Real-Time qualitative PCR (RT-qPCR) with specific probes targeting the mcrA gene (additional file 1).
Materials and methods
Two pilot reactors were fed pre-treated organic fractions of municipal solid waste (OFMSW) and wastewater sludge. The pre-treated methods used in this study included a pressure-extrusion (A) and a turbo mixing (B) system. In method A, the separation was achieved through a specially designed extruder press (280 bar) that separated the input waste into two fractions: a dry one to be sent to thermal conversion and a semi-solid one. The pressure-extruded dry fraction of the OFMSW was then diluted with wastewater sludge. By contrast, method B (the turbo-mixing system) was a wet process that works with a total solids (TS) content lower than 8%. The mixing and treating actions are performed by a rotating plate with hummers placed at the bottom of the turbo-mixing chamber that, when rotating at high velocity, induce the suspension to shear and crush. The particles weighing more than water precipitate to the bottom, where they are picked up by a screw and collected in an external vessel. The organic fraction remains in suspension and is pumped into a storage basin after passing through a shredding pump. In this case, OFMSW was directly turbo-mixed with wastewater sludge (about 1:3 proportion). The main physical-chemical characteristics of each kind of feed used in this work, just before entrance into the reactor, are shown in Table 1. The anaerobic co-digestion tests were conducted using a reactor with a total volume capacity of 15 L and a working volume of 10 L (Figure 1). The temperature was mesophilic and maintained at 38 ± 2°C using a water recirculation system connected to a thermostatic valve. The biogas produced was collected and measured in a calibrated gasometer and a mixing system containing the recirculated biogas produced during the anaerobic digestion process. The reactors were equipped with two openings, one at the top for feeding and one below to collect effluent discharge, as showed on Figure 1. Every day, 500 ml of digestate was removed from each reactor before adding another 500 ml of fresh feed. The parameters analysed three times a week in accordance with standard methods (APHA, 1995) included pH, total solids (TS), total volatile solids (TVS), alkalinity, acidity, nitrogen (N), and total carbon. Daily biogas production was measured using a liquid displacement system that was connected to the digester. The biogas volume was corrected using standard temperature and pressure conditions. The biogas composition (in terms of methane and carbon dioxide percentage) was analysed once a week with a portable analyser and confirmed by gas chromatography analysis.
Table 1.
Characteristics of the pretreated inputs with the two different method used in the anaerobic co-digestion processes
| Pre-treatment A | Pre-treatment B | |
|---|---|---|
| pH | 4.4 ± 0.3 | 6.0 ± 0.7 |
| TS (%) | 9.9 ± 0.7 | 4.6 ± 1.1 |
| TVS (%) | 8.7 ± 0.7 | 3.3 ± 1.1 |
| TSV/TS (%) | 86.8 ± 0.2 | 70.6 ± 4.9 |
| C (%TS) | 46.0 ± 0.9 | 37.0 ± 3.4 |
| N (%TS) | 3.1 ± 0.2 | 3.5 ± 0.3 |
| C/N | 15.2 ± 1.1 | 10.4 ± 1.5 |
Figure 1.

The pilot hardware description is illustrated. The same reactor, in different six-month fermentation sessions, with two different pre-treated feedings was used during this research study.
The reactors were operated at a constant organic loading rate of 4,5 ± 0,3 kg TVS/m3 per day when OFMSW pressure-extruded was used and at an average organic loading rate of 1,7 ± 0,5 kg TVS/m3 per day when OFMSW with pulper pretreatment was used. The tests were run over two consecutive hydraulic retention times of 20 days for each organic loading rate: one to ensure the highest replacement parts of the material inside the reactors and the other to analyse the process in a stable condition once all the feed had replaced the inoculum content. The main control parameters for pretreatments A and B are displayed in Table 2. Methanogen subgroups were determined using samples with the highest biogas production rate. These included 15 from pretreatment A and 10 from pretreatment B. The samples were collected during 2009 in 50 ml sterile tube and frozen at -20°C until the extraction session.
Table 2.
Main relevant evaluation parameters of the co-digestion processes divided by pre-treatment method
| Parameters | Pre-treatment A | Pre-treatment B |
|---|---|---|
| Daily biogas production (L/die) | 27.08 ± 3.01 | 4.87 ± 2.46 |
| Specific Biogas production (m3/kg VS added) | 0.64 ± 0.07 | 0.30 ± 0.13 |
| TS reduction (%) | 64.44 ± 7.57 | 31.67 ± 6.23 |
| TSV reduction (%) | 73.84 ± 5.87 | 38.13 ± 6.70 |
| pH | 7.36 ± 0.34 | 6.82 ± 0.52 |
| Ac./Alc. ratio | 0.37 ± 0.18 | 2.47 ± 2.41 |
| CH4 (%) | 60.60 ± 2.90 | 57.50 ± 6.10 |
| CO2 (%) | 37.70 ± 3.20 | 41.00 ± 6.44 |
DNA extraction and purification
The digestate aliquots were thawed at 4°C overnight and centrifuged at 4000 g for 10 minutes. After removing the supernatant, semi-dry aliquots were used for the following steps. Total DNA was extracted from 0.25 g of this particulate matter (residue humidity was equal to 31 ± 5%) using the PowerSoil DNA Isolation Kit following by UltraClean Soil DNA Kit (MoBio Laboratories). The average DNA quantity extracted was 3.51 ± 1.53 ng/μl, and DNA quality was evaluated by gel electrophoresis before the chain reaction. Only samples with a DNA quantity above 1 ng/μl and of sufficient quality were used for the following step.
qRT-PCR analysis
After DNA extraction and purification, different methanogens were quantified using methanogen-specific short primers for a mcrA sequence (Steinberg and Regan 2008) and synthesised by ThermoBiopolymer and previously described specific probes (Steinberg and Regan 2009).
Methanosarcina, Methanobacterium, Methanocorpusculum and Methanosaeta were determined with the respective following probes: msar, mrtA, mcp and msa (Steinberg and Regan 2009). The reactions were conducted in singleplex with a standard super mix (Bio-Rad iQ™ Multiplex Powermix) using RT-PCR Chromo4 (Bio-Rad) and Opticon Monitor 3 Software. The reaction conditions have been previously described (Steinberg and Regan 2009, 2008).
Standard references were available only for the Methanosarcina and Methanobacterium. The references were a Methanosarcina acetivorans mcrA sequence and a Methanobacterium thermoautotrophicum mrtA sequence. Each plasmid is included in pCR21 vector (Invitrogen) supplied by L.M. Steinberg and J.M. Regan, Pennsylvania State University. These plasmids were amplified, transforming Escherichia coli Top10 cells according to the manufacturer's instructions. Transformed cells were selected on LB agar with ampicillin, and the plasmid was extracted using a plasmid DNA purification kit (NucleoSpin Plasmid, Macherey-Nagel). The standard curve had six points, and it was calculated using the threshold cycle method with the highest standard amplified being 2.3 ng of plasmid (~4.5*108 plasmid copies). Between each following standard curve point, there is a 1:10 dilution. Standards and samples were tested in triplicates. The triplicate averages were accepted only if the coefficient of variation was below 20%. Example of regression curves with correlation coefficient and PCR efficiency were showed on Table 3. Resolution limit of the method was settled to 4.5*103 copies of mcrA. The PCR products are about 500 base pairs long.
Table 3.
qRT-PCR probe and reaction descriptions
| Target group | Probe name target | Example of regression curve | r2 | PCR efficiency (%) | Acceptable data (%) |
|---|---|---|---|---|---|
| Methanosarcina | msar | y = -0.2547x +11.34 | 0.997 | 80 | 75 |
| Methanobacteriaceae | mrtA | y = -0.2691x+12.21 | 0.995 | 86 | 4 |
| Methanocorpusculaceae | mcp | y = -0.2627x+12.38 | 0.987 | 83 | 88 |
| Methanosaetaceae | msa | y = -0.2380x+10.27 | 0.943 | 73 | 52 |
There is a standard reference curve only for the Methanosarcina and Methanobatecteriaceae, making it possible to establish the gene copies in the extracted DNA. The last column indicates the percentage of determinable sample on the total 25 tested samples.
For Methanocorpusculaceae and Methanosaetaceae, there was no standard reference available; therefore, quantification could only be considered between samples in the same analytical session. The efficiency of the PCR reactions was determined with serial 1:10 dilution of a sample and are showed on Table 3. The results for these groups were expressed as cycle threshold (Ct) or as 1/Ct, where relative abundance was discussed for each reaction, instead of real quantification, as for the Methanosarcinaeae and Methanobacterium, where results could be expressed as gene copies per microliter of DNA extract.
We used 2 μl of a 1:5 dilution of DNA extracts for amplification. This quantity of sample was evaluated as the best among various tested quantities for obtaining quantifications within the standard curve range and with acceptable PCR efficiency. The 1:5 dilution is sufficient to avoid the effect of inhibition substances present in this kind of sample. Only a percentage of the 25 total samples were acceptable as detailed on the table 3, and values ranged by methanogen group from 4 to 88. In many samples, evaluation of the Ct was not determinable (above 40).
To evaluate precision, we began with the same two samples re-extracted 10-fold. The results of the successive PCR-determination showed a variation coefficient below 6% for msar amplification and below 15% for msa, mrtA and mcp amplifications.
Statistics
Statistical analyses were performed using the SPSS Package, version 17.0, for Windows. A Spearman correlation coefficient was used to assess the relationships between variables. A T-test of independent variables was used to test mean evaluations. The differences and correlations were considered significant at p < 0.05 and highly significant at p < 0.01.
Results
The detected level of various methanogen groups is displayed in Table 4. Groups varied largely in quantity during the digestion processes and were often not present at all. Methanosarcina was not detected in some samples, this happened when the pH was around 6.5 and the production rate was lower than 0.5 m3/kg VSadded. The number of msar copies in the sample can be explained by the relevant level of acetate, the substrate of this group, and the high biogas production rate recorded from the reactor. As described in the literature, an anaerobic digester typically contains more than 1012 cells/μl with an average of 108 methanogens (Amani et al. 2010). Methanobacteriaceae mrtA resulted undetectable nearly in all the samples (table 3) while the Methanomicrobiales resulted prevalent, in particular acetoclastic methanogens (Methanosarcina and Methanosaeta). Furthermore, their presence increased along with the specific biogas production rate (Table 5). Methanocorpusculaceae seemed to have a similar behaviour as showed in table 5 and their presence is highly correlated both to Methanosarcina and Methanosaeta. Methanosarcina was significantly correlated with all the control parameters (positively with the pH, specific biogas production and % TSV; negatively with the acidity/alkalinity ratio) as showed on table 4. With increases in the TVS, there was also an increase in Methanocorpusculaceae and Methanosaetaceae. A significant, positive correlation with the pH was also observed for the other acetoclastic group, Methanosaetaceae (Table 4).
Table 4.
Descriptive analysis of the acceptable data by each probe
| Target (measure unit) | Min | Max | Mean | Dev. std. |
|---|---|---|---|---|
| Methanosarcina (gene copies/μl) | 4.77E+04 | 6.03E+07 | 1.19E+07 | 1.51E+07 |
| Methanobacteriaceae (gene copies/μl) | 1.52E+05 | 1.52E+05 | 1.52E+05 | - |
| Methanocorpusculaceae (1/Ct) | 2.52E-02 | 3.98E-02 | 2.966E-02 | 3.6E-03 |
| Methanosaetaceae (1/Ct) | 2.56E-02 | 3.74E-02 | 2.969E-02 | 3.7E-03 |
Table 5.
Spearman's rho correlation between the detected methanogen groups and the monitored control parameters
| pH | Ac/Alc ratio | % TVS added | Biogas production (m3/kg VS added) | msar (gene copies/μl) | msa (1/Ct) | |
|---|---|---|---|---|---|---|
| msar (gene copies/μl) | 0.630** | -0.589** | 0.744** | 0.673** | 1 | 0.782** |
| msa (1/Ct) | 0.847** | - | 0.641* | 0.576* | 0.782** | 1 |
| mcp (1/Ct) | - | - | 0.449* | - | 0.719** | 0.868** |
Significant correlation at p < 0.05 is identified with a single asterisk while highly significant at p < 0.01 with a double asterisk. The hyphen is introduced when no significant correlations (n.c.) were observed.
The significant correlations among the various methanogen groups and control parameters are displayed on Table 5. In Figure 2, the Methanosarcina loads were differentiated in relation to the pre-treatment of the input material (A and B). The difference between the mean of the Methanosarcina levels, during the digestion with the pressure-extrusion input, is significantly higher than the turbo-mixing one (1.68E7 vs 2.55E5, F = 6.821, p = 0.018).
Figure 2.
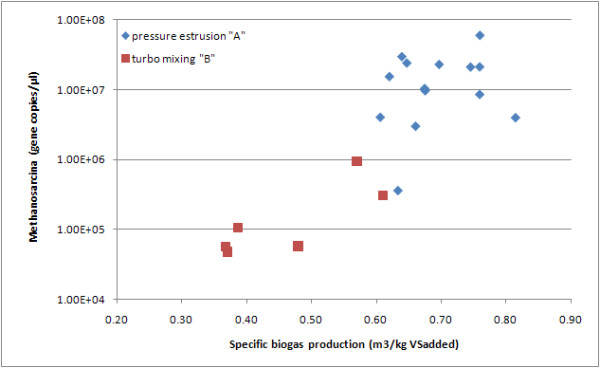
The quantification of Methanosarcina during the two monitored processes in relation to specific biogas production rate subdivided by pre-treatment.
Moreover the figure 2 illustrates as all the samples, collected during the process conducing after pressure-estrusion pre-treatment, showed a biogas production rate above or near to 0.6 m3/kg TSVadded. This cut-off is a suitable division between optimal and suboptimal digestion conditions as has been documented in the literature (Amani et al. 2010).
Discussion
Anaerobic digestion is among the most complicated and unknown biological processes in the environment (Schink 1997). Different aspects attract operational, chemical and biological criticisms. Moreover, these aspects are strictly interconnected with one another. A wide number of papers in this field have been published in recent years (Khalid et al. 2011). Most of these studies, however, didn't include methanogens characterization or they have been based on a metagenomic approach in which a small subunit of ribosomal RNA was used (Pycke et al. 2011; Supaphol et al. 2011). Methanogen studies using the mcrA-based method have become more common in recent years (Narihiro and Sekiguchi 2011).
Over 90% of the detected methanogenic Archaea in the mesophilic reactor fed swine slurry belonged to the hydrogenotrophic methanogens. These were predominantly Methanobacteriales followed by Methanomicrobiales (Zhu et al. 2011). On the other hands always in mesophilic biogas plant but fed with cattle manure, 84% of all detected methanogens were affiliated with the Methanomicrobiales, whereas only 14% belonged to the Methanosarcinales and 2% to the Methanobacteriales (Bergmann et al. 2010a, b) and in other plant always running on cattle manure, the methanogen community presented the following composition: 41.7% of clones were affiliated with Methanomicrobiales, 30% with Methanosarcinales, and 19% with Methanobacteriales; at temperatures lower than 25°C, the Methanomicrobiales became most prevalent (> 90%) (Rastogi et al. 2008).
In reactor fed leachate and OFMSW, various orders of hydrogenotrophic methanogens belonging to Methanomicrobiales and Methanobacteriales were identified (Cardinali-Rezende et al. 2009). However, during mesophilic digestion of wastewater sludge, Methanosarcina and Methanosaeta were most abundant, comprising up to 90% of the total Archaea present or more (Narihiro et al. 2009; Das et al. 2011). This data confirms the results of our work and the ability of Methanosarcina species to form multicellular aggregates that may resist inhibitions in the reactor (Vavilin et al. 2008).
Despite the data variability such bio-molecular approach can improve the available knowledge of anaerobic digestion, as demonstrated in this work, the biogas production efficiency is significantly and positively correlated to two methanogen groups (Methanosarcina and Methanosaetaceae). Most importantly, this method can represent a way to introduce useful bioindicators into the reactors for early diagnosis of an unbalance or a sufferance situation in the microbiologic community. Establishing an efficiency cut-off during the anaerobic digestion process - optimal production that for our set up is around 0.6 CH4 m3/kg SVadded - it makes possible to observe a role for certain groups of methanogens, primarily the Methanosarcina as useful Archaea bioindicators in the digestion process. On the other hands the produced data shows a clear advantage in the pressure-extrusion respect to turbo-mixing pre-treatment as production rate moreover also the cost of the two pre-treatment plants are very different, against the pressure-extrusion. After a validation process with different digestion processes, the definition of a threshold of alarm seems to be possible.
Finally, it is critical that this kind of approach be utilised and that knowledge in this scientific field be increased. The methanogen diversity in the reactor is widely influenced by the feeding. During anaerobic digestion in which input is mainly cattle manure, the presence of hydrogenotroph methanogens is favoured. However, when other feedings are involved, as in this experimental activity, the methanogen community structure differs in terms of the prevalence of Methanosarcineae such as Methanosarcina and Methanosaeta. This family presents a prevalent acetoclastic methane production. A closer examination is needed for substrate and product analysis. A profile of the substrates, such as butyrate, propionate, H2 and CO2, could be useful in understanding the microbiologic dynamics and the consequent methanogen modulations.
Competing interests
The authors declare that they have no competing interests.
Supplementary Material
Graphical abstract. During mesophilic anaerobic co-digestion, biomolecular methanogen determinants in the reactor vary among groups in different biochemical pathways, indicating that variation in biogas yield supplies early bioindicators of methane production.
Contributor Information
Deborah Traversi, Email: deborah.traversi@unito.it.
Silvia Villa, Email: silvia.villa@unito.it.
Marco Acri, Email: marco.acri@smatorino.it.
Biancamaria Pietrangeli, Email: biancamaria.pietrangeli@ispesl.it.
Raffaella Degan, Email: raffaella.degan@unito.it.
Giorgio Gilli, Email: giorgio.gilli@unito.it.
Acknowledgements
The authors wish to thank the Piedmont Region and ISPESL for funding support. The work was part of a large project called DigestedEnergy, which was founded in response to the 2006 call for pre-competitive development and industrial research. It includes ten different public and private organisations. Special acknowledgments are due to L. Steinberg and J. Regan for the plasmid standard supply. Finally the authors thank all the numerous collaborators employed in each of the involved institutions: Università degli Studi del Piemonte Orientale "A. Avogadro", Politecnico di Torino, SMAT S.p.A., Amiat S.p.A., Ansaldo FC S.p.A., Acsel Susa S.p.A., VM-press s.r.l., Federsviluppo, E.R.A.P.R.A Piemonte, and Università degli Sudi di Torino.
References
- Amani T, Nosrati M, Sreekrishnan TR. Anaerobic digestion from the viewpoint of microbiological, chemical, and operational aspects - a review. Environ Rev. 2010;18:255–278. doi: 10.1139/A10-011. [DOI] [Google Scholar]
- Barber RD, Zhang L, Harnack M, Olson MV, Kaul R, Ingram-Smith C, Smith KS. Complete genome sequence of Methanosaeta concilii, a specialist in aceticlastic methanogenesis. J Bacteriol. 2011;193(14):3668–3669. doi: 10.1128/JB.05031-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann I, Mundt K, Sontag M, Baumstark I, Nettmann E, Klocke M. Influence of DNA isolation on Q-PCR-based quantification of methanogenic Archaea in biogas fermenters. Sys Appl Microbiol. 2010;33(2):78–84. doi: 10.1016/j.syapm.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Bergmann I, Nettmann E, Mundt K, Klocke M. Determination of methanogenic Archaea abundance in a mesophilic biogas plant based on 16S rRNA gene sequence analysis. Can J Microbiol. 2010;56(5):440–444. doi: 10.1139/W10-021. [DOI] [PubMed] [Google Scholar]
- Boe K, Batstone DJ, Steyer JP, Angelidaki I. State indicators for monitoring the anaerobic digestion process. Water Res. 2010;44(20):5973–5980. doi: 10.1016/j.watres.2010.07.043. [DOI] [PubMed] [Google Scholar]
- Cardinali-Rezende J, Debarry RB, Colturato LFDB, Carneiro EV, Chartone-Souza E, Nascimento AMA. Molecular identification and dynamics of microbial communities in reactor treating organic household waste. Appl Microbiol Biot. 2009;84(4):777–789. doi: 10.1007/s00253-009-2071-z. [DOI] [PubMed] [Google Scholar]
- Das KC, Garcia SL, Jangid K, Whitman WB. Transition of microbial communities during the adaption to anaerobic digestion of carrot waste. Bioresource Technol. 2011;102(15):7249–7256. doi: 10.1016/j.biortech.2011.04.098. [DOI] [PubMed] [Google Scholar]
- De Baere L. Will anaerobic digestion of solid waste survive in the future? Water Sci Technol. 2006;53(8):187–194. doi: 10.2166/wst.2006.249. [DOI] [PubMed] [Google Scholar]
- Dolan T, Cook MB, Angus AJ. Financial appraisal of wet mesophilic AD technology as a renewable energy and waste management technology. Sci Total Environ. 2011;409(13):2460–2466. doi: 10.1016/j.scitotenv.2011.03.011. [DOI] [PubMed] [Google Scholar]
- Ferry JG. The chemical biology of methanogenesis. Planet Space Sci. 2010;58(14-15):1775–1783. doi: 10.1016/j.pss.2010.08.014. [DOI] [Google Scholar]
- Ferry JG. CO in methanogenesis. Ann Microbiol. 2010;60(1):1–12. doi: 10.1007/s13213-009-0008-5. [DOI] [Google Scholar]
- Galagan JE, Nusbaum C, Roy A, Endrizzi MG, Macdonald P, FitzHugh W, Calvo S, Engels R, Smirnov S, Atnoor D, Brown A, Allen N, Naylor J, Stange-Thomann N, DeArellano K, Johnson R, Linton L, McEwan P, McKernan K, Talamas J, Tirrell A, Ye WJ, Zimmer A, Barber RD, Cann I, Graham DE, Grahame DA, Guss AM, Hedderich R, Ingram-Smith C, Kuettner HC, Krzycki JA, Leigh JA, Li WX, Liu JF, Mukhopadhyay B, Reeve JN, Smith K, Springer TA, Umayam LA, White O, White RH, de Macario EC, Ferry JG, Jarrell KF, Jing H, Macario AJL, Paulsen I, Pritchett M, Sowers KR, Swanson RV, Zinder SH, Lander E, Metcalf WW, Birren B. The genome of M-acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 2002;12(4):532–542. doi: 10.1101/gr.223902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalid A, Arshad M, Anjum M, Mahmood T, Dawson L. The anaerobic digestion of solid organic waste. Waste Manag. 2011;31(8):1737–1744. doi: 10.1016/j.wasman.2011.03.021. [DOI] [PubMed] [Google Scholar]
- Knittel K, Boetius A. Anaerobic Oxidation of Methane: Progress with an Unknown Process. Annu Rev Microbiol. 2009;63:311–334. doi: 10.1146/annurev.micro.61.080706.093130. [DOI] [PubMed] [Google Scholar]
- Lantz M, Svensson M, Bjornsson L, Borjesson P. The prospects for an expansion of biogas systems in Sweden - Incentives, barriers and potentials. Energ Policy. 2007;35(3):1830–1843. doi: 10.1016/j.enpol.2006.05.017. [DOI] [Google Scholar]
- Liu YC, Whitman WB. Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann Ny Acad Sci. 2008;1125:171–189. doi: 10.1196/annals.1419.019. [DOI] [PubMed] [Google Scholar]
- Mata-Alvarez J, Dosta J, Mace S, Astals S. Codigestion of solid wastes: A review of its uses and perspectives including modeling. Crit Rev Biotechnol. 2011;31(2):99–111. doi: 10.3109/07388551.2010.525496. [DOI] [PubMed] [Google Scholar]
- Narihiro T, Sekiguchi Y. Oligonucleotide primers, probes and molecular methods for the environmental monitoring of methanogenic archaea. Microb Biotechno. 2011;l4(5):585–602. doi: 10.1111/j.1751-7915.2010.00239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narihiro T, Terada T, Ohashi A, Wu JH, Liu WT, Araki N, Kamagata Y, Nakamura K, Sekiguchi Y. Quantitative detection of culturable methanogenic archaea abundance in anaerobic treatment systems using the sequence-specific rRNA cleavage method. Isme Journal. 2009;3(5):522–535. doi: 10.1038/ismej.2009.4. [DOI] [PubMed] [Google Scholar]
- Nayak BS, Levine AD, Cardoso A, Harwood VJ. Microbial population dynamics in laboratory-scale solid waste bioreactors in the presence or absence of biosolids. J Appl Microbiol. 2009;107(4):1330–1339. doi: 10.1111/j.1365-2672.2009.04319.x. [DOI] [PubMed] [Google Scholar]
- Nunoura T, Oida H, Miyazaki J, Miyashita A, Imachi H, Takai K. Quantification of mcrA by fluorescent PCR in methanogenic and methanotrophic microbial communities. Fems Microbiol Ecol. 2008;64(2):240–247. doi: 10.1111/j.1574-6941.2008.00451.x. [DOI] [PubMed] [Google Scholar]
- Pognani M, D'Imporzano G, Scaglia B, Adani F. Substituting energy crops with organic fraction of municipal solid waste for biogas production at farm level: A full-scale plant study. Process Biochem. 2009;44(8):817–821. doi: 10.1016/j.procbio.2009.03.014. [DOI] [Google Scholar]
- Pycke BF, Etchebehere C, Van de Caveye P, Negroni A, Verstraete W, Boon N. A time-course analysis of four full-scale anaerobic digesters in relation to the dynamics of change of their microbial communities. Water Sci Technol. 2011;63(4):769–775. doi: 10.2166/wst.2011.307. [DOI] [PubMed] [Google Scholar]
- Rastogi G, Ranade DR, Yeole TY, Patole MS, Shouche YS. Investigation of methanogen population structure in biogas reactor by molecular characterization of methyl-coenzyme M reductase A (mcrA) genes. Bioresource Technol. 2008;99(13):5317–5326. doi: 10.1016/j.biortech.2007.11.024. [DOI] [PubMed] [Google Scholar]
- Rozzi A, Remigi E. Methods of assessing microbial activity and inhibition under anaerobic contitions: a literature review. Rev Environ Sci Biotechnol. 2004;3:93–115. doi: 10.1007/s11157-004-5762-z. [DOI] [Google Scholar]
- Schievano A, D'Imporzano G, Adani F. Substituting energy crops with organic wastes and agro-industrial residues for biogas production. J Environ Manage. 2009;90(8):2537–2541. doi: 10.1016/j.jenvman.2009.01.013. [DOI] [PubMed] [Google Scholar]
- Schievano A, Scaglia B, D'Imporzano G, Malagutti L, Gozzi A, Adani F. Prediction of biogas potentials using quick laboratory analyses: Upgrading previous models for application to heterogeneous organic matrices. Bioresource Technol. 2009;100(23):5777–5782. doi: 10.1016/j.biortech.2009.05.075. [DOI] [PubMed] [Google Scholar]
- Schink B. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol R. 1997;61(2):262–280. doi: 10.1128/mmbr.61.2.262-280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi Y, Kamagata Y, Syutsubo K, Ohashi A, Harada H, Nakamura K. Phylogenetic diversity of mesophilic and thermophilic granular sludges determined by 16S rRNA gene analysis. Microbiol-Sgm. 1998;144:2655–2665. doi: 10.1099/00221287-144-9-2655. [DOI] [PubMed] [Google Scholar]
- Steinberg LM, Regan JM. Phylogenetic Comparison of the Methanogenic Communities from an Acidic, Oligotrophic Fen and an Anaerobic Digester Treating Municipal Wastewater Sludge. Appl Environ Microb. 2008;74(21):6663–6671. doi: 10.1128/AEM.00553-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg LM, Regan JM. mcrA-Targeted Real-Time Quantitative PCR Method To Examine Methanogen Communities. Appl Environ Microb. 2009;75(13):4435–4442. doi: 10.1128/AEM.02858-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supaphol S, Jenkins SN, Intomo P, Waite IS, O'Donnell AG. Microbial community dynamics in mesophilic anaerobic co-digestion of mixed waste. Bioresource Technol. 2011;102(5):4021–4027. doi: 10.1016/j.biortech.2010.11.124. [DOI] [PubMed] [Google Scholar]
- Tambone F, Genevini P, D'Imporzano G, Adani F. Assessing amendment properties of digestate by studying the organic matter composition and the degree of biological stability during the anaerobic digestion of the organic fraction of MSW. Bioresource Technol. 2009;100(12):3140–3142. doi: 10.1016/j.biortech.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Vavilin VA, Qu X, Mazeas L, Lemunier M, Duquennoi C, He PJ, Bouchez T. Methanosarcina as the dominant aceticlastic methanogens during mesophilic anaerobic digestion of putrescible waste. Antonie Van Leeuwenhoek Int J Gen Mol Microbiol. 2008;94(4):593–605. doi: 10.1007/s10482-008-9279-2. [DOI] [PubMed] [Google Scholar]
- Weiss A, Jerome V, Freitag R, Mayer HK. Diversity of the resident microbiota in a thermophilic municipal biogas plant. Appl Microbiol Biot. 2008;81(1):163–173. doi: 10.1007/s00253-008-1717-6. [DOI] [PubMed] [Google Scholar]
- Whitman WB, Bowen TL, Boone DR. The Methanogenic Bacteria. Prokaryotes. 2006;3(Chapter 9):165–207. [Google Scholar]
- Woese CR. Archaea: molecular and cellular biology. Washington: ASM press; 2007. The Archaea: an invitation to evolution. [Google Scholar]
- Zhu CG, Zhang JY, Tang YP, Xu ZK, Song RT. Diversity of methanogenic archaea in a biogas reactor fed with swine feces as the mono-substrate by mcrA analysis. Microbiol Res. 2011;166(1):27–35. doi: 10.1016/j.micres.2010.01.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Graphical abstract. During mesophilic anaerobic co-digestion, biomolecular methanogen determinants in the reactor vary among groups in different biochemical pathways, indicating that variation in biogas yield supplies early bioindicators of methane production.


