Abstract
Sphingosine kinases (SK) regulate the balance between pro-apoptotic ceramides and mitogenic sphingosine-1-phosphate (S1P); however, the functions of the two isoenzymes (SK1 and SK2) in tumor cells are not well defined. Therefore, RNA interference was used to assess the individual roles of SK1 and SK2 in tumor cell sphingolipid metabolism, proliferation and migration/invasion. Treatment of A498, Caki-1 or MDA-MB-231 cells with siRNA specific for SK1 or SK2 effectively suppressed the expression of the target mRNA and protein. Ablation of SK1 did not affect mRNA or protein levels of SK2, and reduced intracellular levels of S1P while elevating ceramide levels. In contrast, ablation of SK2 elevated mRNA, protein and activity levels of SK1, and increased cellular S1P levels. Interestingly, cell proliferation and migration/invasion were suppressed more by SK2-selective ablation than by SK1-selective ablation, demonstrating that the increased S1P does not rescue these phenotypes. Similarly, exogenous S1P did not rescue the cells from the anti-proliferative or anti-migratory effects of the siRNAs. Consistent with these results, differential affects of SK1- and SK2-selective siRNAs on signaling proteins including p53, p21, ERK1, ERK2, FAK and VCAM1 indicate that SK1 and SK2 have only partially overlapping functions in tumor cells. Overall, these data indicate that loss of SK2 has stronger anticancer effects than does suppression of SK1. Consequently, selective inhibitors of SK2 may provide optimal targeting of this pathway in cancer chemotherapy.
Keywords: Sphingosine Kinase, Isoenzyme, siRNA, Proliferation, Migration, Anticancer
Introduction
A large body of data demonstrates that sphingolipids, in particular ceramides, sphingosine and sphingosine-1-phosphate (S1P), act as signaling molecules to regulate biological processes including apoptosis, proliferation and stress responses (1–3). In contrast to pro-apoptotic ceramides and sphingosine, S1P induces cell proliferation and migration/invasion, and promotes survival (4), acting through cell surface S1P receptors (5) or intracellularly as a second messenger (6). Therefore, the enzymes that interconvert sphingolipids have promise as new targets for cancer therapy.
As the key enzymes that catalyze the phosphorylation of sphingosine to S1P, sphingosine kinases (SK) are the checkpoint for S1P production (7). Two isoenzymes, SK1 and SK2, have been identified in human cells (8–9). Although they share homology and catalyze the production of S1P, it is unclear if their cellular functions are redundant in tumor cells. SK1 is mainly localized in the cytosol and migrates to the plasma membrane upon activation (10), which is a critical step for its oncogenic effects (11). SK1 is frequently over-expressed in tumor tissues compared to normal tissues (12), and over-expression in NIH3T3 cells promotes growth (4). Conversely, down-regulation of SK1 in colon adenocarcinoma cells decreases the expression of Cox-2 which is a pathogenic factor in colon carcinogenesis (13). Reports indicate that SK1 regulates motility, growth, and chemoresistance of MCF-7 cells (14), and progression through the cell cycle (15). Furthermore, depletion of SK1 in HEK 293 cells abrogates EGF-induced migration (16). Serum S1P levels in SK1−/− mice are decreased by 50% and tissue S1P levels are normal, suggesting that SK1 deficiency can be at least partially compensated for by SK2 (17).
The roles of SK2 in cancer cells are not fully understood. Over-expression of SK2 was suggested to promote apoptosis (18); however, this likely reflects increased expression of its BH3 domain because a kinase-inactive mutant is similarly pro-apoptotic (19). Interestingly, apoptosis results in S1P production by SK2 which promotes macrophage survival allowing clearance of dead cells (20). This is associated with caspase-1-mediated cleavage of SK2 resulting in its secretion and generation of extracellular S1P (21). In tumor cells, down-regulation of SK2 inhibits the proliferation of glioblastoma cells (22), and eliminates EGF-induced migration of MDA-MB-453 cells (16). Also, the growth of SK2-deficient MCF-7 tumor xenografts is significantly delayed in mice (23). A nuclear localization sequence in SK2 likely accounts for its presence in the nucleus, cytosol and ER under different circumstances (19). These different subcellular localizations may allow unique functions of SK1 and SK2 (24–25). In SK2−/− mice, plasma S1P levels are reduced by only 25%, indicating a greater contribution from SK1 (26).
Small molecule SK inhibitors have been described (12, 27), and a SK2-selective inhibitor ABC294640 has been shown to have antitumor activity in mouse tumor models (28–30). Because SK are being increasingly considered as targets for new anticancer drugs, it is critical that the roles of SK1 and SK2 in cancer cells are defined. Therefore, we have utilized isoenzyme-selective siRNAs to assess the roles of SK1 and SK2 in sphingolipid metabolism, proliferation and migration/invasion in human tumor cells.
Materials and Methods
Cell Lines and Reagents
A498 kidney adenocarcinoma, Caki-1 kidney clear cell carcinoma or MDA-MB-231 breast adenocarcinoma cells were from the ATCC and cultured in MEM, McCoy’s 5A and DMEM, respectively, supplemented with 10% FBS, 50 μg/ml gentamicin in a 37°C, humidified, 5% CO2, 95% air environment. Silencer® validated siRNAs were purchased from Ambion Inc. as follows: SK1 (siSK1, #1181 targeting exon 3 of NM_021972), SK2 (siSK2, #1587 targeting exon 7 of NM_021972), ERK1 (siERK1, #142304 targeting exon 7 of NM_002746) and non-targeted siRNA (negative control, siNC). Additional siRNAs were purchased from Santa Cruz Biotechnology, Inc. and consisted of pools of 3 siRNAs targeting different coding regions of SK1 (sc-44114) or SK2 (sc-39225). All media, supplements and primers were from Invitrogen. S1P and omega(7-nitro-2-1,3-benzoxadiazol-4-yl)(2S,3R,4E)-2-aminooctadec-4-ene-1,3-diol (NBD-sphingosine) were purchased from Avanti Polar Lipids, Inc. All other chemicals were from Sigma-Aldrich. Polyclonal chicken antibodies were raised against a unique SK1 peptide sequence (CVEPPPSWKPQQMPPPEE) and a unique SK2 sequence (EAEEQQDQRPDQELT), respectively, by Aves Lab Inc.
siRNA Transfection
Briefly, 18 μl of Lipofectamine-2000 (Invitrogen) was mixed with 1000 μl Opti-MEM reduced serum media and incubated for 15 min at room temperature. siRNA was added to a final concentration of 50 nM in 6 ml of medium, incubated for 20 min and then added to 5 ml antibiotic-free growth media in 10 cm dishes along with 6×105 cells.
Quantitative PCR (qPCR)
Total RNA was extracted and purified using RNeasy columns (Qiagen), and 1 μg was used to synthesize cDNA (total volume 20 μl) with the SuperScript III kit (Invitrogen) following manufacture’s protocol. Human 18S ribosomal RNA or GAPDH were used as endogenous control transcripts for normalization of the target transcripts. Aliquots (2 μl) of the reverse transcription reaction were used in qPCR (20 μl) in the presence of the following target-specific primers for amplification:
SK1: 5′TGAGCAGGTCACCAATGAAG3′; 5′TGTGCAGAGACAGCAGGTTC3′.
SK2: 5′GGAGGAAGCTGTGAAGATGC3′; 5′GCAACAGTGAGCAGTTGAGC3′
ERK1: 5′CAACATGAAGGCCCGAAACTACC3′; 5′TAACATCCGGTCCAGCAGGTCAAG3′
ERK2: 5′TACACCAACCTCTCGTACATCG3′; 5′CATGTCTGAAGCGCAGTAAGATT3′
18S: 5′TTGGAGGGCAAGTCTGGTG3′; 5′CCGCTCCCAAGATCCAACTA3′
GAPDH: 5′TGCACCACCAACTGCTTAGC3′; 5′GGCATGGACTGTGGTCATGAG3′
SYBR Green Supermix and a MyiQ Real-Time PCR System (Bio-Rad) were used for qPCR with cycling parameters of: 95°C for 5 min; 45 cycles of 95°C for 45 sec and 60°C for 1 min. A dissociation profile was generated for each run to verify specificity of the amplification. The average cycle threshold (Ct) value for each group was determined and normalized by the endogenous control Ct value. The relative percentage expression was calculated using the equation: , where ΔCt represents the difference in normalized Ct for the knockdown groups vs. control (siNC).
Immunofluorescence Microscopy
At room temperature, cells grown on glass coverslips were washed with PBS, fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100 in PBS, and then incubated sequentially with: 10% goat serum for 1 hr; chicken SK1 or SK2 antibodies in 1% goat serum for 1 hr; and fluorophore-conjugated chicken secondary antibody (Invitrogen) for 1 hr. Coverslips were mounted with a DAPI-containing reagent, and examined with a Nikon Eclipse E800 microscope and NIS-Elements software.
SK1 Activity Assay
Cellular SK1 activity was measured using an HPLC-based assay described previously (28). Briefly, cell lysates were incubated with NBD-sphingosine in SK1-selective assay buffer, and the product (NBD-S1P) was resolved by reversed phase HPLC with fluorescence detection. The area-under-the-curve for the NBD-S1P peak was compared with quantified as a measure of SK1 activity.
Protein Isolation and Western Blot Analyses
Cells were washed 3 times with ice-cold PBS, harvested and centrifuged at 1000 × g for 5 min. Cell pellets were lysed with 100 μl of lysis buffer (50 mM Tris, 150 mM NaCl, 5 mM EDTA, 5 mM EGTA, 1% NP-40, pH 7.4, protease and phosphatase inhibitors) by vortexing. Supernatants were prepared by centrifugation at 5,000×g for 30 min at 4°C, and protein concentrations were determined using the BCA kit (Pierce). Equal amount of protein (30 μg/lane) were fractionated by SDS-PAGE and transferred onto PVDF membranes. Immunoblotting was carried out with the following primary antibodies (Cell Signaling except noted): anti-AKT, anti-pAKT, anti-ERK1/2, anti-pERK1/2, anti-p21, anti-p53, anti-FAK, anti-pFAK(Y397), anti-vascular cell adhesion molecule 1 (VCAM1, SantaCruz), anti-β-actin; and HRP-conjugated secondary antibodies. The immunocomplexes were visualized by the ECL chemiluminescence method. Protein expression was quantified by densiometric scanning of the BIOMAX XAR films after normalization to β-actin using the NIH Image J program.
Cell Proliferation Assays
Cells were transfected with siRNAs in 96-well plates (5,000 cell/well), and then at varying times, the cells were washed with PBS; fixed with 10% trichloroacetic acid; washed with PBS; stained with sulforhodamine-B; and destained with 1% acetic acid. The cell-bound dye was then dissolved in 10 mM Tris, and the absorbance at 560 nm was measured using a Spectramax M5 spectrophotometer (Molecular Devices). In certain experiments, 1 μM (final) S1P was added to the culture medium 48 hr after transfection.
Cell Cycle Analyses
Cells were transfected as described above and then harvested by trypsinization, washed and fixed with 75% ethanol at 4°C. The fixed cell were washed with PBS, resuspended and stained with DNase-free RNase A (0.4 mg/ml) and PI (0.05 mg/ml) in PBS. Cell cycle analyses were performed using a FACStarplus flow cytometer (BD Biosciences).
Sphingolipid Mass Measurements
At 72 hr post-transfection, cells were harvested and washed three times with PBS. The cell pellets were subjected to sphingolipid profiling by HPLC-MS by the Lipidomics Core facility at MUSC as described elsewhere (31).
Migration and Invasion Assays
Cells were harvested by trypsinization 48 hr after siRNA transfection, washed with PBS, and suspended in serum-free media. For migration assays, 50,000 cells were placed in a transwell insert (24-well format, 8 μm pore, BD Biosciences). For invasion assays, matrigel precoated inserts were used. In both cases, the inserts were placed into the 24-well plates with serum-containing growth medium and incubated under normal culture conditions for 4 hr (migration) or 24 hr (invasion). To assess the ability of exogenous S1P to rescue cell migration, 1 μM S1P was added to the culture medium of the siRNA-treated cells for the final 24 hr before harvest. The assays were then conducted as described above, except that 1 μM S1P and 0.1% fatty acid-free BSA were added to both the top and bottom compartments of the transwells. The non-migrating cells were removed using cotton swabs, and cells that had migrated to the underside of the inserts were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Cells were counted in five random squares for each well using a light microscope, and results are presented as the average number of migrated/invaded cells per field.
Results
SK1 and SK2 are Selectively Depleted by Isoenzyme-specific siRNAs
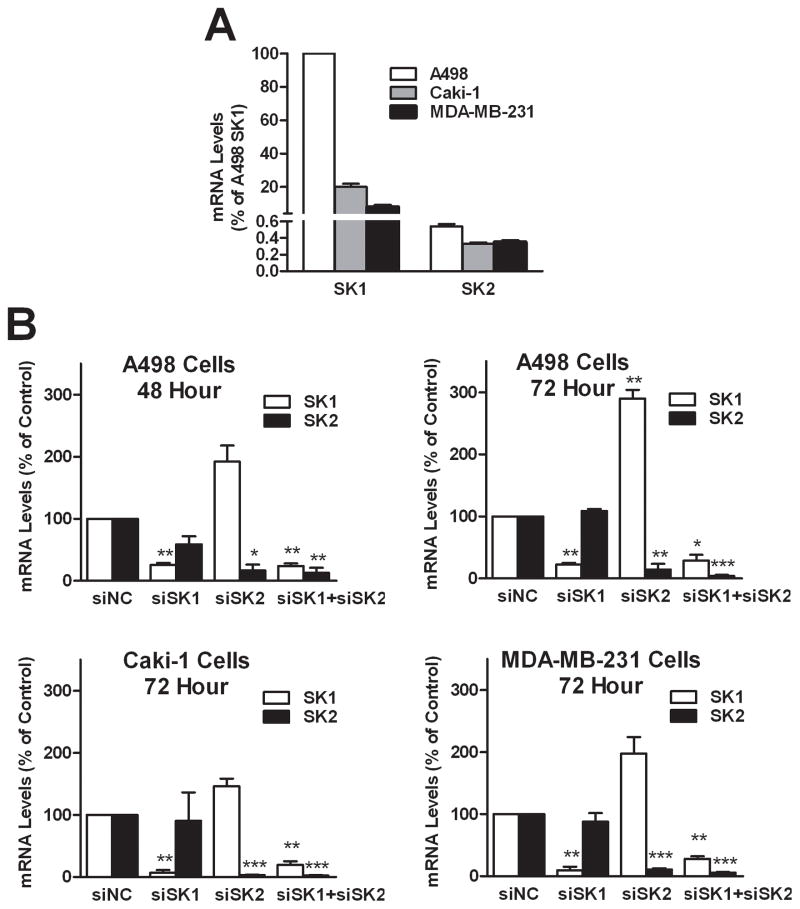
To compare the functions of the endogenous SK1 and SK2, we used siRNAs to knockdown the expression of SK1 and/or SK2 in three human cancer cell lines, A498, Caki-1 and MDA-MB-231. The relative expression levels of SK1 and SK2 measured by qPCR are expressed relative to SK1 in A498 cells (Figure 1A). The data indicate that A498 cells have the highest levels of mRNA encoding SK1 and SK2, and that transcript levels of SK1 are considerably higher than those for SK2 in all three cell lines. For all of the following knockdown experiments, cells transfected with siNC are defined as the control, i.e. 100% expression. As shown in Figure 1B, mRNA levels of the targeted SK isoenzyme in A498 cells were substantially decreased by 48 hr after transfection and remained suppressed for at least 72 hr. For A498 cells at 72 hr post-transfection, SK1 mRNA levels in cells treated with either siSK1 or siSK1+siSK2 were reduced approximately 70%, while siSK1 treatment did not affect levels of SK2 mRNA. Conversely, mRNA for SK2 was reduced approximately 85% in cells treated with either siSK2 or siSK1+siSK2. Surprisingly, cells treated with siSK2 contained dramatically elevated levels of mRNA for SK1. Expression analyses in Caki-1 and MDA-MB-231 cells provided similar results in that the targeted SK isoenzymes were effectively depleted. Importantly, knockdown of SK2 resulted in elevated expression of SK1 mRNA in Caki-1 and MDA-MB-231 cells, although the magnitudes of the responses were less than that observed with the A498 cells. In all of the transfection experiments, the cells remained attached to the plates for at least 72 hr.
Figure 1. Effects of SK siRNA transfection on the mRNA levels of SK1 and SK2.
A) Relative expression of SK mRNAs in 3 cancer cell lines. Data are presented as percentage of SK1 in A498 cells. B) Cells were transfected with siRNAs as indicated, and qPCR was performed after the indicated times to determine the relative expression of SK1 (open bars) and SK2 (filled bars), compared with control (siNC). Data represent the mean ± SEM of three independent experiments. *p<0.05, **p<0.01, ***p<0.001 versus control.
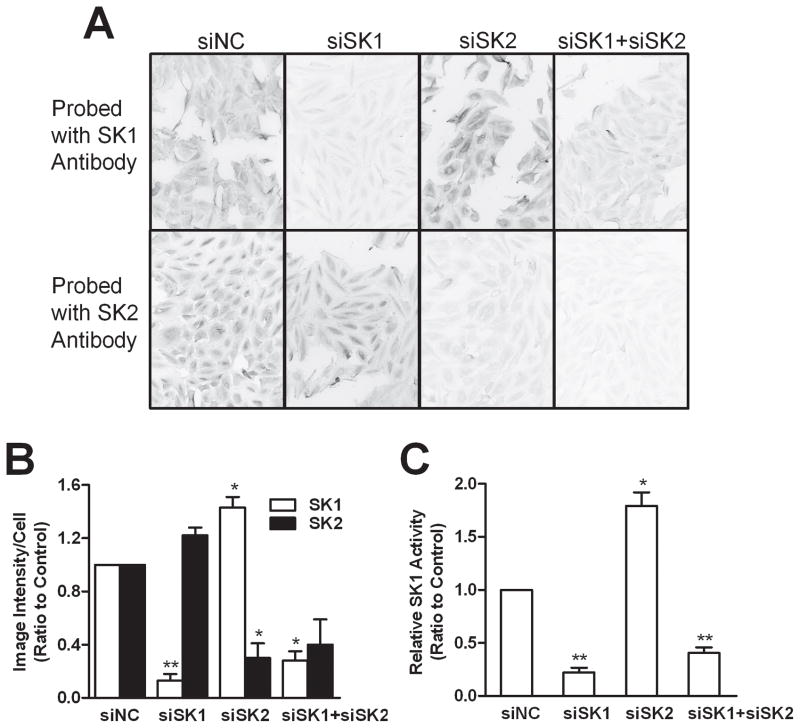
Chicken antibodies against peptides specific for human SK1 and SK2 were made to analyze their protein levels. This was necessary because several commercially available SK antibodies failed to provide us with interpretable data by immunoblotting or immunocytochemical analyses. The newly developed antibodies were poor for immunoblotting, but provided excellent selectivity and sensitivity in immunofluorescence staining experiments (Supplemental Figure 1), suggesting a need for epitope structure for immunorecognition. Using these antibodies, we quantified and confirmed the 70–85% decreases in the protein level of the targeted SK isoenzymes in siRNA-transfected A498 cells (Figures 2A and 2B). Importantly, the expression of SK1 protein was elevated approximately 40% in cells treated with siSK2. To further confirm the effects of the siRNA, SK1 enzymatic activity was measured in lysates from siRNA-transfected A498 cells using a fluorescence-based HPLC assay that we have previously described (28). As indicated in Figure 2C, siSK1 strongly reduced SK1 activity whereas siSK2 treatment increased SK1 activity by approximately 70%. The activity of SK2 was below the detection limit for the assay for all samples. Overall, the data confirm that the siRNAs are effective for selective depletion of the targeted SK isoenzymes, and that SK1 expression appears to be sensitive to the expression level of SK2.
Figure 2. Effects of SK siRNA transfection on the expression of SK1 and SK2 protein.
At 72 hr after transfection, A498 cells were probed with antibodies against SK1 (A, top row) or SK2 (A, bottom row) and visualized with a fluorescent secondary antibody. Images were acquired with identical exposure parameters, and are representative of at least 5 areas from each sample. B) Expression above was quantified by imaging pixel intensity/cell, and the relative protein expression levels of SK1 (open bars) and SK2 (filled bars) were calculated relative to control (siNC). C) The enzymatic activity of SK1 was determined relative to control (siNC-treated) cells. Data represent the mean ± SEM of three independent experiments. *p<0.05, **p<0.01, ***p<0.001 versus control.
Effects of SK Knockdown on Sphingolipids
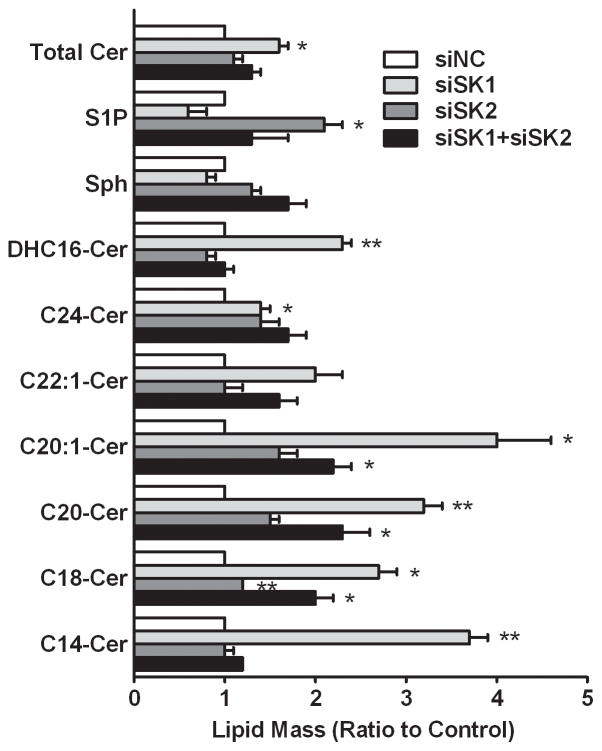
SK convert sphingosine to S1P; however, it is well established that the metabolism of sphingolipids is a dynamic process (32), so that an altered flux through SK may modulate several sphingolipids. As shown in Figure 3, SK1-selective knockdown resulted in elevated levels of total ceramides (total Cer, approximately 70% increase from control) and all of the individual ceramide species measured, i.e. dihydro-C16-ceramide (DHC16-Cer), C24-ceramide (C24-Cer), C20:1-ceramide (C20:1-Cer), C20-ceramide (C20-Cer), C18-ceramide (C18-Cer), C14-ceramide (C14-Cer). Also, the level of S1P was decreased as expected, but there was a small decrease in sphingosine, indicating that the sphingosine was likely being converted to ceramides by ceramide synthase. In marked contrast, the selective knockdown of SK2 did not substantially affect the levels of most ceramide species, causing only a slight increase in total ceramide levels. Interestingly, selective knockdown of SK2 resulted in significantly elevated S1P levels, and this was largely abrogated when SK1 was simultaneously suppressed in the double knockdown cells. The medium from cells grown in serum-free medium was analyzed by HPLC-MS; however, the levels of extracellular S1P were below the detection limit of the assay (data not shown). Combined with the qPCR results described above, it seems likely that the elevated S1P level arises from the increased expression of SK1 in the SK2-selectively-depleted cells. These data indicate that SK1 is the dominant isoenzyme in A498 cells for the synthesis of S1P (on a total S1P mass basis).
Figure 3. Effects of SK siRNA transfection on sphingolipid profiles.
A498 cells were transfected with siRNAs as indicated, and sphingolipid analyses were performed 72 hr later. Bars indicate the ratio of the lipid mass relative to control (siNC). Data are mean ± SEM of three independent experiments. *p<0.05, **p<0.01 versus control.
Knockdown of SK Suppresses Cell Proliferation
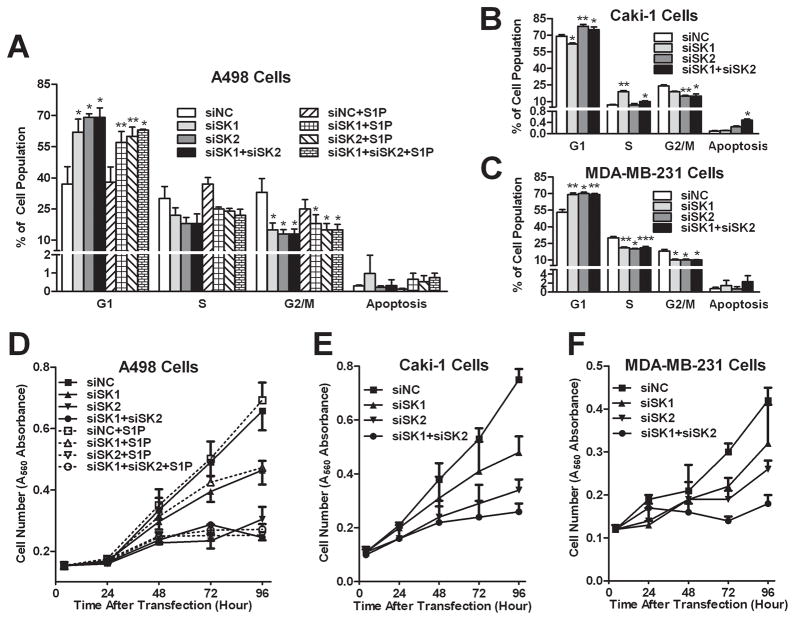
Because SK catalyze the production of mitogenic S1P, we evaluated the effects of their knockdown on cell cycle progression and proliferation. As shown in Figure 4A, SK1 and/or SK2 knockdown resulted in an increase in the percentage of A498 cells in the G1 phase of the cell cycle, with corresponding decreases in the S and G2/M phases. Responses to siSK2 were somewhat greater than responses to siSK1, and combined addition of both siRNA did not further arrest the cells beyond what occurred with siSK2 alone. Moreover, the addition of 1 μM S1P to the siRNA-transfected cells did not overcome the cell cycle arrest (Figure 4B). These results suggest that both SK1 and SK2 promote cell cycle progression, although removal of SK2 appears to be more suppressive. Interestingly, knockdown of SK1 and/or SK2 did not promote a significant level of apoptosis in these A498 cells (< 1% of cells in all cases). Therefore, the accumulation of ceramides, particularly in the SK1-selectively-depleted cells, is not sufficient to drive these cells into apoptosis. Similar responses were seen in Caki-1 and MDA-MB-231 cells (Figure 4B and 4C). It is notable that responses of MDA-MB-231, which have mutated p53, indicate that cell cycle arrest induced by SK knockdown is not dependent on p53.
Figure 4. Effects of SK siRNA transfection on the cell cycle profile and proliferation.
A498 (A, D), Caki-1 (B, E) or MDA-MB-231 (C, F) cells were harvested 72 hr after siRNA transfection (A, B, C) for cell cycle distribution determination, or cell-number-quantified to plot growth curves (D, E, F). For A498 cells (A, D), 1 μM S1P was added to certain samples as indicated. Data (A, B and C) are mean ± SEM of three independent experiments. *p<0.05, **p<0.01, ***p<0.001 versus siNC control. Data (D, E and F) are mean ± SD and representative of at least two independent experiments.
Consistent with the cell cycle data, knockdown of either SK1 or SK2 reduced the proliferation of A498 cells for at least 96 hr after transfection (Figure 4D), with inhibition being much stronger as a result of SK2-selective ablation. Similar results were obtained upon siRNA transfection of Caki-1 and MDA-MB-231 cells (Figure 4E and 4F). It is noteworthy that selective depletion of SK2 completely blocked the proliferation of the cells in spite of the elevated expression of SK1 and the concomitant elevation of S1P levels in these cells. As with the cell cycle studies, addition of exogenous S1P did not rescue A498 cells from inhibition of proliferation caused by SK1 and/or SK2 depletion, indicating that intracellular S1P is involved in these processes. In contrast with extracellular S1P, intracellular S1P functions as a secondary messenger independent of S1P GPCRs (33). While the mechanisms have not been fully characterized, potential targets for intracellular S1P have been discussed (34–35). Because of the physical properties of S1P, extracellular S1P penetrates into cells only very poorly (36), which prevents it from contributing to the intracellular S1P pool. Therefore, it is not surprising that exogenous S1P did not rescue SK-ablated cells, and this is consistent with other recent reports (37–38). This is a critical point in terms of drug development in that it predicts that pharmacologic inhibition of S1P production in tumor cells will block proliferation even in the presence of plasma S1P. The combined results indicate that SK2 is necessary for optimal proliferation of tumor cells, while SK1 plays a lesser role.
Signaling Effects of SK Knockdown
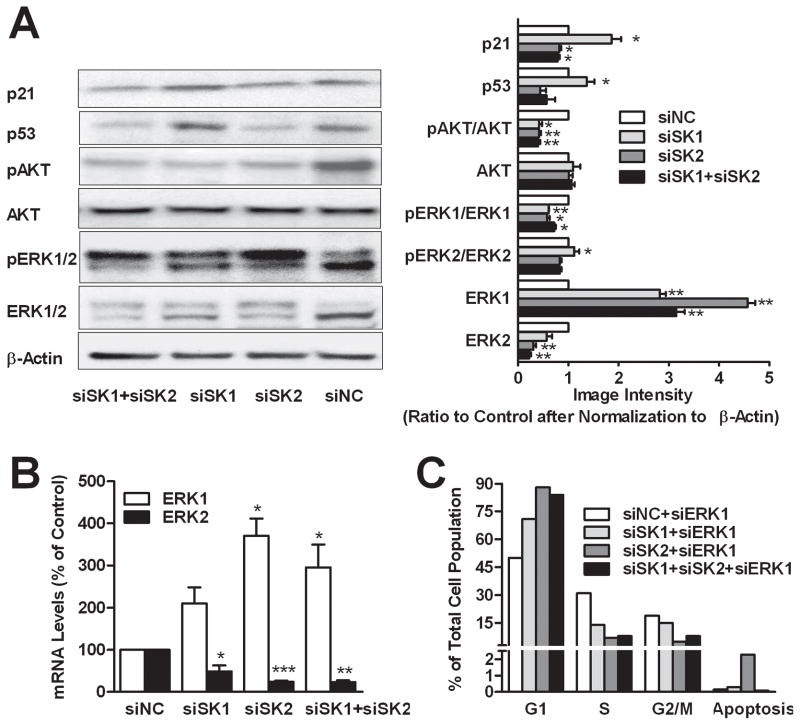
The roles of SK1 and SK2 in signaling pathways that regulate cell cycle progression were assessed in a series of immunoblots using samples prepared from siRNA-treated A498 cells. As shown in Figure 5A, at 72 hr post-transfection, SK1-selectively-ablated cells had elevated p21 levels, which slows progression through the cell cycle at the G1/S-phase transition (39), whereas the SK2-selective and dual knockdown cells showed slight decreases in p21 levels. A regulator of p21, p53 protein levels revealed expression patterns identical to those for p21. These results suggest that cell cycle arrest by selective depletion of SK2 is p53-independent, which is consistent with cell cycle arrest in cell lines with either wild-type or mutated p53.
Figure 5. Effects of SK siRNA transfection on signaling proteins.
A498 cells were transfected with siRNA as indicated for 72 hr. A) Cells were harvested and immunoblotting was conducted with the indicated antibodies and quantified by densitometry. The expression of the indicated proteins is normalized to beta-actin. B) qPCR was performed to determine the expression of mRNA for ERK1 (open bars) or ERK2 (filled bars) relative to control (siNC) after normalization to GAPDH. C) Cell cycle histogram of siERK1 and siSK co-transfected cells. Data are mean ± SEM of three independent experiments, except that C represents a single experiment. *p<0.05, **p<0.01, ***p<0.001 versus control.
As markers of cell survival and proliferation, AKT and ERK were also analyzed. Levels of pAKT were decreased dramatically by depletion of either SK1 or SK2 (Figure 5B), while no further decrease was observed in the double knockdown group. None of the siRNA treatments affected the AKT protein levels. Thus, SK1 and SK2 appear to redundantly regulate signaling through AKT. The effects of SK depletion of on ERK were complex. Surprisingly, compared to controls, all three knockdown groups had greatly elevated pERK1 and decreased pERK2 levels, with the response to siSK2 being quantitatively larger than the response to siSK1. Interestingly, treatment with either siSK1 or siSK2 increased levels of ERK1 protein, while decreasing levels of ERK2 protein. qPCR analyses confirmed that SK2-selective knockdown increased ERK1 mRNA to a larger degree than did selective knockdown of SK1 (Figure 5B). Similarly, the reductions in ERK2 protein levels were accompanied by decreases in expression of ERK2 at the message level. To determine the requirement for ERK1 for cell cycle arrest in the SK knockdown cells, siRNA targeting ERK1 was co-transfected with siSK1 and/or siSK2 into A498 cells. Data shown in Figure 5C indicate that ablation of ERK1 did not alter responses to either SK siRNA. Taken together, the data suggest that SK2 has a greater role than does SK1 in regulating the expression and phosphorylation of ERK1, and promoting the expression and phosphorylation of ERK2.
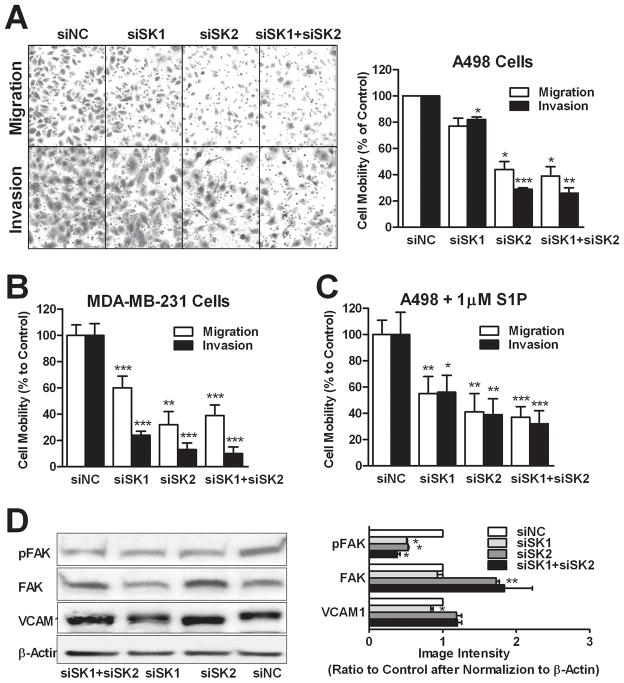
Knockdown of SK Suppresses Cell Migration and Invasion
Because S1P can also induce cell migration (40), we investigated the effects of SK knockdown on FBS-induced migration and invasion (through Matrigel). As shown in Figure 6A, both migration and invasion were more strongly attenuated by SK2-selective and double knockdown than by SK1-selective knockdown in A498 cells. Similar results were obtained in the migration and invasion assays using MDA-MB-231 cells (Figure 6B); however, Caki-1 cells were poorly migratory, so the effects of the SK siRNAs could not be assessed in these cells. Furthermore, exogenous S1P did not rescue the SK1 and/or SK2 knockdown cells from inhibition of migration/invasion (Figure 6C). To examine possible mechanisms for these effects, we assessed the levels of pFAK(Y397), FAK and VCAM1 by immunoblotting. As shown in Figure 6D, all three knockdown groups had decreased levels of pFAK(Y397), which plays a key role in promoting cell migration (41). However, the expression of total FAK protein was markedly increased in the SK2-selective and double knockdown cells. These two groups of cells also demonstrated increased electrophoretic mobility of VCAM1, which may reflect inhibition of its glycosylation – an important step for VCAM1’s pro-migration function (42–43). In contrast, SK1-selective knockdown did not alter VCAM1 electrophoretic mobility, although its expression level was slightly decreased. These results suggest that SK2 may regulate cell migration by altering VCAM1 glycosylation. Overall, the data suggest that SK2 plays a more important role in the regulation of cancer cell migration and invasion than does SK1.
Figure 6. Effects of SK siRNA transfection on cell migration.
A498 (A, C and D) or MDA-MB-231 (B) cells were transfected with siRNAs as indicated and transwell migration and gel invasion assays were performed. In the experiment depicted in panel C, 1 μM S1P was added to both sides of the migration chambers. A) Photos of cells having passed through uncoated (migration) or coated (invasion) filters are representative of at least 5 fields per treatment. Bars in A, B and C represent the percentage of the migratory (open bars) of invasive (filled bars) cells compared with control (siNC). D) Immunoblotting was conducted with the indicated antibodies and quantified. Data are mean ± SEM of three independent experiments except C and D (mean ± SD of triplicates). *p<0.05, **p<0.01, ***p<0.001 versus control.
Discussion
It is well known that S1P production by SK can promote cell proliferation and inhibit apoptosis, making SK potential cancer therapeutic targets. Most of the emphasis has been focused on SK1, particularly since the demonstration that over-expression can transform cells (44). Although SK2 has the same ability as SK1 to produce S1P, different kinetic properties and subcellular localizations of these isoenzymes may provide them with distinct functions in cell signaling. While SK2 has been drawing more attention recently, it is still not clear whether SK1 and SK2 have redundant, overlapping or opposing functions in human cancer cells. Resolution of this ambiguity is essential for the design of SK-targeted drugs, i.e. the preference for isoenzyme-selective or dual SK inhibitors must be defined. To this end, we have utilized RNA interference to assess the activities of SK1 and SK2 in tumor cells. In the present studies, A498 kidney cancer cells were chosen as the primary model because they have relatively high levels of SK1 mRNA (45), and normal kidney tissue has high expression of SK2 mRNA (9). Another kidney cell line (Caki-1) was used to confirm phenotypic effects of SK1 and SK2 ablation in this tissue type. We included the breast cancer cell line MDA-MB-231 to broaden the relevance of the studies and to compare signaling patterns in a cell line with mutated p53 (A498 and Caki-1 cells both contain wild-type p53). Furthermore, MDA-MB-231 cells are reported to be highly metastatic (46).
Transient knockdown of expression of target mRNAs provides an advantage over the use of stably over-expressing cells and cells from transgenic knockout mice in that compensatory changes in the expression of the alternate isoenzyme can be examined. This is particularly important in the study of SK because SK1 expression appears to be regulated by SK2; however, the actions of elevated SK1 do not compensate phenotypically for the loss of SK2. Attempts to make stable knockdowns of SK1 and SK2 using short-hairpin RNAs were unsuccessful in that no clones expressed SK2 mRNA levels lower than approximately 30% of control (data not shown). This implies that survival of the tumor cells requires at least some expression of SK2. Another issue in studies of SK is that commercially-available antibodies for SK1 and particularly for SK2 performed poorly in immunoblotting and immunohistochemical experiments. Therefore, we generated SK isoenzyme-specific chicken antibodies that have excellent specificity and selectivity in immunocytochemistry experiments (Supplemental Figure 1); however, these antibodies also are inadequate for immunoblotting studies.
Our studies suggest a one-way regulation of the expression of SK1 and SK2. Specifically, knockdown of SK1 did not influence levels of SK2 at either the mRNA or protein level in A498 cells. Conversely, knockdown of SK2 significantly induced the expression of SK1 mRNA, protein and enzymatic activity, and this is associated with substantial increases in the cellular levels of S1P. However, it is critical to note that this response did not rescue the cells from inhibition of proliferation, migration or invasion because of ablation of SK2. Furthermore, addition of S1P to the culture medium did not overcome the anti-proliferative or anti-migratory effects of SK2 depletion. This indicates that neither SK1 nor extracellular S1P can functional compensate for loss of SK2 expression and/or activity. The lack of redundancy was also demonstrated by the observations that depletion of SK2 always produced a greater suppression of tumor cell activities (proliferation, migration/invasion). Therefore, it seems clear that SK2 plays unique roles that cannot be replaced by SK1, and this is consistent with recent observations by others (22–23, 47). The mechanism for induction of SK1 expression in response to siSK2 is under further study, and appears to be time-sensitive and quantitatively associated with the level of SK2. For example, SK2-directed siRNAs from Santa Cruz Biotechnology, which target different sequences than the Ambion siRNA, were less efficient in the selective depletion of SK2 and cross-reacted with SK1 to produce a small decrease in its mRNA levels (Supplemental Figure 2A). Although knockdown of SK1 and SK2 were not as complete with the Santa Cruz siRNA, they did confirm that proliferation was much more strongly suppressed by SK2knockdown than by SK1 knockdown (Supplemental Figure 2B).
Lipidomic analyses also indicated differences between SK1 and SK2 knockdown cells. Not only was there reduced S1P following SK1 depletion, but the levels of ceramides were also increased in these cells. Conversely, the selective ablation of SK2 left most ceramide species unchanged. These divergent effects of SK1 and SK2 knockdown on sphingolipid profiles and cell responses may reflect their different subcellular localizations and consequently different subcellular pools of S1P. Unlike SK1 which is predominantly cytosolic, SK2 is localized in nucleus with a small portion in the cytosol or membranes depending on the cell type, as well as ER (19, 48–49). It is possible that when the cytosolic SK1 is ablated, the cytosolic pool of SK2 compensates its loss. However, if SK2 is ablated, SK1 cannot compensate to provide a local organelle (e.g. nuclear) pool of S1P because of lack of targeting. Similarly, exogenous S1P likely does not rescue SK ablated cells because it does not localize correctly in the cells. Thus, the subcellular distribution of S1P, and perhaps ceramides, may provide a mechanism for the lack of redundancy between SK1 and SK2. The data further indicate that the local pool of S1P produced by SK2 is more important for promoting cell proliferation and migration than is cytosolic S1P from SK1.
Current models suggest that S1P activates certain signal pathways, including PI3K/AKT, by binding to the cell surface receptors, (50). Additionally, SK have been shown to activate the RAS/ERK pathway by a GPCR-independent pathway (51). The current data indicate that SK1 and SK2 are both necessary for optimal AKT phosphorylation. Additionally, our results demonstrate an interesting divergence in the regulation of ERK1 and ERK2 by SK, in that depletion of either SK isoenzyme markedly reduced ERK2 expression and activation, but increased ERK1 expression. To investigate the impact of the increased ERK1 on the cell cycle, siERK1 was co-transfected with siSK1 and siSK2. The data indicate that ablation of ERK1 does not affect cell cycle arrest induced by ablation of SK1 or SK2. The observation that ablation of SK2 results in stronger downregulation of ERK2 and proliferation than does ablation of SK1 agrees with the previous studies demonstrating that suppression of ERK1 alone is not sufficient to block cell replication (52–53).
The combined data indicate that SK-isoenzyme-selective ablation differentially affects sphingolipid profiles, signaling, proliferation, migration and invasion. For SK1-selective ablation, reduction of S1P and elevation of ceramides may remove the growth stimulatory signaling through S1P receptors, since the cytosolic pool of S1P is more likely to be secreted. In contrast, selective depletion of SK2 may remove the nuclear pool of S1P, which is quantitatively smaller than the SK1-derived pool, and this down-regulates several key signaling proteins, including ERK2, pAKT, p53 and p21. Based on our results and those of others (18–19), SK2 has two opposing aspects that affect cell proliferation. Specifically, the enzymatic formation of S1P is pro-proliferation; whereas, the BH3 domain is pro-apoptotic. When expressed at a normal level, SK2 makes localized S1P (most likely in the nucleus) which plays an important role in regulating cancer cell proliferation and migration/invasion. Conversely, when SK2 is over-expressed by genetic or pharmacological manipulation, which may happen via a feedback mechanism after cells are treated with a SK2-selective inhibitor (Supplemental Figure 3), the BH3 domain of SK2 provides a magnified pro-apoptotic stimulus that overshadows its proliferative activity. Therefore, blocking the enzymatic activity of SK2 while maintaining its expression, thereby allowing its BH3 domain to promote cell death, may provide the maximum anticancer activity.
Several SK inhibitors have been described in recent years, including ABC294640 which is an SK2-selective inhibitor that competes with sphingosine (28). ABC294640 has anti-proliferative and antitumor activity (28–30), as well as anti-inflammatory activity, in a variety of cellular and animal models, supporting the hypothesis that SK2 is the preferred target over SK1 for new anticancer agents. Because circulating S1P is a major regulator of vascular and immune systems (54), selective inhibition of SK2 should be less toxic in vivo than inhibition of SK1 or dual inhibition of both isoenzymes. Clinical trials of ABC294640 are in progress to test this hypothesis.
Supplementary Material
Acknowledgments
We would like to thank Yuri Peterson helped to design the epitopes of antibodies. This work was supported by NIH 1R01CA122226.
References
- 1.Zhang H, Desai NN, Olivera A, Seki T, Brooker G, Spiegel S. Sphingosine-1-phosphate, a novel lipid, involved in cellular proliferation. The Journal of cell biology. 1991;114:155–67. doi: 10.1083/jcb.114.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hannun YA, Obeid LM. Mechanisms of ceramide-mediated apoptosis. Advances in experimental medicine and biology. 1997;407:145–9. doi: 10.1007/978-1-4899-1813-0_22. [DOI] [PubMed] [Google Scholar]
- 3.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nature reviews. 2008;9:139–50. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 4.Olivera A, Kohama T, Edsall L, Nava V, Cuvillier O, Poulton S, et al. Sphingosine kinase expression increases intracellular sphingosine-1-phosphate and promotes cell growth and survival. The Journal of cell biology. 1999;147:545–58. doi: 10.1083/jcb.147.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pyne S, Pyne NJ. Sphingosine 1-phosphate signalling in mammalian cells. The Biochemical journal. 2000;349:385–402. doi: 10.1042/0264-6021:3490385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenfeldt HM, Hobson JP, Maceyka M, Olivera A, Nava VE, Milstien S, et al. EDG-1 links the PDGF receptor to Src and focal adhesion kinase activation leading to lamellipodia formation and cell migration. Faseb J. 2001;15:2649–59. doi: 10.1096/fj.01-0523com. [DOI] [PubMed] [Google Scholar]
- 7.Spiegel S, Milstien S. Sphingosine 1-phosphate, a key cell signaling molecule. The Journal of biological chemistry. 2002;277:25851–4. doi: 10.1074/jbc.R200007200. [DOI] [PubMed] [Google Scholar]
- 8.Kohama T, Olivera A, Edsall L, Nagiec MM, Dickson R, Spiegel S. Molecular cloning and functional characterization of murine sphingosine kinase. The Journal of biological chemistry. 1998;273:23722–8. doi: 10.1074/jbc.273.37.23722. [DOI] [PubMed] [Google Scholar]
- 9.Liu H, Sugiura M, Nava VE, Edsall LC, Kono K, Poulton S, et al. Molecular cloning and functional characterization of a novel mammalian sphingosine kinase type 2 isoform. The Journal of biological chemistry. 2000;275:19513–20. doi: 10.1074/jbc.M002759200. [DOI] [PubMed] [Google Scholar]
- 10.Johnson KR, Becker KP, Facchinetti MM, Hannun YA, Obeid LM. PKC-dependent activation of sphingosine kinase 1 and translocation to the plasma membrane. Extracellular release of sphingosine-1-phosphate induced by phorbol 12-myristate 13-acetate (PMA) The Journal of biological chemistry. 2002;277:35257–62. doi: 10.1074/jbc.M203033200. [DOI] [PubMed] [Google Scholar]
- 11.Pitson SM, Xia P, Leclercq TM, Moretti PA, Zebol JR, Lynn HE, et al. Phosphorylation-dependent translocation of sphingosine kinase to the plasma membrane drives its oncogenic signalling. The Journal of experimental medicine. 2005;201:49–54. doi: 10.1084/jem.20040559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.French KJ, Schrecengost RS, Lee BD, Zhuang Y, Smith SN, Eberly JL, et al. Discovery and evaluation of inhibitors of human sphingosine kinase. Cancer research. 2003;63:5962–9. [PubMed] [Google Scholar]
- 13.Kawamori T, Osta W, Johnson KR, Pettus BJ, Bielawski J, Tanaka T, et al. Sphingosine kinase 1 is up-regulated in colon carcinogenesis. Faseb J. 2006;20:386–8. doi: 10.1096/fj.05-4331fje. [DOI] [PubMed] [Google Scholar]
- 14.Sarkar S, Maceyka M, Hait NC, Paugh SW, Sankala H, Milstien S, et al. Sphingosine kinase 1 is required for migration, proliferation and survival of MCF-7 human breast cancer cells. FEBS letters. 2005;579:5313–7. doi: 10.1016/j.febslet.2005.08.055. [DOI] [PubMed] [Google Scholar]
- 15.Taha TA, Kitatani K, El-Alwani M, Bielawski J, Hannun YA, Obeid LM. Loss of sphingosine kinase-1 activates the intrinsic pathway of programmed cell death: modulation of sphingolipid levels and the induction of apoptosis. Faseb J. 2006;20:482–4. doi: 10.1096/fj.05-4412fje. [DOI] [PubMed] [Google Scholar]
- 16.Hait NC, Sarkar S, Le Stunff H, Mikami A, Maceyka M, Milstien S, et al. Role of sphingosine kinase 2 in cell migration toward epidermal growth factor. The Journal of biological chemistry. 2005;280:29462–9. doi: 10.1074/jbc.M502922200. [DOI] [PubMed] [Google Scholar]
- 17.Allende ML, Sasaki T, Kawai H, Olivera A, Mi Y, van Echten-Deckert G, et al. Mice deficient in sphingosine kinase 1 are rendered lymphopenic by FTY720. The Journal of biological chemistry. 2004;279:52487–92. doi: 10.1074/jbc.M406512200. [DOI] [PubMed] [Google Scholar]
- 18.Liu H, Toman RE, Goparaju SK, Maceyka M, Nava VE, Sankala H, et al. Sphingosine kinase type 2 is a putative BH3-only protein that induces apoptosis. The Journal of biological chemistry. 2003;278:40330–6. doi: 10.1074/jbc.M304455200. [DOI] [PubMed] [Google Scholar]
- 19.Maceyka M, Sankala H, Hait NC, Le Stunff H, Liu H, Toman R, et al. SphK1 and SphK2, sphingosine kinase isoenzymes with opposing functions in sphingolipid metabolism. The Journal of biological chemistry. 2005;280:37118–29. doi: 10.1074/jbc.M502207200. [DOI] [PubMed] [Google Scholar]
- 20.Weigert A, Johann AM, von Knethen A, Schmidt H, Geisslinger G, Brune B. Apoptotic cells promote macrophage survival by releasing the antiapoptotic mediator sphingosine-1-phosphate. Blood. 2006;108:1635–42. doi: 10.1182/blood-2006-04-014852. [DOI] [PubMed] [Google Scholar]
- 21.Weigert A, Cremer S, Schmidt MV, von Knethen A, Angioni C, Geisslinger G, et al. Cleavage of sphingosine kinase 2 by caspase-1 provokes its release from apoptotic cells. Blood. 115:3531–40. doi: 10.1182/blood-2009-10-243444. [DOI] [PubMed] [Google Scholar]
- 22.Van Brocklyn JR, Jackson CA, Pearl DK, Kotur MS, Snyder PJ, Prior TW. Sphingosine kinase-1 expression correlates with poor survival of patients with glioblastoma multiforme: roles of sphingosine kinase isoforms in growth of glioblastoma cell lines. Journal of neuropathology and experimental neurology. 2005;64:695–705. doi: 10.1097/01.jnen.0000175329.59092.2c. [DOI] [PubMed] [Google Scholar]
- 23.Weigert A, Schiffmann S, Sekar D, Ley S, Menrad H, Werno C, et al. Sphingosine kinase 2 deficient tumor xenografts show impaired growth and fail to polarize macrophages towards an anti-inflammatory phenotype. International journal of cancer. 2009;125:2114–21. doi: 10.1002/ijc.24594. [DOI] [PubMed] [Google Scholar]
- 24.Hagen N, Van Veldhoven PP, Proia RL, Park H, Merrill AH, Jr, van Echten-Deckert G. Subcellular origin of sphingosine 1-phosphate is essential for its toxic effect in lyase-deficient neurons. The Journal of biological chemistry. 2009;284:11346–53. doi: 10.1074/jbc.M807336200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hait NC, Allegood J, Maceyka M, Strub GM, Harikumar KB, Singh SK, et al. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science (New York, NY. 2009;325:1254–7. doi: 10.1126/science.1176709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kharel Y, Lee S, Snyder AH, Sheasley-O’neill SL, Morris MA, Setiady Y, et al. Sphingosine kinase 2 is required for modulation of lymphocyte traffic by FTY720. The Journal of biological chemistry. 2005;280:36865–72. doi: 10.1074/jbc.M506293200. [DOI] [PubMed] [Google Scholar]
- 27.French KJ, Upson JJ, Keller SN, Zhuang Y, Yun JK, Smith CD. Antitumor activity of sphingosine kinase inhibitors. The Journal of pharmacology and experimental therapeutics. 2006;318:596–603. doi: 10.1124/jpet.106.101345. [DOI] [PubMed] [Google Scholar]
- 28.French KJ, Zhuang Y, Maines LW, Gao P, Wang W, Beljanski V, et al. Pharmacology and antitumor activity of ABC294640, a selective inhibitor of sphingosine kinase-2. The Journal of pharmacology and experimental therapeutics. 333:129–39. doi: 10.1124/jpet.109.163444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beljanski V, Knaak C, Zhuang Y, Smith CD. Combined anticancer effects of sphingosine kinase inhibitors and sorafenib. Invest New Drugs. 2010 doi: 10.1007/s10637-010-9452-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beljanski V, Lewis CS, Smith CD. Antitumor activity of sphingosine kinase 2 inhibitor ABC294640 and sorafenib in hepatocellular carcinoma xenografts. Cancer Biol Ther. 2011;11:524–34. doi: 10.4161/cbt.11.5.14677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pettus BJ, Bielawski J, Porcelli AM, Reames DL, Johnson KR, Morrow J, et al. The sphingosine kinase 1/sphingosine-1-phosphate pathway mediates COX-2 induction and PGE2 production in response to TNF-alpha. Faseb J. 2003;17:1411–21. doi: 10.1096/fj.02-1038com. [DOI] [PubMed] [Google Scholar]
- 32.Le Stunff H, Galve-Roperh I, Peterson C, Milstien S, Spiegel S. Sphingosine-1-phosphate phosphohydrolase in regulation of sphingolipid metabolism and apoptosis. The Journal of cell biology. 2002;158:1039–49. doi: 10.1083/jcb.200203123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shu X, Wu W, Mosteller RD, Broek D. Sphingosine kinase mediates vascular endothelial growth factor-induced activation of ras and mitogen-activated protein kinases. Molecular and cellular biology. 2002;22:7758–68. doi: 10.1128/MCB.22.22.7758-7768.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.zu Heringdorf DM, Vincent ME, Lipinski M, Danneberg K, Stropp U, Wang DA, et al. Inhibition of Ca(2+) signalling by the sphingosine 1-phosphate receptor S1P(1) Cellular signalling. 2003;15:677–87. doi: 10.1016/s0898-6568(03)00011-1. [DOI] [PubMed] [Google Scholar]
- 35.Takasugi N, Sasaki T, Suzuki K, Osawa S, Isshiki H, Hori Y, et al. BACE1 activity is modulated by cell-associated sphingosine-1-phosphate. J Neurosci. 31:6850–7. doi: 10.1523/JNEUROSCI.6467-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kihara A, Ikeda M, Kariya Y, Lee EY, Lee YM, Igarashi Y. Sphingosine-1-phosphate lyase is involved in the differentiation of F9 embryonal carcinoma cells to primitive endoderm. The Journal of biological chemistry. 2003;278:14578–85. doi: 10.1074/jbc.M211416200. [DOI] [PubMed] [Google Scholar]
- 37.Huwiler A, Kotelevets N, Xin C, Pastukhov O, Pfeilschifter J, Zangemeister-Wittke U. Loss of sphingosine kinase-1 in carcinoma cells increases formation of reactive oxygen species and sensitivity to doxorubicin-induced DNA damage. British journal of pharmacology. 162:532–43. doi: 10.1111/j.1476-5381.2010.01053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berdyshev EV, Gorshkova I, Usatyuk P, Kalari S, Zhao Y, Pyne NJ, et al. Intracellular S1P Generation Is Essential for S1P-Induced Motility of Human Lung Endothelial Cells: Role of Sphingosine Kinase 1 and S1P Lyase. PloS one. 6:e16571. doi: 10.1371/journal.pone.0016571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niculescu AB, 3rd, Chen X, Smeets M, Hengst L, Prives C, Reed SI. Effects of p21(Cip1/Waf1) at both the G1/S and the G2/M cell cycle transitions: pRb is a critical determinant in blocking DNA replication and in preventing endoreduplication. Molecular & Cellular Biology. 1998;18:629–43. doi: 10.1128/mcb.18.1.629. [erratum appears in Mol Cell Biol 1998 Mar;18(3):1763] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang F, Van Brocklyn JR, Hobson JP, Movafagh S, Zukowska-Grojec Z, Milstien S, et al. Sphingosine 1-phosphate stimulates cell migration through a G(i)-coupled cell surface receptor. Potential involvement in angiogenesis. Journal of Biological Chemistry. 1999;274:35343–50. doi: 10.1074/jbc.274.50.35343. [DOI] [PubMed] [Google Scholar]
- 41.Sieg DJ, Hauck CR, Ilic D, Klingbeil CK, Schaefer E, Damsky CH, et al. FAK integrates growth-factor and integrin signals to promote cell migration. Nature cell biology. 2000;2:249–56. doi: 10.1038/35010517. [DOI] [PubMed] [Google Scholar]
- 42.Deem TL, Cook-Mills JM. Vascular cell adhesion molecule 1 (VCAM-1) activation of endothelial cell matrix metalloproteinases: role of reactive oxygen species. Blood. 2004;104:2385–93. doi: 10.1182/blood-2004-02-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Devine L, Lightman SL, Greenwood J. Role of LFA-1, ICAM-1, VLA-4 and VCAM-1 in lymphocyte migration across retinal pigment epithelial monolayers in vitro. Immunology. 1996;88:456–62. doi: 10.1046/j.1365-2567.1996.d01-666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xia P, Gamble JR, Wang L, Pitson SM, Moretti PA, Wattenberg BW, et al. An oncogenic role of sphingosine kinase. Curr Biol. 2000;10:1527–30. doi: 10.1016/s0960-9822(00)00834-4. [DOI] [PubMed] [Google Scholar]
- 45.http://dtp.nci.gov/mtweb.
- 46.Seddighzadeh M, Zhou JN, Kronenwett U, Shoshan MC, Auer G, Sten-Linder M, et al. ERK signalling in metastatic human MDA-MB-231 breast carcinoma cells is adapted to obtain high urokinase expression and rapid cell proliferation. Clinical & experimental metastasis. 1999;17:649–54. doi: 10.1023/a:1006741228402. [DOI] [PubMed] [Google Scholar]
- 47.Sensken SC, Bode C, Nagarajan M, Peest U, Pabst O, Graler MH. Redistribution of sphingosine 1-phosphate by sphingosine kinase 2 contributes to lymphopenia. J Immunol. 184:4133–42. doi: 10.4049/jimmunol.0903358. [DOI] [PubMed] [Google Scholar]
- 48.Igarashi N, Okada T, Hayashi S, Fujita T, Jahangeer S, Nakamura S. Sphingosine kinase 2 is a nuclear protein and inhibits DNA synthesis. The Journal of biological chemistry. 2003;278:46832–9. doi: 10.1074/jbc.M306577200. [DOI] [PubMed] [Google Scholar]
- 49.Sankala HM, Hait NC, Paugh SW, Shida D, Lepine S, Elmore LW, et al. Involvement of sphingosine kinase 2 in p53-independent induction of p21 by the chemotherapeutic drug doxorubicin. Cancer research. 2007;67:10466–74. doi: 10.1158/0008-5472.CAN-07-2090. [DOI] [PubMed] [Google Scholar]
- 50.Baudhuin LM, Jiang Y, Zaslavsky A, Ishii I, Chun J, Xu Y. S1P3-mediated Akt activation and cross-talk with platelet-derived growth factor receptor (PDGFR) Faseb J. 2004;18:341–3. doi: 10.1096/fj.03-0302fje. [DOI] [PubMed] [Google Scholar]
- 51.Olivera A, Rosenfeldt HM, Bektas M, Wang F, Ishii I, Chun J, et al. Sphingosine kinase type 1 induces G12/13-mediated stress fiber formation, yet promotes growth and survival independent of G protein-coupled receptors. The Journal of biological chemistry. 2003;278:46452–60. doi: 10.1074/jbc.M308749200. [DOI] [PubMed] [Google Scholar]
- 52.Bessard A, Fremin C, Ezan F, Fautrel A, Gailhouste L, Baffet G. RNAi-mediated ERK2 knockdown inhibits growth of tumor cells in vitro and in vivo. Oncogene. 2008;27:5315–25. doi: 10.1038/onc.2008.163. [DOI] [PubMed] [Google Scholar]
- 53.Fremin C, Ezan F, Boisselier P, Bessard A, Pages G, Pouyssegur J, et al. ERK2 but not ERK1 plays a key role in hepatocyte replication: an RNAi-mediated ERK2 knockdown approach in wild-type and ERK1 null hepatocytes. Hepatology (Baltimore, Md. 2007;45:1035–45. doi: 10.1002/hep.21551. [DOI] [PubMed] [Google Scholar]
- 54.Forrest M, Sun SY, Hajdu R, Bergstrom J, Card D, Doherty G, et al. Immune cell regulation and cardiovascular effects of sphingosine 1-phosphate receptor agonists in rodents are mediated via distinct receptor subtypes. The Journal of pharmacology and experimental therapeutics. 2004;309:758–68. doi: 10.1124/jpet.103.062828. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.