Abstract
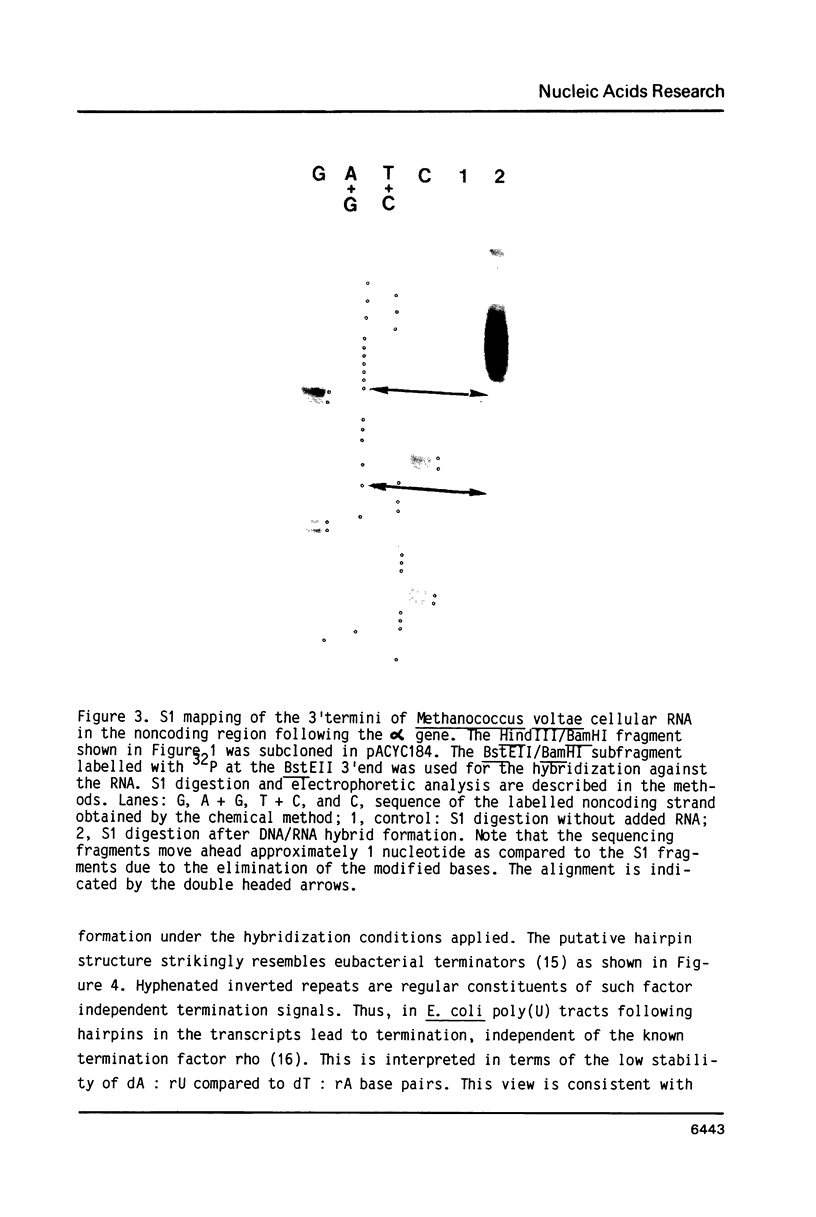
The 3'termini of transcripts originating from genes organized in a highly expressed transcription unit were analyzed in the archaebacterium Methanococcus voltae. The putative termination signals were found in an AT-rich intergenic region following the 3'-terminal gene. The two detected signals both contain oligo(T) sequences. A possible stem/loop structure immediately precedes one of the oligo(T) tracts. This secondary structure is considered to have an additional function in stabilizing the transcripts.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bakken A., Morgan G., Sollner-Webb B., Roan J., Busby S., Reeder R. H. Mapping of transcription initiation and termination signals on Xenopus laevis ribosomal DNA. Proc Natl Acad Sci U S A. 1982 Jan;79(1):56–60. doi: 10.1073/pnas.79.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollschweiler C., Kühn R., Klein A. Non-repetitive AT-rich sequences are found in intergenic regions of Methanococcus voltae DNA. EMBO J. 1985 Mar;4(3):805–809. doi: 10.1002/j.1460-2075.1985.tb03701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citron B., Falck-Pedersen E., Salditt-Georgieff M., Darnell J. E., Jr Transcription termination occurs within a 1000 base pair region downstream from the poly(A) site of the mouse beta-globin (major) gene. Nucleic Acids Res. 1984 Nov 26;12(22):8723–8731. doi: 10.1093/nar/12.22.8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassarma S., Rajbhandary U. L., Khorana H. G. Bacterio-opsin mRNA in wild-type and bacterio-opsin-deficient Halobacterium halobium strains. Proc Natl Acad Sci U S A. 1984 Jan;81(1):125–129. doi: 10.1073/pnas.81.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes W. M., Platt T., Rosenberg M. Termination of transcription in E. coli. Cell. 1983 Apr;32(4):1029–1032. doi: 10.1016/0092-8674(83)90287-8. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Thomm M., Stetter K. O. Transcription in methanogens. Evidence for specific in vitro transcription of the purified DNA-dependent RNA polymerase of Methanococcus thermolithotrophicus. Eur J Biochem. 1985 Jun 3;149(2):345–351. doi: 10.1111/j.1432-1033.1985.tb08932.x. [DOI] [PubMed] [Google Scholar]
- Whitman W. B., Ankwanda E., Wolfe R. S. Nutrition and carbon metabolism of Methanococcus voltae. J Bacteriol. 1982 Mar;149(3):852–863. doi: 10.1128/jb.149.3.852-863.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wich G., Jarsch M., Böck A. Apparent operon for a 5S ribosomal RNA gene and for tRNA genes in the archaebacterium Methanococcus vannielii. Mol Gen Genet. 1984;196(1):146–151. doi: 10.1007/BF00334107. [DOI] [PubMed] [Google Scholar]