Abstract
Next-generation sequencing (NGS) is arguably one of the most significant technological advances in the biological sciences of the last 30 years. The second generation sequencing platforms have advanced rapidly to the point that several genomes can now be sequenced simultaneously in a single instrument run in under two weeks. Targeted DNA enrichment methods allow even higher genome throughput at a reduced cost per sample. Medical research has embraced the technology and the cancer field is at the forefront of these efforts given the genetic aspects of the disease. World-wide efforts to catalogue mutations in multiple cancer types are underway and this is likely to lead to new discoveries that will be translated to new diagnostic, prognostic and therapeutic targets. NGS is now maturing to the point where it is being considered by many laboratories for routine diagnostic use. The sensitivity, speed and reduced cost per sample make it a highly attractive platform compared to other sequencing modalities. Moreover, as we identify more genetic determinants of cancer there is a greater need to adopt multi-gene assays that can quickly and reliably sequence complete genes from individual patient samples. Whilst widespread and routine use of whole genome sequencing is likely to be a few years away, there are immediate opportunities to implement NGS for clinical use. Here we review the technology, methods and applications that can be immediately considered and some of the challenges that lie ahead.
Overview
Over the past six years we have witnessed a revolution in sequencing technologies that has already had a profound impact on our understanding of genetics and genome biology. In a research setting, NGS has been widely implemented for de novo genome sequencing, DNA resequencing, transcriptome sequencing and epigenomics. These research efforts have forged the way in the development of new protocols (both molecular and bioinformatic contexts) and have been instrumental in gaining an understanding of the major strengths and weaknesses of this technology. From a clinical perspective there is great potential for NGS in the management and treatment of human health. Immediate and significant impact will come from either replacement or augmentation of existing technologies for genetic screening purposes. Some striking examples of its clinical use include prenatal testing for the detection of chromosomal aneuploidy in foetal DNA,1 identification of rare genetic variants associated with monogenic Mendelian disorders2–4 and efficient detection of either inherited or somatic mutations in cancer genes.5,6
As cancer is a genetic disease driven by heritable or somatic mutations, new DNA sequencing technologies will have a significant impact on the detection, management and treatment of disease. Next-generation sequencing is enabling worldwide collaborative efforts, such as the International Genome Consortium (ICGC)7 and The Cancer Genome Atlas (TCGA) project,8 to catalogue the genomic landscape of thousands of cancer genomes across many disease types. Several early reports from individual studies contributing to these consortia have already been published.9–11 These discoveries will ultimately lead to a better understanding of disease pathogenesis, bridging to a new era of molecular pathology and personalised medicine.12 It is easy to imagine that soon every patient will have both their constitutional and cancer genomes sequenced, the latter perhaps multiple times in order to monitor disease progression, thus enabling an accurate molecular subtyping of disease and the rational use of molecularly guided therapies. Many molecular pathology laboratories are now considering the sequencing platforms, methods and additional equipment required for making the transition to NGS. Here we review the current sequencing technology, applications and bioinformatics with special consideration given to the development of clinical DNA sequencing.
Next-Generation Sequencing Technology
NGS broadly describes those technologies that share the ability to massively parallel sequence millions of DNA templates. The terms second-generation and third-generation sequencing are also used synonymously to describe the evolution of sequencing technology from the first-generation dideoxy ‘Sanger’ sequencing. To achieve massive parallel sequencing, second-generation platforms employ the clonal amplification of DNA templates on a solid support matrix followed by cyclic sequencing. The shift to single molecule PCR-free protocols and cycle-free chemistry is broadly characteristic of the progression to third-generation platforms.13 The advance of second- and third-generation technology has been enabled by innovation in sequencing chemistries, better imaging, microfabrication and information technology (IT). For the purpose of this review we will not discuss each platform in detail as these have been described extensively elsewhere.14,15 In addition, we will focus on the commercial second-generation platforms that are currently suitable for diagnostic applications, in preference to a detailed description of those platforms offered solely by sequencing service providers (Complete Genomics) or third-generation platforms such as Pacific Biosciences.16 Third-generation sequencing platforms offer many theoretical benefits relating to reduced cost, increased speed and removal of PCR-bias, however, the technology is still maturing and it is likely to be a few years yet before such platforms seriously rival the second-generation instruments and enter mainstream diagnostic use.
Second-Generation Sequencing Platforms
There are currently three companies offering second-generation sequencing platforms: Roche, Illumina and Life Technologies. Each company entered the market with large-scale instruments and maximum output in mind to satisfy a research market that demands a high-throughput technology for discovery-based applications and whole genome sequencing potential. Roche was the first to enter the market, acquiring the company 454 Life Sciences from its founder Jonathan Rothberg. The Roche 454 platform distinguishes itself from the other two large-scale platforms with longer read lengths, which are now approaching those of Sanger sequencing (700–1000 base pairs (bp)). The total sequence output from even the highest capacity 454 instrument (454 FLX+) is, however, far less than that of Illumina (HiSeq) and Life Technologies (SOLiD 5500), which generate many more sequence reads but of a much shorter length. Recently the attention has turned to smaller-scale low cost instruments with the introduction of the Roche 454 Junior, Life Technologies Ion Torrent and the soon to be released Illumina MiSeq, which are well suited to smaller research and diagnostic applications. A brief summary of currently available or near to release instruments and their performance is described in Table 1 but we also refer the reader to a more comprehensive review of current sequencing platforms, their specifications and cost breakdown for further detail.17
Table 1.
Current platform options for second-generation sequencing.
| Company/Platform | Sequencing | Amplification | Read length | Max. Output | Run time | Pros/Cons |
|---|---|---|---|---|---|---|
| Roche 454 GS FLX+ | SBS Pyro | emPCR | 700 bp* (SE, PE) | 700 Mb | 10–23 h |
Pro: Long reads, short run time Con: High Mb cost, homopolymer errors |
| Roche 454 GS Junior | SBS Pyro | emPCR | 400 bp* (SE, PE) | 35 Mb | 10 h | Same as GS FLX+ Additional Con: Lowest output of small scale instruments |
| Illumina HiSeq 1000 | SBS RDT | Bridge PCR | 36–101 bp (SE, PE) | ≤150 Gb | 1.5–8.5 days |
Pro: Ultra high output, ease of use Con: No run scalability like SOLiD 5500 |
| Illumina HiSeq 2000 | SBS RDT | Bridge PCR | 36–101 bp (SE, PE) | ≤300 Gb | 2.5–11 days | Same as HiSeq 1000 |
| Illumina GAIIx | SBS RDT | Bridge PCR | 36–151 bp (SE, PE) | ≤95 Gb | 2–14 days |
Pro: Mature platform Con: Superseded by HiSeq, higher Mb cost |
| Illumina MiSeq | SBS RDT | Bridge PCR | 36–151 bp (SE, PE) | >1 Gb | 4–27 h |
Pro: Proven chemistry, fully automated workflow Con: Unproven instrument |
| Illumina HiScanSQ | SBS RDT | Bridge PCR | 100 bp (SE, PE) | ≤150 Gb | 1.5–8.5 days |
Pro: Dual use instrument (microarray) Con: Higher Mb cost than HiSeq |
| Life Technologies 5500 | SBL | emPCR | 35–75 bp (SE, PE) | 77 Gb | 2–7 days |
Pro: Ultra high output, scalable runs allow sequencing on part flow cell Con: Shorter reads than other platforms, longer time to clonal template prep than Illumina |
| Life Technologies 5500XL (4hp) | SBL | emPCR | 35–75 bp (SE, PE) | 155 Gb | 2–7 days | Same as 5500 |
| Life Technologies Ion Torrent | SBS H+ | emPCR | 316+318 chip >100 bp (SE) |
316– >100 Mb 318– >1 Gb |
2 h+ |
Pro: Label-free chemistry– cheap and fast, highly scalable, long read length potential Con: Homopolymer errors, no PE yet, laborious template preparation but semi-automatable |
Note: Specifications for all platforms were derived from company websites.
Mode read length: the individual fragment read length is variable. SBS = Sequencing-bysynthesis; Pyro = Pyrosequencing; RDT = reverse dye terminator chemistry; H+ = Hydrogen ion detection; SBL = Sequencing-by-ligation; emPCR = emulsion PCR; SE = Single-end read; PE = Paired-end read; Mb = Megabases; Gb = Gigabases; bp = base pairs.
Second-generation sequencers rely upon two principles: polymerase-based clonal replication of single DNA molecules spatially separated on a solid support matrix (bead or planar surface) and cyclic sequencing chemistries. Each platform is defined by the methods used to achieve these two processes. All platforms have similar front-end library preparation methods involving the addition of universal adapter sequences to the terminal ends of the DNA fragment. These oligonucleotide adapters are complementary to PCR primers used to amplify the library and oligonucleotides immobilised to a solid support for clonal DNA amplification. Both Roche (454) and Life Technologies (SOLiD 5500 and Ion Torrent) use emulsion PCR (emPCR) to generate clonal DNA fragments on beads.18 A water and oil emulsion is created where beads and template are added at a precise concentration such that each emulsion droplet is likely to contain a single bead and single DNA molecule. Following emPCR the emulsion is broken, the template carrying beads are enriched and then deposited into separate ‘pico-wells’ or bound to a derivatised glass flow cell.19,20 Illumina uses an alternative strategy by creating DNA clusters directly on the flow cell by bridge PCR.21 From a practical perspective there are advantages and disadvantages to each approach. The process of emPCR is labour intensive although each company has developed automation to partially reduce the labour burden of this process. Cluster generation by bridge PCR has been fully automated and is therefore more streamlined. The MiSeq instrument, in fact, will require no user intervention from cluster generation to data analysis, which is highly attractive from a process point of view. The potential downside to Illumina bridge PCR is that the success of cluster generation is not known until sequencing has begun, an expensive exercise if cluster generation fails. From our experience, cluster generation is typically quite robust provided the sequencing libraries are of high quality and the concentration of the library is accurately measured by quantitative PCR.
Each of the available platforms uses different sequencing chemistries and methods for signal detection. All 454 platforms employ pyrosequencing, whereby chemiluminescent signal indicates base incorporation and the intensity of signal correlates to the number of bases incorporated through homopolymer reads.22 Ion Torrent uses a similar sequencing-by-synthesis (SBS) strategy but detects signal by the release of hydrogen ions resulting from the activity of DNA polymerase during nucleotide incorporation. In essence, the Ion Torrent chip is a very sensitive pH meter. Each ion chip contains millions of ion-sensitive field-effect transistor (ISFET) sensors that allow parallel detection of multiple sequencing reactions.23 There have been recent reports that Roche will adopt a similar detection method to Ion Torrent through a licence from the British company DNA Electronics, which would make the 454 and Ion Torrent platforms essentially identical.24 The virtues of semi-conductor technology are that the instrument, chips and reagents are very cheap to manufacture, the sequencing process is fast (although off-set by emPCR) and the system is scalable, although this may be somewhat restricted by the bead size used for emPCR.25 The SBS chemistry used by both 454 and Ion Torrent is also conducive to longer reads. Ion Torrent is currently restricted to fragments much shorter than that of Roche 454 but this will likely improve with future versions. Both have the common issue of homopolymer sequence errors manifesting as false insertions or deletions (indels). Whether fine-tuning the detection and analytical software can improve this issue is yet to be seen.
The Illumina and Life Technology SOLiD 5500 platforms are both considered short read sequencers but employ very different sequencing chemistries. Illumina uses reversible dye terminator SBS chemistry involving iterative cycles of single base incorporation, imaging and cleavage of the terminator chemistry. SOLiD uses sequencing by ligation (SBL) involving iterative rounds of oligo ligation extension, which is where the name originates (Sequencing by Oligo Ligation Detection). The principle of SBL in the context of massively parallel sequencing was originally described by Church and colleagues.26 The SBL process essentially measures every base twice by dinucleotide encoding, which is translated into ‘colour space’ rather than conventional base space. The new ‘Exact Call Chemistry’ offered with the 5500 line instruments uses a three base encoding, allowing even greater accuracy and actual base calls. Illumina SBS has a slight advantage over SOLiD SBL in terms of read length, now up to 100 bp on HiSeq and 150 bp with other Illumina instruments. The SOLiD SBL chemistry has a maximum read length of 75 bp but the two or three base encoding provides higher accuracy over the Illumina chemistry. Both Illumina and SOLiD platforms have a paired-end or mate-paired capability enabling reads to be generated from both ends of a single clonal fragment, as do the Roche 454 platforms.
Choosing a Platform
In choosing a platform there can be many considerations. Principally, one may be concerned with performance metrics such as read length, accuracy and total sequence output. In general, all second-generation platforms produce data of a similar accuracy (98–99.5%), relying upon adequate sequence depth or ‘coverage’ to make higher accuracy consensus base calls (>99.9% accuracy). Some systematic biases have been reported when comparing between NGS platforms and with other orthologous technologies such as Sanger sequencing. For example, significant non-uniformity of sequence coverage has been reported with short read instruments, whilst systematic errors have been reported for all NGS platforms.27–29 Systematic errors rather than random errors are more problematic as they cannot be overcome with higher read coverage. Modifications to protocols suggested by large genome centres may help to improve some uniformity issues,30 whilst newer read alignment and variant calling strategies have helped to reduce systematic errors caused by multi-mapping of reads or presence of indels (see Bioinformatics section below). Read length may be important for specific applications, such as identification of complex structural rearrangements or mapping across repetitive sequences. There is also some advantage in longer reads for direct amplicon sequencing.
Additional process-related considerations such as sequence output and running cost need to be balanced against speed and hardware costs. The larger more expensive instruments tend to have longer run times but generate orders of magnitude more data at a fraction of the price. Molecular barcoding or indexing of samples can be used with all platforms, allowing pooling or multiplexing of samples for high-throughput and translation of lower sequencing cost to targeted sequencing applications requiring a relatively small number of reads. Very high-throughput, however, is required to fill runs on the larger sequencing platforms in order to take advantage of the capacity and reduced cost, although platforms such as the SOLiD 5500 have more flexibility in this regard. If fast turnaround and flexibility is paramount then the smaller-scale instruments are likely to be preferable. Usability and reliability of the equipment are important and often cannot be ascertained from the company marketing the product. A large online NGS community has emerged with user forums, news and blog sites often being useful for getting first hand experience and early insights into new equipment and methods (Table 2). Informatics is also a very important consideration. Informatics hardware cost is largely dependent on the throughput of the sequencer. Larger sequencers require Linux servers with multiple cores and large amounts of RAM at a significant capital cost and require dedicated human resources to maintain, whilst smaller-scale sequencers can be run from high-powered Windows or Linux-based desktop systems.
Table 2.
Internet genomics news, forums and blog sites.
| Name | URL | Type |
|---|---|---|
| GenomeWeb | http://www.genomeweb.com | News |
| SEQanswers | http://seqanswers.com | Forum |
| MassGenomics | http://www.massgenomics.org | Blog |
| Next-Gen Sequencing | http://nextgenseq.blogspot.com | Blog |
| Omics! Omics! | http://omicsomics.blogspot.com | Blog |
| Genetic Future | http://www.wired.com/wiredscience/geneticfuture | Blog |
| Blue Collar Bioinformatics | http://bcbio.wordpress.com | Blog |
| Genetic Inference | http://www.genetic-inference.co.uk/blog/ | Blog |
Sequencing Applications
Whole Genome Sequencing
The predominant application of NGS in a clinical setting will undoubtedly be resequencing of genomic DNA. Whole genome sequencing (WGS) simply provides the ultimate genetic survey of an individual’s genome or cancer genome where a detailed map of single nucleotide variations (SNV), indels, complex structural rearrangements and copy number changes can be attained in a single assay.31 As the sequencing technology has advanced there have been major improvements to data quality and throughput, driving the cost of WGS towards $1000/genome, a threshold widely considered to be the tipping point for widespread clinical implementation. However, generating sufficient data for WGS remains a relatively expensive exercise for most laboratories. Strong competition between large service providers means that outsourcing WGS is currently a more affordable option for most and perhaps one that will persist until the third-generation technologies mature and become mainstream. An important consideration for sequencing whole genomes is that generating the actual sequence data is only a fraction of the total cost and does not take into account the expense associated with data storage, analysis and interpretation.32 Sequencing an entire genome reveals an enormous amount of genetic information, some of which can be interpreted and actionable, but a significant amount will be either novel and/or of unknown clinical importance. Additionally, there are significant ethical issues concerning privacy of data and incidental findings that will need to be resolved. For these reasons a more targeted approach to genome sequencing seems to be the logical first step towards widespread clinical implementation of the technology.
DNA Library Preparation
Regardless of the technology, all NGS platforms follow similar molecular protocols for the preparation of sequencing libraries. Although standard library preparation does not necessitate any specialised equipment, there are a number of auxiliary instruments that can aid in the library preparation process (Table 3). For the preparation of ‘shotgun’ fragments, DNA is sheared either mechanically or by enzymatic digestion to create fragment sizes in a required size range. A series of enzymatic steps are then used to repair library DNA ends and ligate common adapters that are complementary to oligonucleotides on beads or flow cells. Library preparation reagents are provided in kit form by the major NGS vendors and are also available through third-party companies. A relatively new alternative to the conventional fragment library preparation involves an in vitro transposition method (Nextera, Epicentre/Illumina), which removes the need for mechanical shearing and multiple enzymatic and purification steps. PCR is commonly used to amplify libraries prior to sequencing, however, the number of cycles is often limited to avoid excess PCR duplicates that can contribute to false-positive sequencing error. The use of PCR and common adapter sequences introduces a high risk of cross contamination between libraries therefore standard principles of pre- and post-PCR work areas are essential. Size selection of DNA libraries may be necessary to aid in analysis and standardise cluster size. Traditionally, size selection has been done using either agarose or polyacrylamide gel electrophoresis (PAGE), however, new automated methods are also available. Finally, high-throughput library preparation can be automated and there are several commercial solutions currently available.
Table 3.
Auxiliary laboratory instrumentation for next-generation sequencing library preparation.
| Equipment | Description/Use | Vendors and Instruments |
|---|---|---|
| Shearing Device | Used for shearing of DNA or RNA in creation of shotgun sequencing libraries | Covaris S-Series E-series L-series (focal acoustic) Epigentek EpiSonic Multi-Functional Bioprocessor 1000 (sonication) Diagenode Bioruptor (sonication) Digilab Hydroshear (PointSink Shearer) |
| Automated Microfluidic Electrophoresis | Automated size separation and analysis for quality assessment of RNA, DNA and library preparation | Agilent Bioanalyzer 2100 Caliper LabChip GX/GXII Qiagen QIAxcel Shimadzu MCE-202 MultiNA |
| Automated Fragment Size selection | For size selection of libraries as automated alternative to agarose gel or PAGE DNA extraction | Sage Science Pippen Prep Caliper LabChip XT/XTe |
| Liquid Handling Automation | Automated library preparation, hybridisation capture and other enrichment methods | Agilent Bravo Caliper Sciclone NGS Workstation Qiagen EpMotion Diagenode SX-8G IP-Star (Principally epigenetic applications) Beckman SPRI and NX |
Sequencing Requirements
To overcome the higher error rate of NGS platforms compared to traditional Sanger sequencing a high level of redundancy or sequence coverage is required to accurately call bases. Typically, a 30–50x coverage is required for accurate base calling, although this can vary based on the accuracy of the sequencing platform, variant detection methods, and the material being sequenced.33 Using the Illumina HiSeq instrument approximately 100 Gb of sequence data is required to sequence a diploid genome or about three lanes of a flow cell using the new V3 sequencing reagents. Less coverage may be required on the SOLiD 5500 platforms owing to the higher read accuracy enabled by the two base encoding. Greater depth may be necessary for interrogating cancer genomes where normal tissue contamination and the heterogeneity of some cancers can reduce variant allele representation in sequence data well below the 50% frequency expected for a diploid heterozygous call.
Targeted enrichment before sequencing can reduce costs, allow higher coverage over regions of interest and potentially simplify the bioinformatic interpretation of NGS data. The amount of sequencing required for targeted applications will ultimately depend on the method and target region size. As an example, for whole exome sequencing (targeting all annotated coding genes) approximately 10–12 Gb of data is required, achieving an average of 100-fold coverage and at least 20-fold coverage for 80–90% of targeted bases. At current specification this means up to 32 exomes can be run per flow cell on the HiSeq instrument with similar throughput likely to be possible on SOLiD 5500. The smaller sequencing instruments such as MiSeq generate substantially less data (∼1 Gb) and therefore are suited to smaller targeted sequencing applications where, at most, a few hundred genes could be sequenced in a single run.
Targeted DNA Sequencing
Targeted enrichment strategies feeding into NGS are finding traction in both research and clinical diagnostic fields. An assortment of methods and technologies has been described in the literature, most of which can now be purchased as commercial products (Table 4). When comparing these approaches there are several factors that need to be considered. From a technical perspective the fidelity of capture is important. Off-target enrichment and low uniformity of capture can mean more sequencing is required to attain adequate sequence depth for all targeted regions. Different capture methods can also be affected by sample quality and the presence of variants within the capture region. Scalability, throughput and ease of use are important for high-throughput, whilst the targeted region size may dictate what method is most appropriate. Finally, the need for specialised equipment and the reagent price are also key considerations.
Table 4.
Targeted enrichment methods for next-generation sequencing.
| PCR | Fluidigm Access Array | RainStorm | MIP† | TruSeq Amplicon | Hybridisation Capture | |
|---|---|---|---|---|---|---|
| Sensitivity | High | High | High | High | High | High |
| Specificity | High | High | High | High | High | 70–80% on target depending on design |
| Uniformity | Variable-high with normalisation of PCR products | 90% TB within 2-fold mean coverage* | >90% TB within 10-fold mean coverage | 58% TB within 10-fold mean coverage | Unknown | 80–90% TB within 5 fold mean coverage |
| Run speed | Small region-high Large region-low |
High | High | High | High | Low |
| Max. Capture Size | Low | Dependent on amplicon length and multiplexing | 1.63 Mb demonstrated | 1.7 Mb demonstrated | 384 x 250 bp amplicons | 42–62 Mb |
| DNA Amount | ∼5 ng per amplicon | 50 ng | 2 μg | >200 ng | 200 ng | 500 ng–3 μg |
| Sample Throughput | Moderate | High | Low-moderate | Low-moderate | High | Low-moderate |
| Reagent Cost/Mb | High | Moderate | Moderate | Low | Moderate | Low |
| Automation | Yes | Yes | Yes | NA | Possible | Yes |
| Design service | NA | Yes | Yes | NA | Yes | Yes |
| Company | NA | Fluidigm | RainDance | NA | Illumina | Agilent Roche Nimblegen Life Technologies Illumina Rivia Febit |
Targeted enrichment methods fall broadly into two categories: PCR-amplicon and hybridisation capture approaches. As PCR-based approaches are already used routinely in diagnostic laboratories they fit well with existing diagnostic workflows. PCR is highly specific and has the advantage of generating more uniform coverage than comparative hybridisation approaches, provided the concentrations of individual PCR products are adequately normalised before pooling and sequencing. Different strategies have been used to generate PCR amplified libraries. Some use concatenation of PCR products to generate fragment libraries; shearing PCR concatamers and feeding into shotgun library preparation. A more straightforward protocol that is compatible with long-read sequencing instruments is to incorporate the sequence adaptors into the 5′ -end of the PCR primer enabling pooling of amplicons and direct sequencing. Conventional PCR methods are obviously better suited to targeting a small number of regions as the logistics, cost and amounts of DNA required to assay larger regions can be prohibitive.
Long-range PCR (LR-PCR) can reduce the burden of generating tens of PCR primer sets to amplify across regions of interest and has been employed to target contiguous regions or to amplify across several exonic regions. Uneven coverage can be an issue using LR-PCR although some remedies to this have been described.34 Generation of long amplicons can also be prone to reproducibility issues and is inherently not suited to the use of degraded DNA such as from formalin fixed paraffin embedded (FFPE) material. Additionally, the need to generate shotgun sequencing libraries post-PCR, regardless of sequencing platform, creates further work, expense and potential risk for failure and contamination.
The key to scaling PCR-based sequence enrichment involves automation, miniaturisation and multiplexing of PCR reactions. All of these methods aim to increase the scale of PCR enabling hundreds to thousands of PCR reactions whilst minimising reagent use, labour burden and amount of DNA template required. Two commercially available platforms enable miniaturised PCR by microfluidics. Fluidigm is a microfluidics-based method that uses multilayer soft lithography (MSL).35 A microfluidic circuitry is fabricated from a soft rubber composite that allows the controlled flow of reagents by using pressure to create tiny valves in the circuitry and reaction chambers for PCR. Fluidigm was originally developed for real-time quantitative PCR and single nucleotide polymorphism (SNP) genotyping applications but more recently the Access Array has been released, allowing retrieval of PCR product for targeted resequencing applications. The current Access Array system is capable of parallel PCR reactions for 48 samples by 48 single-plex assays. An attractive aspect of this platform is that relatively small quantities of template are required (∼50 ng/sample). Assays can also be multiplexed to improve throughput. Furthermore, as with many targeted approaches, index or barcoding tags can be incorporated into the universal adapter regions of the PCR product enabling the pooling of samples before direct sequencing.
A second platform, RainStorm (Raindance Technologies36), involves the generation of microdroplets in an oil emulsion, which then act as miniaturised reaction chambers for PCR.37 Highly uniform microdroplets containing reaction components (PCR primers and DNA template) are created using a combination of microfluidic chip design and high-pressure pumps. Thousands of reagent-laden microdroplets are loaded into a microfluidic device with a steady oil stream where they can be merged and manipulated through channels using electromagnetic fields before emPCR amplification and then retrieval of amplified product. Recently, this method has been marketed for the use of DNA extracted from FFPE tissue. The current limitations of this technology are the relatively large amount of DNA required and the sequential processing of individual samples.
The alternative to miniaturisation and microfluidic manipulation is to use methods enabling highly multiplexed PCR. One such approach uses molecular inversion probes (MIP). A MIP is a long oligonucleotide composed of sequence specific primer ends tethered by a universal linker sequence. Target specific primer ends hybridise to complementary DNA flanking the region of interest. Polymerase extension and then ligation results in the circularisation of the MIP. Captured regions are then amplified either by rolling circle amplification or by PCR from universal PCR priming sites within the linker region.38,39 The assay was originally described for amplification of exons targeting a relatively small number (10) of genes. However, the method was later shown to be highly scalable, and by using programmable microarrays, MIP pools targeting up to 50,000 exonic regions could be generated.40,41 The downsides to MIPs, however, are that they have been shown to provide inferior capture uniformity compared to hybridisation-based enrichment and they can be expensive for low throughput or custom applications as currently there are no commercial reagents available.
Illumina has developed a method that is similar in principle to MIP and is due for release in 2011 (personal communication, Illumina Australia). The ‘TruSeq Amplicon’ approach is derived from the method used for the Illumina ‘Golden Gate Genotyping’ assay. Instead of using MIP, two independent left and right flanking oligonucleotides are hybridised to a genomic DNA template enabling polymerase extension and ligation. Like MIP, the flanking oligonucleotides contain universal sequences for step-out PCR and incorporation of universal barcoded Illumina adapters. According to the vendor (Illumina) up to 384 targets can be amplified in a single reaction. The capture is theoretically very scalable, as all steps can be performed in a 96-well PCR plate and can be automated by liquid handling. The detailed performance specifications of this method are currently unknown.
Targeted enrichment by hybridisation capture has been extensively employed in a research setting and is typically suited for capture of larger target regions and exons from hundreds of genes. Oligonucleotides designed against complementary target regions are used as probes or ‘baits’ to hybridise and capture target DNA or ‘prey’ from pre-prepared shotgun libraries.42 The majority of methods employ microarray in situ oligonucleotide synthesis to generate the bait libraries (e.g. Agilent, Roche Nimblegen, Rivia), whilst Illumina uses its massive oligonucleotide production facilities to generate long oligonucleotides by conventional column-based synthesis. The microarray itself can be used as the capture device or the sequences can be cleaved from the array to generate in-solution bait libraries.43 The solution capture method has been more widely utilised owing to the scalability of the process, better performance for larger capture regions and no requirement for specialised equipment.44 The advantage of hybridisation enrichment is the ability to easily capture large regions in a single tube assay and it has become the mainstay of ‘exome’ enrichment resequencing for research.45–48 All companies now offer services for design of custom bait libraries in addition to off-the-shelf reagents for common applications, meaning fast turnaround and a price competitive market. The downside to hybridisation capture is the lack of specificity (compared to PCR) owing to cross hybridisation and lack of uniformity in capture, which relates to GC content of target sequence. Improvements to the method have been described, helping to overcome some of these issues, in addition to optimised methods for high throughput automation.49
RNA Sequencing
The application of RNA sequencing (RNA-seq) is fast superseding microarray technology by providing a superior digital readout whilst also enabling discovery of novel spliceforms, transcripts and RNA-editing.50 Quantitative gene expression data derived from RNA-seq has been shown to be comparable to that of microarrays but has better dynamic range and lower detection limit for lowly expressed transcripts.51 Methods for analysing differential expression from RNA-seq data, however, are still being resolved and are reminiscent of the initial issues encountered for normalisation and statistical analysis of microarray data.52 Clinical applications of gene expression microarrays as a cancer diagnostic and prognostic tool have been demonstrated with examples including identification of tissue of origin for cancer of unknown primary and prediction of recurrence in early stage breast and colorectal cancer.53–55 Whilst it remains to be seen whether RNA-seq could replace microarrays or quantitative PCR as a clinical assay, it would seem intuitive to think that this could occur if the technology becomes cheaper and more robust.
RNA-seq has also been applied to mutation detection and has proven especially useful for the detection of gene fusions resulting from genomic translocations.56 A virtue of calling variants from RNA-seq data over genomic DNA sequencing is that the data simultaneously provides functional information about a variant or fusion product. The large dynamic range of expression between genes, however, makes it difficult to acquire sufficient sequence coverage to accurately call variants without generating large amounts of data per sample. Targeted hybridisation capture of cDNA libraries, similar to that used for DNA sequencing, has recently been shown to improve the sensitivity to detect sequence variants and fusion products from RNA-seq data.57 Any advantages of sequencing RNA in preference to DNA for mutation detection are yet to be demonstrated. The larger number of RNA transcript copies that can be derived from a single cell, however, may suggest RNA-based methods could have greater sensitivity over those interrogating DNA.
Epigenomics
There is growing appreciation for the role of epigenetics in cancer.58 The burgeoning field of epigenetics has been advanced by the development of NGS methods for surveying DNA methylation, mapping of transcription factor occupancy, modified histones and epigenetic regulators. The most well studied epigenetic mark, DNA methylation, can be interrogated at the whole genome level by bisulphite sequencing.59 Methods for reduced representation bisulphite sequencing (RRBS) and targeted enrichment followed by bisulphite treatment can also be used to reduce sequencing cost whilst maintaining the nucleotide resolution and qualitative measurement.60,61 Other methods employ affinity enrichment assays using methyl cytosine-specific antibodies (MeDIP-seq) and recombinant methyl binding domains of proteins such as MBD2.62 These methods work in a similar way to chromatin immunoprecipitation (ChIP-seq) where enriched methylated DNA or an immunoprecipitated protein-DNA chromatin complex are aligned to the genome to reveal peaks, marking the averaged distribution of the epigenetic mark of interest.63–65 Like RNA-seq, these methods are superior to array-based interrogation for discovery applications as they make no a priori assumptions of where to interrogate the genome. The potential diagnostic and prognostic utility of DNA methylation has been demonstrated.66 Single gene DNA methylation assays, such as those assaying MGMT methylation to predict response to DNA alkylating agents in glioma, are now being incorporated into clinical management,67 whilst single gene assays for detection of lung and colorectal cancer are also in development.68 However, to our knowledge, no multi-gene or genome-wide DNA methylation assays have yet been clinically adopted.
Bioinformatics for Next-Generation Sequencing
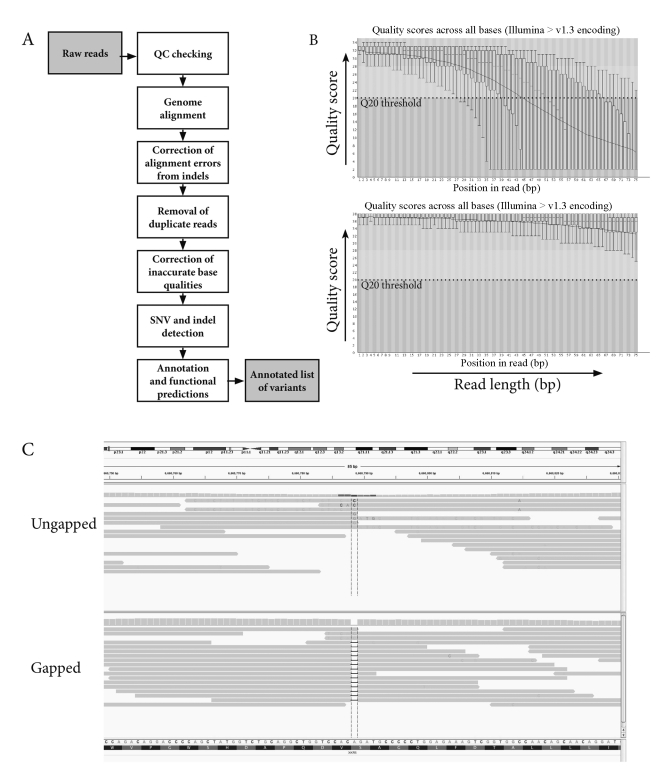
Whilst sequencing technologies have developed at a rapid rate and robust commercial platforms have become readily available, the analysis of NGS data remains a significant challenge. For experienced users of Sanger sequencing, the general concepts of analysing NGS data can be easily digested. However, the sheer scale of analysis, terminology and need for command line computer languages can be a formidable challenge. To overcome the latter issue there are several commercial solutions for NGS analysis, some of which are provided by the companies distributing NGS instruments. These programs aim to make the analysis simpler by providing easy-to-use graphical user interfaces (GUI). Such software tools may be a suitable entry point for small-scale laboratories, especially for analysis of simple datasets, but are generally limited in their flexibility and scalability and often do not adequately resolve issues around data handling and management. It is also important to remember that many challenges around NGS analysis are still being resolved and that commercial software packages are not exempt from common issues experienced with analysing NGS data and may not be as advanced as the open-source tools being developed by large genome centres. In a research setting, most groups have invested in bioinformatics support by employing computer programmers and IT specialists for developing sequencing analysis pipelines. This partially reflects the complexity and scale of discovery-based applications but is also indicative of the continuous advances being made in this area. For the purpose of this review we will discuss the basic concepts of analysing NGS data for DNA resequencing that are common to analysis pipelines (Figure 1A).
Figure 1.
(A) A typical pipeline schematic for variant detection with next-generation sequencing, showing the steps from raw reads to production of a final list of variants. (B) Box plots created by the tool FastQC showing the base qualities of reads for two samples. The box plot on the top shows a ‘bad’ result, where the quality deteriorates rapidly from the middle of the reads to the ends, while the box plot on the bottom shows a ‘good’ result with just a slight decrease in quality towards the ends of the reads. Trimming of the reads in the sample on the top is recommended as it would lead to more accurate variant detection. (C) The effect of using an ungapped (MAQ) versus a gapped aligner (BWA). With the ungapped aligner the presence of a deletion in the sample leads to misalignment of the parts of the read next to the deletion resulting in false positive single nucleotide variation (SNV) calls.
Aspects of the Analysis Pipeline
Primary data output from each platform essentially consists of a text file containing raw sequence reads plus the quality scores for each base. Each sequencing platform comes with its own proprietary analysis software to call the bases and generate the associated quality metric. Independent base calling algorithms and software tools have also been developed to improve upon base calling accuracy and reduce systematic errors,69 although for the average user these third-party softwares are unlikely to be considered. While quality scores cannot be directly compared between sequencing platforms they generally all use a Phred-like score, which is logarithmically related to the base-calling error probabilities.70,71 A quality score assigned to a base is relative to the confidence in detection, with individual factors affecting the base quality being determined by the nuances of each sequencing technology. Base quality tends to deteriorate towards the ends of reads and hence low quality ends may need to be trimmed to improve the overall data quality (Figure 1B). The software provided with each platform will typically provide an overall run quality report, whilst quality assessment using third-party tools (e.g. FastQC72) are also frequently used.
Shorter reads, higher error rates and the sheer scale of data mean that specific methods are required for NGS read alignment. A number of alignment algorithms exist and some of the most popular ones are BWA, MAQ, Bowtie and Novoalign,73–76 whilst most commercial softwares have developed their own proprietary algorithms. Gapped aligners such as BWA and Novoalign are better suited to variant detection compared with non-gapped aligners like MAQ and Bowtie as these enable indel detection and are less prone to calling false-positive SNVs around indels (Figure 1C). Performance differences exist between algorithms and there is typically a trade-off between speed and accuracy.77 Once alignment is complete, sequence alignment maps (SAM or binary format BAM) are generated that can be used to visualise sequence reads in genome browsers such as IGV.78 Commercial solutions are likely to have a genome browser built into the software package and the intermediate files generated during analysis are often hidden from the user.
As in the case of alignment, there are many programs available for variant calling, the most commonly used of which are Samtools, GATK Unified Genotyper and SOAPsnp.79–81 The challenge is to separate real variations from sequencing noise and most variant callers use Bayesian algorithms which incorporate the probability of seeing a variant at a particular location given the known polymorphism rate and sequencing errors. Detection of variants has become increasingly sophisticated and further refinement steps are now commonly carried out post-alignment to increase the accuracy of variant detection. For whole genome and exome analysis, these steps include realignment of reads around indels, removal of duplicate reads, and recalibration of base quality scores prior to variant calling.79 Phred-like quality scores are generated for the consensus variant calls, however, like the quality scores generated for individual reads, these scores cannot be directly compared between different variant calling programs.
Once variant detection is complete, results are then typically annotated to add gene and transcript identifiers and to predict the functional significance (i.e. non-synonymous or frameshift changes). Variants may be compared with databases to identify known associations with disease and also there are a number of tools available to predict the functional consequence of novel missense changes.82 Genome databases such as ENSEMBL provide an application program interface (API) facilitating automated annotation of variants.83 Commercial programs often require the user to upload individual annotation files separately, which can be laborious and inherently more difficult to manage.
Structural Variation and Copy Number Analysis
Structural variation, which includes copy number alterations, is responsible for a large proportion of the variation between individuals84 and is a frequent event within cancer. The identification of structural variants from NGS data, especially short read platforms, is often considered more challenging than for SNV and small indels owing to the complex types of rearrangements that can take place, such as translocations, inversions and tandem duplications. Structural variations are also more common in repetitive regions, which creates mapping issues for short read alignment. Methods for detecting structural variations from NGS data are still under active research and development and four different approaches are currently being used; read-pair, read depth, split-read and assembly. These methods can produce widely different results with little overlap.85 The majority of studies that have applied this type of analysis have used data from WGS to identify structural variation, however, it has been shown that structural variation analysis can also be applied to targeted resequencing data. This could have new application in a clinical setting to replace methods for identification of common translocation fusion partners or for identifying viral integration sites.86–88
Common Errors/False Positives and How to Overcome Them
False positives can still be a major problem using NGS but steps can be taken to minimise this issue. Common errors occur due to ambiguity in short read sequence alignment and sequencing errors. Multi-mapping can frequently occur if a read aligns to paralogous or repetitive regions within the genome. Furthermore, it is known that the human genome assembly is not perfect, with gaps and misassemblies present, which can also lead to misalignments.89,90 With short reads, misalignments to the reference genome are a more likely occurrence. Longer reads, such as those produced by Roche 454 technology, enable reads to be aligned with higher confidence, whilst using paired-end reads can also help to mitigate the multi-mapping issue.
Another major source of false positive variants arises through either mis-incorporation of bases during PCR amplification or sequence detection errors. Duplicate reads contribute to false positives derived from PCR-associated errors and are therefore routinely removed during analysis. Paired-end sequencing from shotgun fragment libraries are useful in this context as there is a high likelihood that two reads are duplicates if paired-ends from two independent fragments have the same start sites. When using a PCR-amplicon approach, this problem is not so easily resolved as all reads will align with the same start location based on primer design. Designing overlapping PCR-amplicons and accepting only consensus variant calls between amplified intervals may help to reduce this problem. Recently, an alternative approach called SafeSeq has been described to reduce false positives from PCR amplicons.91 The method, in principle, is similar to molecular barcoding (indexing) of samples. Instead of using a known 4–6 base index to tag individual samples during library preparation, every molecule is given a unique barcode by incorporating a degenerate 12–16 base index into the universal adapter sequence. Samples are subject to PCR and deep sequencing whereby only variants identified in 95% of duplicate reads are kept. The application is well suited to detection of rare variants but could be applied to any PCR-based diagnostic application.
Cancer-Specific Challenges
Sequencing tumour samples presents an additional set of challenges for bioinformatic analysis. Challenges specific for variant detection in cancer datasets result from the inherent characteristics of tumour samples: aneuploidy, tumour heterogeneity and contamination with normal tissue. Most of the widely-used variant callers such as GATK Unified Genotyper and Samtools assume a diploid genome and are thus not well suited to tumour samples, where diploidy is not guaranteed and copy number alterations are common. Efforts have been made to develop tools specific for cancer variant detection, such as SNVmix, addressing the challenge of low frequency variant detection.92
Current Opportunities for Cancer Diagnostics
There are a number of exciting possibilities for the implementation of NGS in a clinical setting and, not surprisingly, there is a lot of interest and activity in this space. Some of these methods will simply replace existing Sanger sequencing or PCR-based assays for genetic testing within genes linked to familial cancer syndromes or for detection of mutations in genes of therapeutic importance within cancer cells or tissues. A real driver for these applications will be the ability to rapidly screen numerous gene targets at minimal cost. Notwithstanding existing legal battles concerning infringements over gene patents, this will hopefully translate to more individuals being allowed access to genetic testing and a positive outcome for patients and their families. The emergence of small molecule inhibitors and antibodies against druggable gene targets is revolutionising cancer treatment. Many of these agents are considered most effective when used in combination with companion diagnostic assays. Other novel clinical applications are also emerging, including the monitoring of disease burden though minimally invasive detection of tumour DNA in the peripheral blood of cancer patients.93,94 However, these more novel applications still remain in development and will not be discussed in greater detail here.
Familial Genetic Testing
Genetic testing for high penetrance familial cancer genes, such as BRCA1, BRCA2, APC and the mismatch repair genes to name a few, is provided to individuals deemed to be at high risk due to their family and clinical history. The introduction of NGS should translate to significant cost savings through simultaneous sequencing of multiple targets and multiple samples. One immediate and positive impact of reduced-cost sequencing for genetic testing is that individuals with diseases such as breast and ovarian cancer, who don’t meet the current stringent criteria for recommending genetic testing, may become eligible for screening, since a major factor in determining the stringency of such guidelines is the cost of genetic testing and available resources. It has been reported that around 30–50% of individuals with a mutation will not have a significant family history to warrant testing.95,96 Therefore, these individuals would only be tested if other more local guidelines are used such as young age of onset or triple negative breast tumour pathology. These groups are likely to benefit from a more readily available NGS approach. There is a similar situation in high-grade serous ovarian cancer in that around 50% of women with serous ovarian cancer who had a BRCA1 or BRCA2 mutation did not have a significant family history.97
Furthermore, lower frequency genes not routinely tested but implicated in familial cancer syndromes could be included in a standard genetic screen if disease risks could be attributed to mutations in such genes. The selection criteria for testing could then be based on whether variants within a particular gene can be used to improve risk estimates given by the clinic to the patient rather than resource limitations. An additional positive impact would likely be a reduction in the timeframe for genetic testing from months to weeks while at the same time increasing the potential throughput for testing. This would increase the utility of BRCA1 and BRCA2 testing in the context of enrolment in trials open to carriers of BRCA1 or BRCA2 mutations such as Poly (ADP ribose) polymerase (PARP) inhibitor trials and as an aid to surgical management decisions.98
There have been several recent publications describing the use of NGS for the purpose of familial cancer genetic screening.99–102 As a first example, Morgan et al. used LR-PCR to amplify and then sequence across all exons of BRCA1, BRCA2 and TP53 in a series of cell lines and patient samples.99 A relatively small target size enabled pooling of samples for sequencing on a single lane of an Illumina Genome Analyzer (IIx) flow cell. They found that all known pathogenic variants (determined by Sanger sequencing in the same sample set) could be found, including deletions up to 16 bp, with zero false positives using either the commercial software NextGene (SoftGenetics) or custom developed software for analysis.
A second study by Walsh et al. used a hybridisation capture approach to sequence 21 genes known to be associated with predisposition to breast and ovarian cancer.102 They designed bait libraries against coding and intronic regions in addition to 10 Kb upstream of each genic region totalling approximately 1.0 Mb capture size. A panel of samples with known point mutations, deletions and duplications were sequenced attaining greater than 1200-fold average coverage and at least 20-fold coverage for every targeted base. Every known pathogenic change was identified in the samples tested including up to 100 Kb deletions that could be determined from inferred copy number using depth-of-coverage from the capture enrichment and sequencing. This study demonstrates that lower uniformity of coverage from using hybridisation capture could be overcome with simply greater sequencing depth, whilst the ability to derive copy number information from the data and sequence a larger number of genes is a clear advantage over PCR-based approaches. However, the specificity of hybridisation capture still remains as a point for further investigation. This is especially important when dealing with diagnostic assays for genes with highly homologous pseudogenes, such as PMS2, BRAF and CHEK2, associated with colorectal cancer, melanoma and breast cancer respectively, since this could theoretically lead to misinterpretation of mutation status or copy number data.
Identification of Somatic Mutations in Cancers
Recently, a number of targeted therapies have become available for various cancers, most notably melanoma, lung cancer and colorectal cancer. The success of such treatments relies to a very large extent on the genetic profile of the individual tumour being treated. Melanoma tumours harbouring the p.Val600Glu mutation in the BRAF gene respond dramatically in the first instance to treatment with vemurafinib103 whilst non-small cell lung carcinomas respond to treatment with gefitinib or erlotinib if they harbour one of a range of activating mutations in the EGFR gene.104 Conversely, most exon 12 and 13 mutations in the KRAS gene are predictive of a lack of response to monoclonal antibody therapies, such as cetuximab and panitumumab, directed at the EGFR protein.105
Whilst single targets for single therapies are currently the norm, it is very likely that future treatments will rely more on therapies directed to multiple targets to avoid relapses common to these treatment modalities. Furthermore, it is likely that somatic mutations, either acquired or primary, may contribute as a mechanism for disease relapse. Indeed, the p.Thr790Met mutation in the EGFR gene has been described as a resistance mutation for targeted therapies with potential for predicting the likelihood of disease relapse following treatment.106 Therefore, monitoring of disease response to therapy to detect emerging resistance and concomitant genetic testing to detect de novo mutations will likely form part of many future treatment strategies.
Initial reports of using NGS to clinically sequence multi-gene panels in patient tumour samples are now emerging. For example, a recent study by Wagle et al. has reported the use of targeted resequencing in melanoma to identify a previously unknown mechanism of acquired resistance to the BRAF inhibitor PLX4032 (vemurafinib).107 Hybridisation capture and resequencing of 138 cancer genes in tumour samples taken from a single patient before and after relapse revealed a p.Cys121Ser mutation in the MEK1 kinase that was only found in the relapse sample. Experimental validation involving ectopic expression of the MEK1 p.Cys121Ser allele in a melanoma cell line harbouring BRAF p.Val600Glu mutation, which is normally highly sensitive to BRAF inhibition, confirmed the resistance phenotype of the MEK1 mutant. This is the first report of an activating mutation occurring downstream of the BRAF kinase, adding to a growing list of other known mechanisms of acquired resistance to BRAF inhibition.108–110 It would seem unlikely that novel mutations such as this would have been found using conventional sequencing or genotyping approaches. However, it also raises the issue of how to interpret any new findings in clinical practice. Building a knowledge-base of genes and mutations that are likely to be conferring resistance to a target therapeutic agent will be important for the interpretation of sequencing results and future deployment of effective combination therapies.
Sequencing tumour DNA from diagnostic tumour tissue can present specific technical challenges including the use of heterogeneous tumour samples and the use of small amounts of degraded and fixative-affected DNA. A major difficulty encountered when attempting to detect mutations in gene targets arises from a trade-off between sensitivity and the detection of de novo sequence variations. As an example, standard Sanger sequencing is capable of detecting the majority of mutations within a targeted region, however mutations present at less than 10% are unlikely to be detected. Specific allelic discrimination assays have been reported with sensitivities of 99% or greater, however highly multiplexed allelic discrimination assays such as the Oncomap panel have average mutation detection sensitivities approaching 89% or less for FFPE samples.111 NGS appears ideally suited to address both challenges of sensitivity and coverage. A recent study by Querings et al. benchmarked the sensitivity of detecting EGFR and KRAS mutations in lung cancer specimens comparing PCR enrichment followed by NGS against Sanger sequencing and pyrosequencing.112 They found that NGS provided sensitivity superior to the other two methods, detecting 100% of patients with response to an EGFR inhibitor.
Undoubtedly one of the major technical challenges to using NGS for cancer diagnostics will be the use of DNA extracted from FFPE material. Very few published studies have yet assessed the ability to use DNA extracted from FFPE material for NGS resequencing.86,113 We and others have begun to test the sequence enrichment methodologies on FFPE pathology specimens, using either PCR-based or hybridisation capture enrichment. Anecdotal and unpublished reports are that this appears quite feasible, although overall success and sequencing accuracy will likely depend on the DNA quality, which can be highly variable from FFPE samples. Further studies will be required to validate the robustness of NGS for distinguishing between sequencing artefacts that may be introduced during sample processing and rare low frequency tumour mutations that are likely to be detected when benchmarked against other high sensitivity assays such as mass spectrometry genotyping.
Conclusion
There is much hope given to the promise of genomics and the impact that technology platforms like NGS will have on our understanding, diagnosis and treatment of common diseases. However, it should be remembered that the sequencing technology platform is only one aspect of a routine diagnostic molecular pathology laboratory. DNA sequencing technologies have been used for some time as a clinical diagnostic tool and it has taken at least a decade for the support systems required for data analysis and interpretation to be developed, scrutinised and validated for clinical diagnostic use. In some aspects they still fall short of the requirements of a diagnostic laboratory for a number of applications. Nevertheless, during this time the limitations of Sanger sequencing systems have become well understood and commercial software tools are readily available to assist with these tasks.
NGS will present specific problems from a technical perspective, including higher error rates, fundamental platform differences, the selection of appropriate quality values and data handling. In many respects these issues are not unique to NGS but are likely to be exaggerated by its use. The validation of new sequencing platforms will be achieved through retrospective comparison to previously analysed samples and will also require validation of the software used by the platforms and/or supplied by third party vendors or open access software. Exhaustive prospective comparisons will be required since retrospective testing is unlikely to correlate entirely with data obtained from newly designed NGS panels.
The demands on clinical data interpretation will be much greater both within the laboratory and in the clinic. Protocols for dealing with NGS data that guide what and how particular information will be reported and conveyed to the clinician will need to be established. These issues are currently of high importance to regulatory organisations such as the Therapeutic Goods Administration (TGA) in Australia. In Australia, the use of in vitro diagnostic devices (IVDs) is governed by NATA/RCPA laboratory accreditation and National Pathology Accreditation Advisory Council (NPAAC) guidelines are available at the Australian Government Department of Health and Ageing.114 The TGA have recently introduced a register for IVDs where each Class 3 IVD, including the majority of the tests described in this review, must be registered by 1 July 2014 if used in a diagnostic setting in Australia.115 This accreditation will cover all aspects of the IVD including the NGS platform, targeting methodologies and data interpretation and reporting.
Acknowledgments
We would like to thank Dr Alison Trainer and Associate Professor Alex Dobrovic at the Peter MacCallum Cancer Centre for their helpful comments on the manuscript and Derek Campbell and Brett Kennedy from Illumina Australia for providing early release information regarding Illumina products.
Footnotes
Competing Interests: Dr Meldrum has received honoraria/expenses from Roche. The other authors declare no competing interests.
References
- 1.Chiu RW, Lo YM. Non-invasive prenatal diagnosis by fetal nucleic acid analysis in maternal plasma: the coming of age. Semin Fetal Neonatal Med. 2011;16:88–93. doi: 10.1016/j.siny.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Kuhlenbaumer G, Hullmann J, Appenzellerm S. Novel genomic techniques open new avenues in the analysis of monogenic disorders. Hum Mutat. 2010;32:144–51. doi: 10.1002/humu.21400. [DOI] [PubMed] [Google Scholar]
- 3.Sobreira NL, Cirulli ET, Avramopoulos D, Wohler E, Oswald GL, Stevens EL, et al. Whole-genome sequencing of a single proband together with linkage analysis identifies a Mendelian disease gene. PLoS Genet. 2010;6:e1000991. doi: 10.1371/journal.pgen.1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lupski JR, Reid JG, Gonzaga-Jauregui C, Rio Deiros D, Chen DC, Nazareth L, et al. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med. 2010;362:1181–91. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Welch JS, Westervelt P, Ding L, Larson DE, Klco JM, Kulkarni S, et al. Use of whole-genome sequencing to diagnose a cryptic fusion oncogene. JAMA. 2011;305:1577–84. doi: 10.1001/jama.2011.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Link DC, Schuettpelz LG, Shen D, Wang J, Walter MJ, Kulkarni S, et al. Identification of a novel TP53 cancer susceptibility mutation through whole-genome sequencing of a patient with therapy-related AML. JAMA. 2011;305:1568–76. doi: 10.1001/jama.2011.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.International Cancer Genome Consortium. Hudson TJ, Anderson W, Artez A, Barker AD, Bell C, et al. International network of cancer genome projects. Nature. 2010;464:993–8. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The Cancer Genome Atlas http://cancergenome.nih.gov (Accessed 4 October 2011).
- 9.Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Puente XS, Pinyol M, Quesada V, Conde L, Ordóñez GR, Villamor N, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–5. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris TJ, Mccormick F. The molecular pathology of cancer. Nat Rev Clin Oncol. 2010;7:251–65. doi: 10.1038/nrclinonc.2010.41. [DOI] [PubMed] [Google Scholar]
- 13.Schadt EE, Turner S, Kasarskis A. A window into third-generation sequencing. Hum Mol Genet. 2010;19(R2):R227–40. doi: 10.1093/hmg/ddq416. [DOI] [PubMed] [Google Scholar]
- 14.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–45. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 15.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 16.Pacific Biosciences http://www.pacificbiosciences.com (Accessed 4 October 2011).
- 17.Glenn TC. Field guide to next-generation DNA sequencers. Mol Ecol Resour. 2011;11:759–69. doi: 10.1111/j.1755-0998.2011.03024.x. [DOI] [PubMed] [Google Scholar]
- 18.Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc Natl Acad Sci USA. 2003;100:8817–22. doi: 10.1073/pnas.1133470100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–80. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKernan KJ, Peckham HE, Costa GL, McLaughlin SF, Fu Y, Tsung EF, et al. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 2009;19:1527–41. doi: 10.1101/gr.091868.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–9. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ronaghi M, Uhlén M, Nyrén P. A sequencing method based on real-time pyrophosphate. Science. 1998;281:363–5. 365. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- 23.Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M, et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475:348–52. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 24.Roche Partners with DNA Electronics to Help Migrate 454 Platform to Electrochemical Detection. Nov 2, 2010. http://www.genomeweb.com/sequencing/roche-partners-dna-electronics-help-migrate-454-platform-electrochemical-detecti (Accessed 4 October 2011).
- 25.Robison K. Semiconductors charge into sequencing. Nat Biotechnol. 2011;29:805–7. doi: 10.1038/nbt.1965. [DOI] [PubMed] [Google Scholar]
- 26.Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–32. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 27.Harismendy O, Ng PC, Strausberg RL, Wang X, Stockwell TB, Beeson KY, et al. Evaluation of next generation sequencing platforms for population targeted sequencing studies. Genome Biol. 2009;10:R32. doi: 10.1186/gb-2009-10-3-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura K, Oshima T, Morimoto T, Ikeda S, Yoshikawa H, Shiwa Y, et al. Sequence-specific error profile of Illumina sequencers. Nucleic Acids Res. 2011;39:e90. doi: 10.1093/nar/gkr344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, et al. A large genome center’s improvements to the Illumina sequencing system. Nat Methods. 2008;5:1005–10. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–6. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mardis ER. The $1,000 genome, the $100,000 analysis? Genome Med. 2010;2:84. doi: 10.1186/gm205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koboldt DC, Ding L, Mardis ER, Wilson RK. Challenges of sequencing human genomes. Brief Bioinform. 2010;11:484–98. doi: 10.1093/bib/bbq016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harismendy O, Frazer K. Method for improving sequence coverage uniformity of targeted genomic intervals amplified by LR-PCR using Illumina GA sequencing-by-synthesis technology. Biotechniques. 2009;46:229–31. doi: 10.2144/000113082. [DOI] [PubMed] [Google Scholar]
- 35.Fluidigm http://www.fluidigm.com (Accessed 4 October 2011).
- 36.RaindanceTechnologies http://www.raindancetechnologies.com (Accessed 4 October 2011).
- 37.Tewhey R, Warner JB, Nakano M, Libby B, Medkova M, David PH, et al. Microdroplet-based PCR enrichment for large-scale targeted sequencing. Nat Biotechnol. 2009;27:1025–31. doi: 10.1038/nbt.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dahl F, Stenberg J, Fredriksson S, Welch K, Zhang M, Nilsson M, et al. Multigene amplification and massively parallel sequencing for cancer mutation discovery. Proc Natl Acad Sci U S A. 2007;104:9387–92. doi: 10.1073/pnas.0702165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fredriksson S, Banér J, Dahl F, Chu A, Ji H, Welch K, et al. Multiplex amplification of all coding sequences within 10 cancer genes by Gene-Collector. Nucleic Acids Res. 2007;35:e47. doi: 10.1093/nar/gkm078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Porreca GJ, Zhang K, Li JB, Xie B, Austin D, Vassallo SL, et al. Multiplex amplification of large sets of human exons. Nat Methods. 2007;4:931–6. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- 41.Turner EH, Lee C, Ng SB, Nickerson DA, Shendure J. Massively parallel exon capture and library-free resequencing across 16 genomes. Nat Methods. 2009;6:315–6. doi: 10.1038/nmeth.f.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–9. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hodges E, Rooks M, Xuan Z, Bhattacharjee A, Benjamin Gordon D, Brizuela L, et al. Hybrid selection of discrete genomic intervals on custom-designed microarrays for massively parallel sequencing. Nat Protoc. 2009;4:960–74. doi: 10.1038/nprot.2009.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mamanova L, Coffey AJ, Scott CE, Kozarewa I, Turner EH, Kumar A, et al. Target-enrichment strategies for next-generation sequencing. Nat Methods. 2010;7:111–8. doi: 10.1038/nmeth.1419. [DOI] [PubMed] [Google Scholar]
- 45.Comino-Méndez I, Gracia-Aznárez FJ, Schiavi F, Landa I, Leandro-García LJ, Letón R, et al. Exome sequencing identifies MAX mutations as a cause of hereditary pheochromocytoma. Nat Genet. 2011;43:663–7. doi: 10.1038/ng.861. [DOI] [PubMed] [Google Scholar]
- 46.Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–6. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei X, Walia V, Lin JC, Teer JK, Prickett TD, Gartner J, et al. NISC Comparative Sequencing Program Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011;43:442–6. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan X-J, Xu J, Gu Z-H, Pan C-M, Lu G, Shen Y, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43:309–15. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 49.Fisher S, Barry A, Abreu J, Minie B, Nolan J, Delorey TM, et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 2011;12:R1. doi: 10.1186/gb-2011-12-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–17. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oshlack A, Robinson MD, Young MD. From RNA-seq reads to differential expression results. Genome Biol. 2010;11:220. doi: 10.1186/gb-2010-11-12-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tothill RW, Kowalczyk A, Rischin D, Bousioutas A, Haviv I, van Laar RK, et al. An expression-based site of origin diagnostic method designed for clinical application to cancer of unknown origin. Cancer Res. 2005;65:4031–40. doi: 10.1158/0008-5472.CAN-04-3617. [DOI] [PubMed] [Google Scholar]
- 54.van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 55.Salazar R, Roepman P, Capella G, Moreno V, Simon I, Dreezen C, et al. Gene expression signature to improve prognosis prediction of stage II and III colorectal cancer. J Clin Oncol. 2011;29:17–24. doi: 10.1200/JCO.2010.30.1077. [DOI] [PubMed] [Google Scholar]
- 56.Shah SP, Köbel M, Senz J, Morin RD, Clarke BA, Wiegand KC, et al. Mutation of FOXL2 in granulosacell tumors of the ovary. N Engl J Med. 2009;360:2719–29. doi: 10.1056/NEJMoa0902542. [DOI] [PubMed] [Google Scholar]
- 57.Levin JZ, Berger MF, Adiconis X, Rogov P, Melnikov A, Fennell T, et al. Targeted next-generation sequencing of a cancer transcriptome enhances detection of sequence variants and novel fusion transcripts. Genome Biol. 2009;10:R115. doi: 10.1186/gb-2009-10-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–22. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hodges E, Smith AD, Kendall J, Xuan Z, Ravi K, Rooks M, et al. High definition profiling of mammalian DNA methylation by array capture and single molecule bisulfite sequencing. Genome Res. 2009;19:1593–605. doi: 10.1101/gr.095190.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–77. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26:779–85. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y, Shin H, Song JS, Lei Y, Liu XS. Identifying positioned nucleosomes with epigenetic marks in human from ChIP-Seq. BMC Genomics. 2008;9:537. doi: 10.1186/1471-2164-9-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, et al. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods. 2008;5:829–34. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–7. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 66.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–59. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 67.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 68.Epigenomics http://www.epigenomics.com (Accessed 4 October 2011).
- 69.Ledergerber C, Dessimoz C. Base-calling for next-generation sequencing platforms. Brief Bioinform. 2011;12:489–97. doi: 10.1093/bib/bbq077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–85. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 71.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–94. [PubMed] [Google Scholar]
- 72.Andrews S. FastQC. http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/ (Accessed 14 September 2011).
- 73.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–8. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Novoalign http://www.novocraft.com (Accessed 14 September 2011).
- 77.Ruffalo M, Laframboise T, Koyutürk M. Comparative analysis of algorithms for next-generation sequencing read alignment. Bioinformatics. 2011;27:2790–6. doi: 10.1093/bioinformatics/btr477. [DOI] [PubMed] [Google Scholar]
- 78.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li R, Li Y, Fang X, Yang H, Wang J, Kristiansen K, et al. SNP detection for massively parallel whole-genome resequencing. Genome Res. 2009;19:1124–32. doi: 10.1101/gr.088013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Karchin R. Next generation tools for the annotation of human SNPs. Brief Bioinform. 2009;10:35–52. doi: 10.1093/bib/bbn047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–70. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Conrad DF, Bird C, Blackburne B, Lindsay S, Mamanova L, Lee C, et al. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat Genet. 2010;42:385–91. doi: 10.1038/ng.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12:363–76. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Duncavage EJ, Magrini V, Becker N, Armstrong JR, Demeter RT, Wylie T, et al. Hybrid capture and next-generation sequencing identify viral integration sites from formalin-fixed, paraffin-embedded tissue. J Mol Diagn. 2011;13:325–33. doi: 10.1016/j.jmoldx.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nord AS, Lee M, King MC, Walsh T. Accurate and exact CNV identification from targeted high-throughput sequence data. BMC Genomics. 2011;12:184. doi: 10.1186/1471-2164-12-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Talkowski ME, Ernst C, Heilbut A, Chiang C, Hanscom C, Lindgren A, et al. Next-generation sequencing strategies enable routine detection of balanced chromosome rearrangements for clinical diagnostics and genetic research. Am J Hum Genet. 2011;88:469–81. doi: 10.1016/j.ajhg.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Church DM, Schneider VA, Graves T, Auger K, Cunningham F, Bouk N, et al. Modernizing reference genome assemblies. PLoS Biol. 2011;9:e1001091. doi: 10.1371/journal.pbio.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kidd JM, Sampas N, Antonacci F, Graves T, Fulton R, Hayden HS, et al. Characterization of missing human genome sequences and copy-number polymorphic insertions. Nat Methods. 2010;7:365–71. doi: 10.1038/nmeth.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci USA. 2011;108:9530–5. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Goya R, Sun MG, Morin RD, Leung G, Ha G, Wiegand KC, et al. SNVMix: predicting single nucleotide variants from next-generation sequencing of tumors. Bioinformatics. 2010;26:730–6. doi: 10.1093/bioinformatics/btq040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med. 2010;2:20ra14. doi: 10.1126/scitranslmed.3000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McBride DJ, Orpana AK, Sotiriou C, Joensuu H, Stephens PJ, Mudie LJ, et al. Use of cancer-specific genomic rearrangements to quantify disease burden in plasma from patients with solid tumors. Genes Chromosomes Cancer. 2010;49:1062–9. doi: 10.1002/gcc.20815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.de Sanjosé S, Léoné M, Bérez V, Izquierdo A, Font R, Brunet JM, et al. Prevalence of BRCA1 and BRCA2 germline mutations in young breast cancer patients: a population-based study. Int J Cancer. 2003;106:588–93. doi: 10.1002/ijc.11271. [DOI] [PubMed] [Google Scholar]
- 96.Møller P, Hagen AI, Apold J, Maehle L, Clark N, Fiane B, et al. Genetic epidemiology of BRCA mutations—family history detects less than 50% of the mutation carriers. Eur J Cancer. 2007;43:1713–7. doi: 10.1016/j.ejca.2007.04.023. [DOI] [PubMed] [Google Scholar]
- 97.Risch HA, McLaughlin JR, Cole DE, Rosen B, Bradley L, Fan I, et al. Population BRCA1 and BRCA2 mutation frequencies and cancer penetrances: a kin-cohort study in Ontario, Canada. J Natl Cancer Inst. 2006;98:1694–706. doi: 10.1093/jnci/djj465. [DOI] [PubMed] [Google Scholar]
- 98.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 99.Morgan JE, Carr IM, Sheridan E, Chu CE, Hayward B, Camm N, et al. Genetic diagnosis of familial breast cancer using clonal sequencing. Hum Mutat. 2010;31:484–91. doi: 10.1002/humu.21216. [DOI] [PubMed] [Google Scholar]
- 100.Schroeder C, Stutzmann F, Weber BH, Riess O, Bonin M. High-throughput resequencing in the diagnosis of BRCA1/2 mutations using oligonucleotide resequencing microarrays. Breast Cancer Res Treat. 2010;122:287–97. doi: 10.1007/s10549-009-0639-z. [DOI] [PubMed] [Google Scholar]
- 101.Summerer D, Wu H, Haase B, Cheng Y, Schracke N, Stähler CF, et al. Microarray-based multicycle-enrichment of genomic subsets for targeted next-generation sequencing. Genome Res. 2009;19:1616–21. doi: 10.1101/gr.091942.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Walsh T, Lee MK, Casadei S, Thornton AM, Stray SM, Pennil C, et al. Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc Natl Acad Sci U S A. 2010;107:12629–33. doi: 10.1073/pnas.1007983107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. BRIM-3 Study Group Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–57. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 105.De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu D, Siena S, et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA. 2010;304:1812–20. doi: 10.1001/jama.2010.1535. [DOI] [PubMed] [Google Scholar]
- 106.Shih JY, Gow CH, Yang PC. EGFR mutation conferring primary resistance to gefitinib in non-small-cell lung cancer. N Engl J Med. 2005;353:207–8. doi: 10.1056/NEJM200507143530217. [DOI] [PubMed] [Google Scholar]
- 107.Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085–96. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–72. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–95. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.MacConaill LE, Campbell CD, Kehoe SM, Bass AJ, Hatton C, Niu L, et al. Profiling critical cancer gene mutations in clinical tumor samples. PLoS One. 2009;4:e7887. doi: 10.1371/journal.pone.0007887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Querings S, Altmüller J, Ansén S, Zander T, Seidel D, Gabler F, et al. Benchmarking of mutation diagnostics in clinical lung cancer specimens. PLoS One. 2011;6:e19601. doi: 10.1371/journal.pone.0019601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schweiger MR, Kerick M, Timmermann B, Albrecht MW, Borodina T, Parkhomchuk D, et al. Genome-wide massively parallel sequencing of formaldehyde fixed-paraffin embedded (FFPE) tumor tissues for copy-number and mutation-analysis. PLoS One. 2009;4:e5548. doi: 10.1371/journal.pone.0005548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Australian Government Department of Health and Ageing Requirements for the development and use of in-house in vitro diagnostic devices (IVDs) 2007 Edition. http://www.health.gov.au/internet/publications/publishing.nsf/Content/npaac-devivd-toc (Accessed 14 September 2011).
- 115.Therapeutic Goods Administration Medical Devices and IVDs 2011. http://www.tga.gov.au/industry/devices.htm (Accessed 14 September 2011).
- 116.Kaper F, Wang J, Anderson MJ, Chen P, Lin M, Pieprzyk M, et al. Parallel Preparation of Targeted Resequencing Libraries from 480 Genomic Regions Using Multiplex PCR on the Access Array™ System. http://www.fluidigm.com/home/fluidigm/Posters/AACR-2010_MultiplexPCR_Kaper.pdf (Accessed 14 September 2011).