Abstract
Potentials of mean force (PMF) between ionizable amino acid side chains (Arg, Lys, His, Glu) in the headgroup area of a palmitoyl oleoyl phosphatidylcholine lipid bilayer were obtained from all-atom molecular dynamics simulations and the adaptive biasing force method. Simulations in bulk water were also performed for comparison. Side chains were constrained in collinear, stacking and orthogonal (T-shaped) orientations. The most structured and attractive PMFs were observed for hydrogen-bonded side chains. Contact minima occurred at a distance of 2.6–3.1 Å between selected atoms or centers of mass with the most attractive interaction (−9.6 kcal/mol) observed between Arg+ and Glu−. Hydrogen bonds play a significant role in stabilizing these interactions. Interactions between like-charged side chains can also be very attractive, if the charges are screened by surrounding molecules or groups (e.g. the PMF value at the contact minimum for Arg+…Arg+ is −7.6 kcal/mol). Like-charged side chains can have contact minima as close as 3.6 Å. The PMFs depend strongly on the relative orientation of the side chains. In agreement with experimental studies and other simulations, we found the stacking arrangement of like-charged side chains to be the most favorable orientation. Interaction energies and Lennard-Jones energies between side chains, headgroups and water molecules were analyzed in order to rationalize the observed PMFs and their dependence on orientation. In general, the results cannot be explained by simple dielectric arguments.
Keywords: Potential of mean force, interaction free energy, lipid bilayer, all-atom molecular dynamics, adaptive biasing force, arginine-arginine interactions
Introduction
Ionizable residues are abundant in proteins and often play key roles in their function and stability. 1–4 Interactions between them are long-range and often dominate despite the screening by solvent and electrolytes.5 Experimental and theoretical estimates of the contribution of salt bridges to protein stability range from stabilizing, 6–9 to being insignificant 10–12, to being destabilizing 13–16. These interactions are arguably the hardest to model accurately, especially in implicit solvent simulations, as they consist of a large Coulomb contribution modulated by equally large solvation contributions.
In an effort to systematically characterize solvent-mediated electrostatic interactions, several groups have calculated the potential of mean force (PMF) between groups mimicking amino acids side chains (SC) in water. 17–27 The PMF for guanidinium ions in parallel orientation revealed a stable contact pair with a free energy depending on models used for water and guanidinium.17 Rozanska and Chipot18 calculated the PMF between guanidinium and acetate in collinear orientation using umbrella sampling and the AMBER force field. The same system was studied later with the CHARMM27 force field using the adaptive basis force (ABF) method.19 Masunov and Lazaridis20 determined the PMF between a large number of ionizable side chain pairs using a Spherical Solvent Boundary Potential and the CHARMM19 force field. PMFs averaged over all possible orientations and simulations of pairs of ammonium, guanidinium and acetate ions were calculated by Maksimiak et al.21 Hassan22 studied PMFs of hydrogen-bonded amino acid side chains in water. He later studied the effect of salt on the PMFs and showed that the addition of salt may stabilize or destabilize the interactions, depending on the nature of the interacting molecules.23 In another study the PMF of Lys+…Glu− analogues in water was generated by rotating the charged groups away from each other with the Cmethyl-Cmethyl or Ccarboxyl-Namino distance held constant. Addition of salt destabilized all Lys+…Glu− configurations and to a greater extent those in which the charged groups of the ions were close.24 Mandell et al. calculated PMFs between phosphorylated amino acid side chains as well as Glu−, Arg and Lys in water.25 Vaitheeswaran and Thirumalai calculated PMFs between two neutral and charged methane molecules in confined water droplets and found that the PMFs between charged species and the tendency to be pinned at the surface depended strongly on the water droplet size.26 In a later study of confinement effect on PMF between amino acid side chains these authors showed that salt bridge was promoted in a nanopore compared to the bulk water and interactions had an enhanced dependence on side chain relative orientation.27 In a study of interaction between an ion embedded on a hydrophobic surface with another ion in water it was found that the contact between charges was strongly affected by the curvature. The solute ion was most stable in contact with an embedded charge in a surface having negative curvature, which represented a receptor.28 Geney et al. studied salt bridges in a solvated protein environment and found the Arg-Glu pair to be more stable than Lys-Glu. The doubly H-bonded Arg-Glu pair was ~3 times more stable than the singly H-bonded one.29
The interaction between ionizable side chains should depend strongly on solvent environment, but this dependence has not yet been explored by either experiment or theory. In addition to this fundamental physicochemical question, of particular interest to us are such interactions in the vicinity of lipid bilayers, which could play important roles in membrane protein folding or oligomerization. For example, interactions between Arg were observed in the nonnative, left-handed transmembrane dimer of glycophorin A.30 In a different study the juxtamembrane residues were found to affect significantly the association free energy of transmembrane helices.31 Thus, it is critical to model these interactions correctly. So far, only solvation and permeation of single side chains in bilayers have been studied by several groups,32–39 with the exception of a very recent study where the insertion of two Arg has been considered40. In this work we study the interaction between amino acid side chains in the headgroup area of a lipid bilayer. Our goal is to quantify the strength of interactions and determine the most favorable orientations of ionizable amino acid side chains at the lipid bilayer-water interface. We report the PMF between ionizable amino acid side chains in a palmitoyl oleoyl phosphatidylcholine (POPC) lipid bilayer obtained from all-atom explicit solvent molecular dynamics simulations using the ABF approach41,42. For comparison, we also calculated PMFs of amino acid side chains in a box of water using the same protocol and constraints. We compare our results with experimental data and previously reported data from MD simulations in water and solvated proteins. To obtain insights and to understand the differences observed for different ions and different orientations, we analyze and discuss such properties as H-bonds and interaction energies calculated from the obtained MD trajectories. The results could be useful for rationalizing membrane protein structures, predicting membrane protein interactions, and for the development of effective membrane potentials.43–50
Methods
All MD simulations were conducted with NAMD51 using CHARMM2752,53 topology and parameter files. The deprotonated Arg pairs represented by methyl-guanidine were modeled using a recently developed force field.54 VMD55 was used to prepare input files and to analyze the MD trajectories, as well as for visualization.
Construction and equilibration of the starting structures
The membrane builder of VMD was used to construct a POPC membrane consisting of 66 lipid molecules in each monolayer. The SOLVATE facility of VMD was used to hydrate the bilayer with 9980 TIP3P56 water molecules. The resulting system was energy-minimized for 5000 steps and a short (25000 time steps) simulation was run to heat the system up to a constant temperature of 310 K that was followed by 5 ns equilibration at constant temperature and pressure. A water box containing ~3500 water molecules was also constructed in VMD and equilibrated for 1 ns using the same protocol.
The coordinates of the amino acid side chain atoms were extracted from pdb files used in previous work where the backbone was removed by replacing the α-carbon with a hydrogen atom and adjusting the charge on the β-carbon.20 The coordinates of missing H-atoms were generated by the GUESSCOORD application of VMD. Each amino acid side chain pair was inserted into the headgroup region of the pre-equilibrated lipid bilayer at 13.5 Å from the center of the membrane or in the center of the water box. Any overlapping POPC (up to 3 lipids depending on SC pair) or water molecules were removed. The resulting system was then energy minimized for 3000 steps, equilibrated for 0.5–1 ns and then subjected to production phase MD simulations.
Molecular Dynamics
NAMD with its standard potential energy function was employed for MD simulations. Simulations were carried out in the NPAT ensemble57 to ensure the correct value of area per lipid. The constant pressure component normal to the lipid-water interface was set to P= 101.325 kPa. A combination of the Nose-Hoover constant pressure method58 with piston fluctuation control implemented using Langevin dynamics59 was used to control the pressure. The Langevin piston oscillation period and the oscillation decay time were equal to 200 fs and 100 fs, respectively. The temperature was kept constant at 310 K using the Langevin dynamics method with a Langevin damping coefficient of 10/ps applied only to nonhydrogen atoms. The pressure was calculated using the hydrogen-group-based pseudomolecular virial and kinetic energy (useGroupPressure option of NAMD) in conjunction with the SHAKE algorithm. The bond lengths between each hydrogen atom and the atom to which it was covalently bonded were constrained to their equilibrium values using the SHAKE algorithm60, which allowed a time step of 2 fs for the integration of Newton’s equations. A cutoff of 10.5 Å was used for Lennard-Jones and electrostatic interactions with smoothing functions activated at 8.5 Å. The long-range Coulombic forces were updated every four steps. The van der Waals interactions were modified using special 1–4 parameters defined in the parameter file and were truncated smoothly at the cutoff distance. Periodic boundary conditions were used and the long-range electrostatic interactions were calculated by the smooth particle mesh Ewald61 (PME) approach with 1 pm distance between grid points.
We chose not to include any salt or counterions in the system for a number of reasons. First, most previous calculations of PMFs in water were done in the absence of salt, so comparisons with them would have been impossible. In any investigation, results in pure water are the natural starting point. Second, the sampling required in the presence of counterions is substantially more extensive. Early simulations performed with just one or two neutralizing counterions showed that direct interaction with side chains significantly affected the PMF. Therefore, we resorted to the “neutralizing plasma” that PME employs for systems that carry a net charge. While this approach leads to erroneous absolute energies62 it does not appear to affect significantly the calculated PMFs. When we repeated certain calculations with counterions fixed far from the side chains (as did others22,25) we obtained essentially identical results (the same observation was made by Hilder & Chung63).
An additional concern is whether the artificial periodicity imposed on the system will introduce artifacts into the calculated PMFs. Such artifacts have been thoroughly investigated in the case of solvation free energies and PMFs between simple ions64, peptides65, 66 and larger biomolecules67, 68. Most of the early studies were conducted in box sizes up to 40 Å and found that the problems were small at the larger box sizes studied. A recent study of a beta-heptapeptide in methanol found that periodicity artifacts are negligible in a box of 60 Å edge. We believe these artifacts are negligible in our case because a) our simulation box is about 70 Å long, b) the solutes are monovalent and very small compared to the simulation box, c) the computed effect of these artifacts on the PMF between small ions in a 40 Å box was flat below 15 Å69 and d) although the solutes are near a low dielectric region, they interact through the membrane interface, which is a high dielectric medium70.
Calculation of the free energy
The PMF was calculated using the ABF approach41,42,71 implemented as a suite of Tcl routines in NAMD. The ABF method does not require a constraint to be applied on the value of the reaction coordinate ξ. It is based on computing the mean force on ξ and then removing this force to improve sampling. The resulting system randomly samples conformations along ξ with uniform probability. The potential of mean force is obtained from integrating the computed mean force along ξ. ABF removes the need to guess a priori the biasing potential or to refine it iteratively. Instead the biasing force is estimated locally from the sampled conformations of the system and continuously updated as additional samples are gathered and more-accurate averages are obtained.72
The distance between two atoms or the centers of mass of selected atoms was chosen as reaction coordinate. The atoms selected in each case are given in the Figure captions. The pairs were constrained in three orientations: collinear, stacking and orthogonal (see Table 1). Harmonic restraints were applied to the heavy atoms to force the side chains to move parallel to the membrane. These restraints are perpendicular to the reaction coordinate, so that the corresponding forces have no influence upon the calculation of Fξ.71 The width of the bins (5 pm) in which the forces are accumulated has to be small enough to ascertain that the free energy profile is smooth in the ξ-interval and large enough to ensure sufficient sampling and avoid large fluctuations in the average force. Harmonic bias was used to keep the reaction coordinate between 2 and 12 Å with the force Const parameter of NAMD equal to 150–200 kcal/mol/Å2 for POPC and 150 kcal/mol/Å2 for the water box. To reduce force fluctuations, the mean force was averaged over 10 pm (dSmooth parameter of NAMD). The convergence of the PMF calculation was gauged by examining the number of samples collected in each bin. Uniform sampling along the trajectory as well as a high number of samples collected in each bin were required before stopping the simulations. The statistical accuracy of the calculated PMFs was estimated by duplicate simulations for the Arg+…Glu−, His+…His+, Arg+…His+, and Glu−…Glu− pairs in POPC and the Arg+…Arg+ pair in water. The error bars shown in the figures correspond to the difference between the two runs.
Table 1.
Amino acid side chains orientations and PMF data.
| Pair | Orientation | Structure | CPM r [Å] | PMFCPM* [kcal/mol] | SSM r [Å] | PMFSSM*** [kcal/mol] | Simulation Time (ns) |
|---|---|---|---|---|---|---|---|
| POPC (Water) | |||||||
| Arg+…Glu− | Collinear |
|
2.7 | −9.6±0.6 | 5.2 | −3.8±0.3 | 2.5 |
| (2.7) | (−11.6) | (5.2) | (−3.7) | ||||
| Stacking |
|
3.7 | −3.3 | - | - | 3.3 | |
| Lys+…Glu− | Collinear |
|
2.8 | −4.0 | - | - | 3.9 |
| (2.8) | (−1.8) | ||||||
| Stacking |
|
3.7 | −6.3 | - | - | 2.5 | |
| His+…Glu− | Collinear |
|
2.6 | −7.2 | 5.4 | −1.2 | 3 |
| (2.6) | (−5.8) | (5.1) | (−1.5) | ||||
| His0…Glu− | Collinear |
|
2.7 | −3.5 | 5.2 | −2.8 | 3 |
| Arg+…Arg+ | Collinear |
|
5.2 | 0.1 | 2.5 | ||
| Stacking |
|
3.9 | −7.6 | 7.0 | −3.1 | 2.7 | |
| (3.9) | (−4.4±0.02) | ||||||
| Orthogonal |
|
- | - | 6.0 | −2.4 | 5 | |
| Arg0…Arg0 | Collinear |
|
4.3 | −3.6 | 7.6 | ||
| Arg0…Arg0 | Stacking |
|
3.4 | −5.6 | 5 | ||
| His+…His0 | Collinear |
|
2.9 | −6.0 | 5.6 | −3.2 | 4 |
| His0…His0 | Collinear |
|
2.9 | −3.4 | 5.6 | −1.3 | 2.5 |
| His+…His+ | Collinear |
|
5.2 | −0.85 | 3.5 | ||
| Stacking |
|
3.7 | −4.1±0.6 | 6.6 | −1.6±0.4 | 6.8 | |
| (3.6) | (−2.5) | (6.9) | (−1.1) | ||||
| Orthogonal |
|
- | - | 6.1 | −1.4 | 2.5 | |
| Arg+…His+ | Collinear |
|
5.0 | −1.5 | 7.6 | ||
| Stacking |
|
3.6 | −3.1±1.0 | 6.7 | −0.75±0.2 | 4 | |
| (3.6) | (−4.9) | ||||||
| Orthogonal |
|
- | - | 6.3 | −0.24 | 2.5 | |
| Lys+…His0 | Collinear |
|
2.9 | −3.6 | - | - | 13 |
| Lys+…His+ | Collinear |
|
- | - | 6.4 | −1.8 | 2.7 |
| Orthogonal |
|
- | - | 6.0 | −2.7 | 5.3 | |
| (6.8) | (−0.35) | ||||||
| Lys+…Lys+ | Collinear |
|
4.5 | −2.1 | 2.5 | ||
| Lys+…Arg+ | Collinear |
|
- | - | 6.4 | −1.8 | 4.3 |
| Orthogonal |
|
4.0 | −1.95 | 7.3 | −0.57 | 3.7 | |
| (4.0) | (−2.0) | (7.2) | (−0.44) | ||||
| Glu−…Glu0 | Collinear |
|
5.0 | −2.7 | 1 | ||
| (3.1) | (−1.3) | (5.5) | (−1.5) | ||||
| Glu0…Glu0 | Collinear |
|
3.1 | −5.7 | 5.7 | −1.7 | 2.5 |
| Glu−…Glu− | Collinear |
|
4.3 | −1.0 | 6.8 | −0.26 | 2.2 |
| Orthogonal |
|
5.0 | −2.5±0.1 | 4.7 | |||
Calculation of system properties
Interaction energies (IE), Lennard-Jones (LJ) energies and H-bonds were calculated from ABF trajectories. Interaction energies and LJ energies were calculated using the NAMDEnergy plug-in of VMD. H-bonds were calculated using the measure hbonds command with only non-hydrogen atoms considered. A H-bond is said to exist when the distance between donor and acceptor is 3 Å and the angle formed by the donor, hydrogen, and acceptor is less than 60º. The aforementioned commands are available through TCL scripting in VMD. The number of H-bonds, IE, LJ energies were averaged over the ABF trajectories.
Results
We studied in total 29 side chain pairs in three orientations: collinear (head-to-head), stacking (side-to-side) and orthogonal (head-to-side) (Table 1). The side chains considered are Arg, His, Lys and Glu. Asp was not included because in most collinear orientations and in some stacking orientations the additional CH2 group of Glu does not interact with the partner side chain and, therefore, should make no difference in the PMF. For those orientations where the hydrophobic parts of the side chains could come in contact (e.g. His-Glu), some additional stabilization due to hydrophobic interactions is expected for Glu relative to Asp. Lys was simulated only in its protonated form but for the remaining side chains both charged and neutral states were examined.
The calculated PMFs are shown in Figures 1–8 and in Supplementary Material. To isolate the impact of the POPC headgroups on intermolecular interactions, we also obtained PMFs for selected pairs in a box of water. The PMF curves are anchored to 0 at 12 Å. Since they are flat beyond 10 Å (except for the orthogonal Glu−…Glu− and neutral Arg pairs where the curve were flat beyond 11 Å), the figures show the free energy up to that distance.
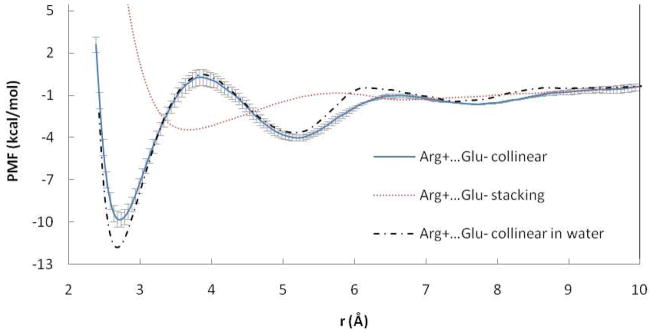
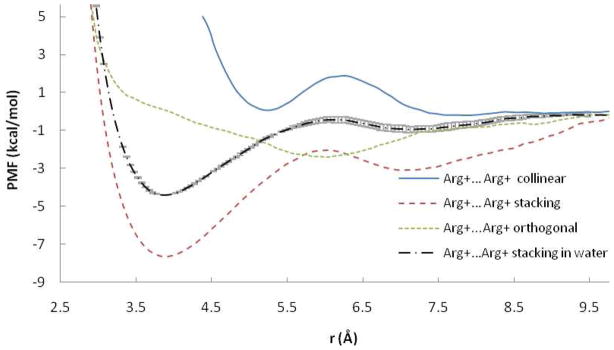
Figure 1.
PMF curves for the Arg+…Glu− ion pairs in POPC and water. r is the distance between COM of NH atoms of Arg and OE atoms of Glu. Error bars are displayed for collinear pair in POPC.
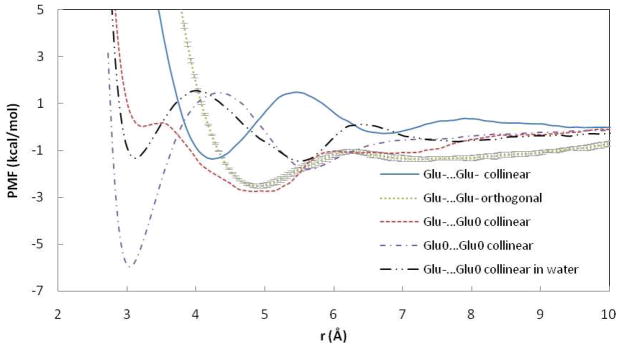
Figure 8.
PMF curves for the Glu…Glu ion pairs in POPC and water. r is the distance between COM of OE atoms of Glu. Error bars are displayed for orthogonal pair.
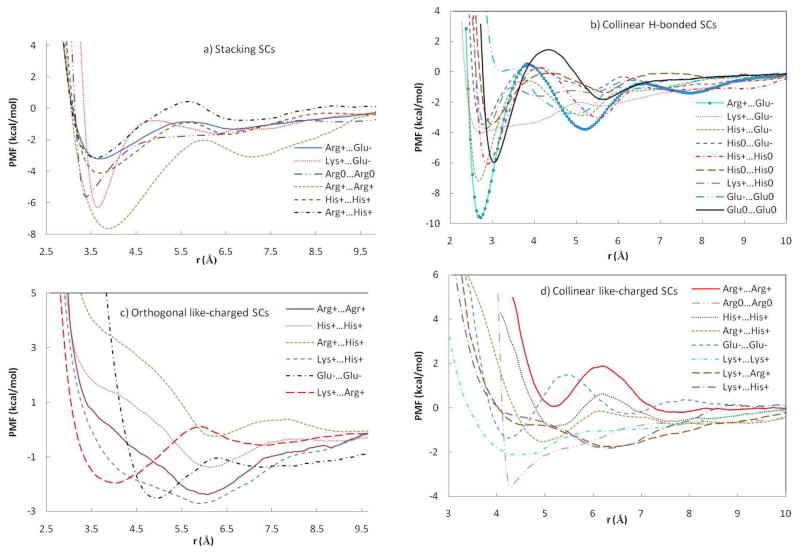
The PMFs between unlike-charged arginine and glutamate amino acid side chains are reported in Figure 1. In collinear orientation, the profile is smooth and structured with a narrow well corresponding to the contact pair (CP) state and a second minimum corresponding to a solvent-separated state. In stacking orientation, the profile has a broader and more shallow contact minimum and lacks a solvent-separated minimum. The same protocol for collinear orientation in water gives a more stable CP than in the POPC bilayer. The value of the contact minimum in water obtained here (−11.6 kcal/mol) is larger than results obtained previously: −8.019, −4.520, −5.922, −8.525, −6.529 kcal/mol. The discrepancy is probably due to the different force field (CHARMM1920, CHARMM2222, OPLS25), the smaller water sphere/box20, different set of constraints 22,25, and the use of a less accurate treatment of long range electrostatics20.
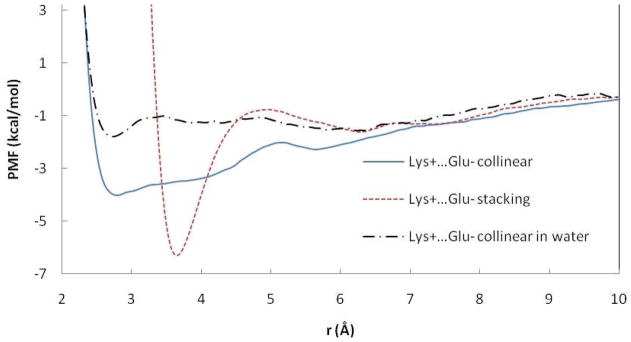
The lysine-glutamate pair behaves differently than arginine-glutamate for the same orientations (Table 1, Figure 2). In collinear orientation, the PMF displays a broad contact minimum and a gradual increase in free energy with distance without a SSM. In contrast, the PMF for stacking orientation is structured with a narrow CP well. The PMF in water for the collinear pair has a minimum at the same distance as in the POPC but the interaction is less attractive. The contact minimum (−1.8 kcal/mol) is in quantitative agreement with previously obtained PMFs in water: −2.3520, −2.3927 and in a solvated protein environment29 (−1.2 kcal/mol). A smaller value (−0.3 kcal/mol) was obtained in Ref. 22.
Figure 2.
PMF curves for the Lys+…Glu− ion pairs in POPC and Lys-Glu. r is the distance between: COM of OE atoms of Glu and NZ atoms of Lys and OE atom and NZ atom for collinear and stacking pairs, respectively.
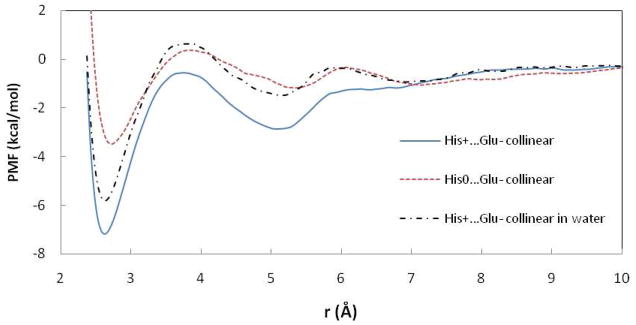
The His+…Glu− pair displays a more attractive CM than His0…Glu− (Table 1, Figure 3). The PMF in water obtained for the His+…Glu− pair is qualitatively similar but less deep than that in POPC. Previously reported results for His+…Glu− in water are less attractive: −1.2220 and −3.922 kcal/mol vs −5.8 kcal/mol obtained here.
Figure 3.
PMF curves for the His…Glu− ion pairs in POPC and water. r is the distance between ND atom of His and OE2 of Glu.
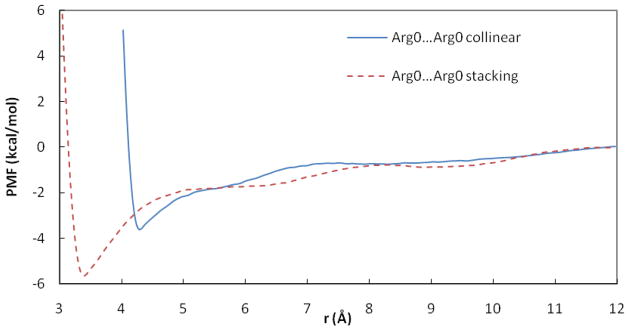
The results for Arg+…Arg+ are shown in Figure 4 and Table 1. A surprisingly strong attractive interaction is observed for the stacking arrangement with a deep CM and a more shallow SSM. In water the PMF for this orientation is similar but much less attractive. The PMF in the water box obtained for this pair differs significantly from the profile obtained in a water sphere20 by having a deeper CM and a more shallow SSM. Another study of two guanidine ions did not produce an attractive well, but in that study only the distance was constrained, not the orientation21. An older study found the depth of the attractive well to be strongly dependent on water model.17 The orthogonal orientation exhibits a shallow minimum at ~ 6 Å and the collinear orientation is repulsive at short distances. Deprotonation of the two Arg at the membrane interface reduces the depth of the CM in stacking orientation and creates a shallow CM in collinear orientation (Table 1, Figure 5). No SSM is observed for the neutral Arg pair.
Figure 4.
PMF curves for the Arg+…Arg+ ion pairs in POPC and water. r is the distance between COM of NH atoms of two Arg residues. Error bars are displayed for stacking pair in water.
Figure 5.
PMF curves for the Arg0…Arg0 ion pairs in POPC and water. r is the distance between COM of NH atoms of two Arg residues.
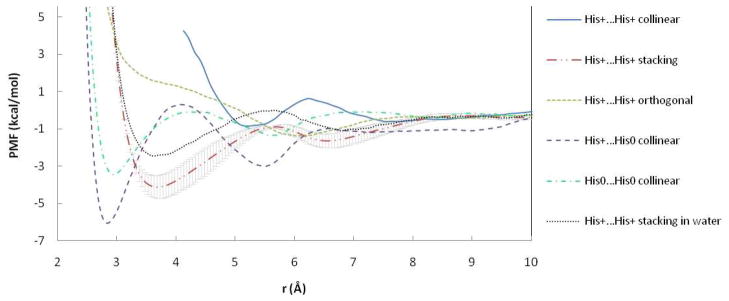
The results for two charged histidine side chains are qualitatively similar to those for the Arg-Arg pairs. The PMF in the water box for the stacking histidine pair is less attractive than in POPC bilayer. Hydrogen-bonded His+…His0 and His0…His0 pairs in collinear orientation displayed structured free energy profiles with stable attractive CM and SSM (Figure 6). These results show the same trends as simulations in water20,22, which also revealed more attractive interactions between neutral and charged His residues.
Figure 6.
PMF curves of His…His pairs in POPC and water. r is the distance between ND atoms of His. Error bars are displayed for stacking pair in POPC.
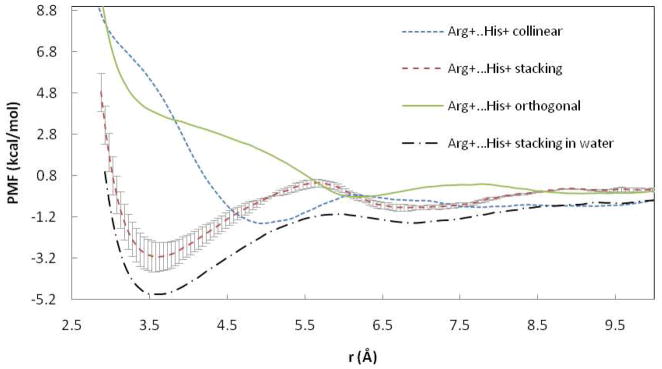
The PMFs for the Arg+…His+ pair in three orientations are shown in Figure 7. As with other cations mentioned above, only the stacking orientation is substantially attractive. However, unlike previously discussed like-charged pairs, the interaction of stacking Arg+…His+ pair in the bilayer is less attractive than in water. Significantly different results were obtained in the water sphere20 with a repulsive contact pair and a slightly attractive SSM.
Figure 7.
PMF curves for the Arg+…His+ ion pairs in POPC and water. r is the distance between COM of NH atoms of Arg and COM of His ring. Error bars are displayed for stacking pair in POPC.
PMF profiles of Lys+…Lys+, Lys+…His+/His0 and Lys+…Arg+ pairs can be found in Supplementary Material in Figures S25 and S29. Interactions between Lys+ and neutral His side chains in collinear orientation are attractive with a well-defined contact pair minimum. Previous simulations in water22 revealed a somewhat weaker free energy of interaction of −2.7 kcal/mol between these residues but a higher activation barrier of ~5 kcal/mol. Collinear but not orthogonal Lys+…His+ and Lys+…Arg+ pairs display similar PMF profiles with shallow minima at ~6 Å. Orthogonal Lys+…Arg+ and collinear Lys+…Lys+ have a CM at shorter distance (~4 Å) with higher activation barrier.
PMFs for two H-bonded Glu ion pairs are shown in Figure 8. The Glu−…Glu0 ion pair displays an unstable shoulder at r=3.1 Å and a shallow contact pair. Glu−…Glu0 side chains in water, unlike in POPC, form a slightly attractive contact pair and a more structured PMF which is similar to previous simulations20. A pair of neutral Glu side chains produced a structured PMF curve with attractive CM and a well-defined SSM. Introduction of two charges (Glu−…Glu− pair) significantly destabilizes the contact pair and shifts the contact minimum to a larger distance (Figure 8). The collinear arrangement of Glu−…Glu− pair is less attractive than that of the orthogonal pair. The free energy of interaction between these ions in w`ater was similar for the two orientations.20
Discussion
This is the first systematic study of interactions between ionizable amino acid side chains in the headgroup area of a lipid bilayer. For comparison, we also calculated selected PMFs in a water box using the same protocol. In this section we first compare the results in water with available earlier calculations and then attempt to gain insights into the molecular origin of the PMFs by calculating interaction energies and hydrogen bonds between the side chains and surrounding headgroups and water molecules using coordinate frames from the simulations (data in Supplementary Material). Although these plots are somewhat “noisy” and do not include entropic effects, they are quite informative.
A number of studies have reported PMFs in water using a variety of methods and force fields19–27. A large number of side chain pairs was studied earlier by our group in a small water sphere using the CHARMM19 force field, umbrella sampling, and an approximate treatment of long range electrostatic interactions20. In the present calculations, unlike-charged and H-bonded pairs show qualitatively similar free energy profiles. The present results confirm previous observations 17,20,73,74 that some interactions between like-charged residues can be attractive. However, some PMF curves for like-charged ions, such as Arg…Arg, His…His, Arg…His, differ significantly from those earlier results20. The observed discrepancies are attributed mainly to the different force fields and simulation protocols. The thoroughly parameterized all-atom CHARMM22 force field52 used here for the side chains should be superior to the older united atom CHARMM19 force field75 used previously. In CHARMM22 all charged sidechains have significantly larger partial charges than in CHARMM19, except for Lys which has about the same partial charges. Other currently used force fields, such as OPLS76 or AMBER77 have somewhat different partial charges from those in CHARMM22. It is impossible at this point to say which one is best. Solvation free energy calculations could help answer this question33. Differences in the precise way the constraints are implemented could also have some effect on the calculated PMFs.19
One difficulty with PMF calculations is that they cannot be validated by direct comparison with experiment. Partial validation, however, can come from comparison with statistical analyses of protein structures. The environment of salt bridges in protein structures varies widely, from entirely aqueous to entirely buried. Because the bilayer interface bears some resemblance to the protein-water interface, comparison with PMFs in the bilayer interface seems most appropriate. Mukherjee et al.73 performed one such statistical analysis within metalloproteins and found collinear Arg-Glu, Lys-Glu, and Glu-Glu interactions. The Arg-Glu interaction was stronger than Lys-Glu and the latter stronger than Glu-Glu, in agreement with observations by Folch et al.78 and experimental mutation data by Tissot et al.79 This is exactly what is observed in our simulations (note, however, that the statistical analysis does not discriminate between charged and uncharged Glu). Attractive interactions were also observed between two Arg, especially in stacking configuration 73,80, but they were not as strong as in our simulations. Lys pairs were repulsive with no well-defined CP73, whereas we observe slightly attractive interactions. Strong interactions were displayed by His side chains in all orientations. Comparison with our simulations is complicated by the unclear protonation states in the statistical analysis, but, in any case, the interactions in the simulations are less strong than those deduced by Mukherjee et al.73, partly due to the influence of metals that many His were coordinating. Marsili et al.81, who computed two-dimensional PMFs from analysis of protein structures, also found attractive interactions between Arg-Arg, His-His, and Arg-His pairs and a preference for stacking orientation, in agreement with our simulations. However, the homomeric and heteromeric interactions between Arg and His were found to be of similar strength, whereas in our simulations the Arg-Arg pair was more attractive than His-His and Arg-His. The Glu-His interaction was weaker in their statistical analysis than in our simulations.
Although a simulation study obtained values of dielectric permittivity in the headgroup region larger than water 82, it is generally accepted that the membrane interface has lower polarity than water 83. Thus, from simple electrostatic arguments one would expect the unlike-charged/H-bonded side chains to be more attractive and the like-charged ones to be more repulsive in the lipid bilayer than in water. Our results do not always conform to this prediction. For example, the Arg-Glu pair is less stable in the bilayer than in water. Even more surprising is the observation of attractive interactions between like-charged side chains becoming stronger in the bilayer. It seems, therefore, that the PMFs are modulated by specific interactions with bilayer components. Below we will attempt to identify such interactions by analyzing interaction energies and hydrogen bonds of the side chains with water and lipid headgroups.
We start with the like-charged interactions, the stronger of which was observed for two stacked Arg (Figure 4). Classical simulations 17,20,84,85, ab initio calculations86,87 and protein structure surveys 17,73,78,88 have found an attraction between Arg side chains. This is attributable to a favorable solvation of an Arg pair by water overcoming the repulsive electrostatic interaction between them. Similar arguments have been used to explain the association of like-charged organic ions in solution or in crystals89,90. An even stronger attraction is observed here in the lipid bilayer interface. It is reasonable to hypothesize that interactions with lipid headgroups, presumably the negatively charged phosphate groups, are involved in overcoming the electrostatic repulsion. Figure S11 shows that a significant contribution to the stacking CM comes from Lennard-Jones interactions between the side chains. The LJ contribution is also substantial for the orthogonal orientation, but is apparently countered by other terms (Figure S11) and fails to create a CM. Side chain interactions with water favor association for all three orientations (Figure S11). Hydrogen bonding to the headgroups is maintained upon approach of the side chains in the stacking orientation, whereas it diminishes in the other orientations (Figure S10). A typical snapshot from the simulations for the Arg pair at the stacking CM (Figure 9) shows hydrogen bonds to the phosphate oxygens. H-bonding to water also slightly increases in the stacking orientation, whereas it decreases for the orthogonal orientation. The stronger interaction in the bilayer vs water can be explained by relative interaction energy: although the side chain-water interactions are, as expected, smaller in magnitude in the bilayer (Figure S11 vs S12), they increase in magnitude upon association more strongly in the bilayer (Figure S13B).
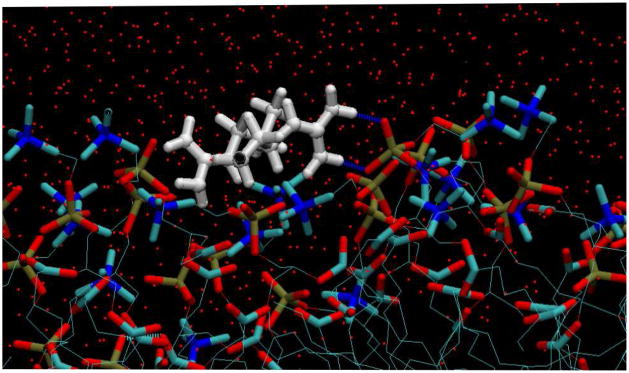
Figure 9.
Snapshot from simulations of stacking Arg+…Arg+ pair in POPC (view from the side of bilayer). Side chains are shown in white ball-and sticks. Lipids are shown in sticks (red-O, blue-N, brown-P, turquoise-C,), water molecules are shown in thin red dots. Blue dotted lines represent H-bonds between side chains and lipids, green dotted lines are H-bonds within the lipid interface.
The analysis of deprotonated Arg allows us to isolate the effect of charge. In the stacking orientation, neutralization of the two Arg reduces the depth of the CM by about 2 kcal/mol. Now the direct interactions between the two side chains are favorable and contribute about 4 kcal/mol, 1 kcal/mol of which is LJ (Figure S16A, D). Interactions with water are no longer favorable to association but those with headgroups are (Figure S16). The attractive minimum for the collinear orientation is more difficult to understand as one can find no favorable contributions in the analysis. It could perhaps be attributed to entropic (packing) effects.
Similar trends were observed for charged His pairs (Figure 6). The less attractive interaction free energy for stacking His+…His+ compared to stacking Arg+…Arg+ is largely due to lower strengthening of interactions with water (Figure S20B vs S13B) upon approach. The direct LJ interactions are about twice as strong for the stacking His pair compared to the orthogonal one (Figure S20D), whereas they are equally strong for two Arg (Figure S13D). Like Arg-Arg, the His-His stacking interaction is stronger in the bilayer than in water, although the difference is smaller than for Arg-Arg. Here the change in side chain-water interactions upon association are similar in the bilayer and bulk water (Figure S20B) but the side chain-headgroup interactions are slightly attractive (Figure S20C), whereas for Arg-Arg they are repulsive (Figure S13C).
The stacking Arg+…His+ pair differs from the above in that it is less attractive in the lipid bilayer than in water (Figure 7). The critical term seems to be side chain-water interactions, which for this pair at the CM are somewhat weaker in the bilayer than in water (Figure S24B). Comparing this term for the stacking Arg+…Arg+, His+…His+, and Arg+…His+ pairs (Figures S13B, S20B, S24B) we observe that water favors association in all three, but to a greater extent for the Arg pair. The side chain-headgroup interaction exhibits the opposite trend (Figures S13C, S20C, S24C). The LJ interaction between side chains makes similar contributions in all three cases (Figures S13D, S20D, S24D).
Comparing collinear like-charged pairs, we see that Lys+…Lys+ exhibits a deeper CM than Arg+…Arg+ and Arg+…Lys+. Interactions with headgroups seem to play a significant role here: Lys+…Lys+ exhibits two H-bonds to the headgroups at CM (Figure S29C) compared to one for Arg+…Arg+ and none for Lys+…Arg+. In addition, only for this pair the interaction energy with headgroups has a deep minimum at CM favoring association (Figure S32C). Collinear Lys+…His+ and Lys+…Arg+ pairs displayed almost identical PMF profiles (Figure S25A, S29A). Considering relative interaction energy values between side chains, side chain-water and side chain-headgroups (Figure S28, S32), the absolute value of interaction energy, after summation of all the components, comes out to be the same for two pairs
The interaction free energy between anionic side chains is much more modest than that between cationic ones (Figure 8). Hydrogen bonding to the headgroups is minimal (Figure S33) and the side chain-headgroup interaction energy is positive throughout (Figure S35). This is because the positive charge of the choline N is sterically inaccessible due to the methyl groups and the most accessible lipid atoms have a negative partial charge. The LJ interaction between the side chains is more favorable for the orthogonal orientation (Figure S35, S36D) and is largely responsible for the more attractive CM for this orientation. However, the interaction with water (Figure S36B) is stronger for the collinear orientation at short distances, which explains the CM at a shorter distance compared to orthogonal orientation.
Among the unlike-charged side chain pairs in collinear orientation, two (Lys+…Glu− and His+…Glu−) displayed more stable CP in the bilayer than in water, as one would expect from dielectric arguments. However, the Arg+…Glu− pair showed the opposite behavior. We note that for the former pair interactions with water are more repulsive in the bilayer interface than in pure water (Figure S3B), whereas the opposite is observed for the latter pairs (S6B, S9B). Also, the side chain-headgroup interactions for this Arg+…Glu− pair are positive and disfavor association (Figure S3C). A different behavior is observed for Lys+…Glu− (Figure S6C) and especially His+…Glu− (S9C). The Lys+…Glu− pair in collinear approach displayed a more shallow CM and a less structured PMF than the other two unlike-charged pairs. This is likely due to the fact that the constraints for the Lys+…Glu− this pair in collinear arrangement do not allow the formation of H-bonds (Figure S4). It is interesting that for the Arg-Glu pair the water-side chain LJ interaction follows quite closely the shape of the PMF (Figure S2D). This is less so for the other two unlike-charged pairs but is observed for some neutralized His and Glu pairs. (Figures S18, S19, S34). It may be that this term captures packing effects more clearly than others and may thus act as a “proxy” for entropic effects. It is interesting that in the case of unlike charged or neutral side chains the side chains have similar interactions with water whether simulated in POPC or water box, as can be seen from the number of H-bonds and their interactions with water (e.g. Figure S1 and S2).
In general, neutralization of one or both side chains destabilizes the PMF for unlike-charged pairs (His-Glu, Figure 3) and stabilizes it for like-charged pairs (His-His, Figure 6, Glu-Glu, Figure 8, Lys-His Figure S25A), as expected. The more attractive interaction free energy between His+ and Glu− compared to His0…Glu− in POPC originates from the stronger direct interactions (Figure S9A) and more favorable interactions with the headgroups (Figure S9C) which are not fully compensated by interactions with water (Figure S9A-B). There is a difference in the behavior of Glu-Glu vs. His-His. For the former the neutral-neutral interaction is stronger than the neutral-charged interaction, whereas for the latter the neutral-charged interaction is the strongest. This is likely due to the possibility of forming two H-bonds at contact in the neutral Glu pair. Indeed, the direct interaction is 1.5 kcal/mol more favorable and the side chain-water interaction less repulsive for the neutral than the neutral-charged Glu pair (Figure S36A and B). Different behavior is observed for the His pair (Figure S20). The Glu−…Glu0 pair forms a stable CP in water but not in the lipid bilayer (Figure 8). This can be explained by considering the interaction energy with the headgroups (Figure S34), which exhibits a minimum at 4.5 Å, longer than the CM, and it increases upon further approach. This apparently destabilizes the clear CM observed in water.
Conclusions
We calculated PMFs between 29 pairs of ionizable side chains in different orientations in the lipid bilayer-water interface. Figure 10 shows all the calculated PMFs grouped according to orientation. Pairs in collinear orientation showed similar PMF profiles with stable attractive contact pair occurring at a short distance, with high activation barrier, and well defined solvent separated minima. Hydrogen-bonded pairs in stacking orientations displayed stable contact pairs at longer distances and no solvent-separated minima. The most attractive CPs between like-charged side chains were obtained for stacked configurations, followed by orthogonal and collinear orientations. For unlike-charged and neutral side chains the orientational differences in the PMF were predominantly a result of H-bond formation. The source of variation is more complex for like-charged side chains. Interactions with headgroups and water play a significant role and, in some cases, LJ interactions between the side chains. The results are in agreement with experimental relative strength of interactions obtained from double mutant cycles and statistical analyses of protein structures. These data could help in the development of improved implicit membrane potentials.
Figure 10.
Comparative plots of PMF curves for side chain pairs in POPC.
The most surprising observation is that the attractive interactions between like-charged amino acids, which have been reported in several previous studies in water, are in some cases stronger in the lipid interface. The interaction is especially strong for a pair of Arg in a stacked configuration. Some of the attraction in this case comes from LJ interactions and the rest from interactions with water which are more favorable in the associated pair. Side chain-headgroup interactions also contribute for other like-charged pairs. When the Arg gets deprotonated the attraction is diminished and the contributions to the PMF change: now side chain-water interactions disfavor association and the attraction is driven by the direct interactions between the side chains.
The interaction free energies of ionizable side chains cannot be explained by simple dielectric arguments. First, the observation of attractive interactions between like-charged ions is incompatible with a screened Coulomb potential. Second, some interactions become weaker and some stronger in the bilayer interface, in a way that cannot be described by a simple change in the value of the effective dielectric constant. Interactions with lipid groups and the way the presence of these groups affects interactions with water seem to play a crucial role.
The results have interesting implications for membrane protein stability and oligomerization, the principles of which are currently poorly understood. Most emphasis so far has been placed on polar and packing interactions in the membrane interior. However, these have been found to make similar contributions in membrane and soluble proteins. 91,92 The present work suggests that ion pairs at the membrane interface, especially some counterintuitive ones, could substantially stabilize membrane protein structure. Such contributions at the interface have been established for aromatic residue pairs93 but have not been explored for ionizable sidechains. Interestingly, the knowledge-based pair score derived by the Baker group for the interface and water-exposed region of membrane proteins (Figure S3 of Ref. 49) shows mild attractions between Arg residues and even stronger attractions between His residues. There are also strong attractions in the membrane interior. It would be interesting to conduct a more detailed survey of currently available structures focusing on the interface region.
The results could also have implications for peptide-membrane interactions and membrane permeabilization. Clusters of Arg have been found in voltage sensors of ion channels94, antimicrobial95 and cell-penetrating96 peptides. Of course, our results may not always apply to side chains that are attached to a common peptide backbone. If they do, they could imply that favorable intramolecular interactions between like-charged side chains could stabilize compact conformations of these peptides. In addition, they could contribute to the aggregation of these peptides which, at least in the case of antimicrobial peptides, seems to play a role in membrane permeabilization97.
Finally, being zwitterionic, the headgroup area of POPC resembles to some extent a hydrated ionic liquid. Thus, the results could also have implications for such liquids. Some PMFs of ion association in ionic liquids have already been reported98 more calculations are needed to characterize their behavior.
Supplementary Material
Acknowledgments
The authors thank Maja Mihajlovic for useful discussions. This research was supported by the National Science Foundation (MCB-0615552) and in part by a grant of computer time from the City University of New York’s High Performance Computing Research Center. Infrastructure support was provided in part by RCMI grant RR03060 from NIH.
Abbreviations
- PMF
potential of mean force
- CP
contact pair
- CM
contact minimum
- SSM
solvent-separated minimum
- ABF
adaptive biasing force
- MD
molecular dynamics
- PME
particle-mesh Ewald
- POPC
palmitoyl oleoyl phosphatidylcholine
- LJ
Lennard-Jones
Footnotes
Supporting Information Available: PMF profiles, H-bonds, total absolute and relative interaction energies and Lennard-Jones energy from ABF simulation trajectories. This material is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Perutz MF. Science. 1978;201:1187–1191. doi: 10.1126/science.694508. [DOI] [PubMed] [Google Scholar]
- 2.Warshel A. Acc Chem Res. 1981;14:284–290. [Google Scholar]
- 3.Sharp KA, Honig B. Annu Rev Biophys Biophys Chem. 1990;19:301–332. doi: 10.1146/annurev.bb.19.060190.001505. [DOI] [PubMed] [Google Scholar]
- 4.Warshel A, Sharma PK, Kato M, Parson WW. Biochim Biophys Acta. 2006;1764:1647–1676. doi: 10.1016/j.bbapap.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Strickler SS, Gribenko AV, Gribenk AV, Keiffer TR, Tomlinson J, Reihle T, Loladze VV, Makhatadze GI. Biochemistry. 2006;45:2761–2766. doi: 10.1021/bi0600143. [DOI] [PubMed] [Google Scholar]
- 6.Horovitz A, Fersht AR. J Mol Biol. 1992;224:733–740. doi: 10.1016/0022-2836(92)90557-z. [DOI] [PubMed] [Google Scholar]
- 7.Marqusee S, Sauer RT. Protein Sci. 1994;3:2217–2225. doi: 10.1002/pro.5560031207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pervushin K, Billeter M, Siegal G, Wuthrich K. Mol Biol. 1996;264:1002–1012. doi: 10.1006/jmbi.1996.0692. [DOI] [PubMed] [Google Scholar]
- 9.Kumar S, Nussinov R. Biophys J. 2002;83:1595 – 1612. doi: 10.1016/S0006-3495(02)73929-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh UC. Proc Natl Acad Sci USA. 1988;85:4280–4284. doi: 10.1073/pnas.85.12.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serrano L, Horovitz A, Avron B, Bycroft M, Fersht AR. Biochemistry. 1990;29:9343–9352. doi: 10.1021/bi00492a006. [DOI] [PubMed] [Google Scholar]
- 12.Barril X, Aleman C, Orozco M, Luque FJ. Proteins: Struct Funct Genet. 1998;32:67–79. doi: 10.1002/(sici)1097-0134(19980701)32:1<67::aid-prot8>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 13.Sun DP, Sauer U, Nicholson H, Matthews BW. Biochemistry. 1991;30:7142–7153. doi: 10.1021/bi00243a015. [DOI] [PubMed] [Google Scholar]
- 14.Dao-Pin S, Anderson DE, Baase WA, Dahlquist FW, Matthews BW. Biochemistry. 1991;30:11521–11529. doi: 10.1021/bi00113a006. [DOI] [PubMed] [Google Scholar]
- 15.Hendsch ZS, Tidor B. Protein Sci. 1994;3:211–226. doi: 10.1002/pro.5560030206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waldburger CD, Schildbach JF, Sauer RT. Nature Struct Biol. 1995;2:122–128. doi: 10.1038/nsb0295-122. [DOI] [PubMed] [Google Scholar]
- 17.Soetens JC, Millot C, Chipot C, Jansen G, Ángyá JG, Maigret B. J Phys Chem. 1997;101:10910–10917. [Google Scholar]
- 18.Rozanska X, Chipot C. J Chem Phys. 2000;112:9691–9694. [Google Scholar]
- 19.Hénin J, Chipot C. J Chem Phys. 2004;121:2904–2914. doi: 10.1063/1.1773132. [DOI] [PubMed] [Google Scholar]
- 20.Masunov A, Lazaridis T. J Am Chem Soc. 2003;125:1722–1730. doi: 10.1021/ja025521w. [DOI] [PubMed] [Google Scholar]
- 21.Maksimiak K, Rodziewicz-Motowidlo S, Czaplewski C, Liwo A, Scheraga HA. J Phys Chem. 2003;107:13496–13504. [Google Scholar]
- 22.Hassan SA. J Phys Chem B. 2004;108:19501–19509. [Google Scholar]
- 23.Hassan SA. J Chem Phys. 2005;109:21989–21996. doi: 10.1021/jp054042r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas AS, Elcock AH. J Am Chem Soc. 2006;128:7796–7806. doi: 10.1021/ja058637b. [DOI] [PubMed] [Google Scholar]
- 25.Mandell DJ, Chorny I, Groban ES, Wong SE, Levine E, Rapp CS, Jacobson MP. J Am Chem Soc. 2007;129:820–827. doi: 10.1021/ja063019w. [DOI] [PubMed] [Google Scholar]
- 26.Vaitheeswaran S, Thirumalai D. J Am Chem Soc. 2006;128:13490–13496. doi: 10.1021/ja063445h. [DOI] [PubMed] [Google Scholar]
- 27.Vaitheeswaran S, Thirumalai D. Proc Natl Acad Sci USA. 2008;105:17636–17641. doi: 10.1073/pnas.0803990105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chorny I, Dill KA, Jacobson MP. J Chem Phys B. 2005:24056–24060. doi: 10.1021/jp055043m. [DOI] [PubMed] [Google Scholar]
- 29.Geney R, Layten M, Gomperts R, Hornak V, Simmerling C. J Chem Theory Comput. 2006;2:115–127. doi: 10.1021/ct050183l. [DOI] [PubMed] [Google Scholar]
- 30.Mottamal M, Zhang J, Lazaridis T. Proteins. 2006;62:996–1009. doi: 10.1002/prot.20844. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J, Lazaridis T. Biophys J. 2009;96:4418–4427. doi: 10.1016/j.bpj.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L, Vorobyov I, Allen TW. J Phys Chem B. 2008;112:9574–9587. doi: 10.1021/jp7114912. [DOI] [PubMed] [Google Scholar]
- 33.Vorobyov I, Li L, Allen TW. J Phys Chem B. 2008;112:9588–9602. doi: 10.1021/jp711492h. [DOI] [PubMed] [Google Scholar]
- 34.Dorairaj S, Allen TW. Proc Natl Acad Sci USA. 2007;104:4943–4948. doi: 10.1073/pnas.0610470104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allen TW. J Gen Physiol. 2007;130:237–240. doi: 10.1085/jgp.200709850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacCallum JL, Bennett WF, Tieleman DP. Biophys J. 2008;94:3393–3404. doi: 10.1529/biophysj.107.112805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johansson AC, Lindahl E. Proteins. 2008;70:1332–1344. doi: 10.1002/prot.21629. [DOI] [PubMed] [Google Scholar]; Proteins. 2008;70:1655–1656. [Google Scholar]
- 38.Johansson AC, Lindahl E. J Phys Chem B. 2009;113:245–253. doi: 10.1021/jp8048873. [DOI] [PubMed] [Google Scholar]
- 39.Johansson AC, Lindahl E. J Chem Phys. 2009;130:185101. doi: 10.1063/1.3129863. [DOI] [PubMed] [Google Scholar]
- 40.Maccallum JL, Bennett WF, Tieleman DP. Biophys J. 2011;101:110–117. doi: 10.1016/j.bpj.2011.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Darve E, Pohorille A. J Chem Phys. 2001;115:6954–6960. [Google Scholar]
- 42.Darve E, Wilson M, Pohorille A. Mol Simul. 2001;28:113–144. [Google Scholar]
- 43.Lazaridis T. Proteins. 2005;52:176–192. doi: 10.1002/prot.10410. [DOI] [PubMed] [Google Scholar]
- 44.Ulmschneider MB, Sansom MSP, Di Nola A. Proteins. 2005;59:252–265. doi: 10.1002/prot.20334. [DOI] [PubMed] [Google Scholar]
- 45.Im W, Feig M, Brooks CL. Biophys J. 2003;85:2900–2918. doi: 10.1016/S0006-3495(03)74712-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanizaki S, Feig M. J Phys Chem B. 2006;110:548–556. doi: 10.1021/jp054694f. [DOI] [PubMed] [Google Scholar]
- 47.Tanizaki S, Feig M. J Chem Phys. 2005;122:124706. doi: 10.1063/1.1865992. [DOI] [PubMed] [Google Scholar]
- 48.Yarov-Yarovoy V, Baker D, Catterall WA. Proc Natl Acad Sci USA. 2006;103:7292–7297. doi: 10.1073/pnas.0602350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yarov-Yarovoy V, Schonbrun J, Baker D. Proteins. 2006;62:1010–1025. doi: 10.1002/prot.20817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lazaridis T. Proteins Struct Funct Genet. 2003;52:176–192. doi: 10.1002/prot.10410. [DOI] [PubMed] [Google Scholar]
- 51.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, et al. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 53.Feller S, MacKerell AD., Jr J Phys Chem B. 2000;104:7510–7515. [Google Scholar]
- 54.Li LB, Vorobyov IV, MacKerell ADJ, Allen TW. Biophys J. 2008;94:L11–L13. doi: 10.1529/biophysj.107.121566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Humphrey W, Dalke A, Schulten K. J Molec Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 56.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 57.Klauda JB, Kucerka N, Brooks BR, Pastor RW, Nagle JF. Biophys J. 2006;90:2796–2807. doi: 10.1529/biophysj.105.075697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feller SE, Zhang YH, Pastor RW, Brooks BR. J Chem Phys. 1995;103:4613–4621. [Google Scholar]
- 59.Feller SE, Zhang Y, Pastor RW, Brooks BR. J Chem Phys. 1995;103:4613–4621. [Google Scholar]
- 60.Ryckaert J, Cicotti G, Berendsen HJC. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 61.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pederson LA. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 62.Bogusz S, Cheatham TE, III, Brooks BR. J Chem Phys. 1998;108:7070–7084. [Google Scholar]
- 63.Hilder TA, Chung SH. Chem Phys Lett. 2011;501:423–426. [Google Scholar]
- 64.Hünenberger PH, McCammon JA. J Chem Phys. 1999;110:1856–1872. [Google Scholar]
- 65.Weber W, Hünenberger PH, McCammon JA. J Phys Chem B. 2000;104:3668–3675. [Google Scholar]
- 66.Reif MM, Kräutler V, Kastenholz MA, Daura X, Hünenberger PH. J Phys Chem B. 2009;113:3112–3128. doi: 10.1021/jp807421a. [DOI] [PubMed] [Google Scholar]
- 67.Hünenberger PH, McCammon JA. Biophys Chem. 1999;78:69–88. doi: 10.1016/s0301-4622(99)00007-1. [DOI] [PubMed] [Google Scholar]
- 68.Kastenholz M, Hünenberger PH. J Phys Chem B. 2004;108:774–788. [Google Scholar]
- 69.Hünenberger PH, McCammon JA. J Chem Phys. 1999;110:1856–1872. [Google Scholar]
- 70.Feller S, MacKerell AD., Jr J Phys Chem B. 2000;104:7510–7515. [Google Scholar]
- 71.Henin J, Chipot C. J Chem Phys. 2004;127:2904–2914. doi: 10.1063/1.1773132. [DOI] [PubMed] [Google Scholar]
- 72.Darve E. Thermodynamic Integration Using Constrained and Unconstrained Dynamics. In: Chipot C, Pohorille A, editors. Free Energy Calculations: Theory and Applications in Chemistry and Biology. Springer-Verlag; Berlin Heidelberg: 2007. pp. 119–170. [Google Scholar]
- 73.Mukherjee A, Bagchi B. Biochem. 2006;45:5129–5139. doi: 10.1021/bi0522899. [DOI] [PubMed] [Google Scholar]
- 74.Heyda J, Mason PE, Jungwirth P. J Phys Chem B. 2010;114:8744–8749. doi: 10.1021/jp101031v. [DOI] [PubMed] [Google Scholar]
- 75.Neria E, Karplus M. J Chem Phys. 1996;105:1902–1921. [Google Scholar]
- 76.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. J Phys Chem B. 2001;105:6474–6487. [Google Scholar]
- 77.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P. J Comp Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 78.Folch B, Rooman M, Dehouck Y. J Chem Inf Model. 2008;48:119–127. doi: 10.1021/ci700237g. [DOI] [PubMed] [Google Scholar]
- 79.Tissot AC, Vuilleumier S, Fersht AR. Biochemistry. 1996;35:6786–6794. doi: 10.1021/bi952930e. [DOI] [PubMed] [Google Scholar]
- 80.Pednekar D, Tendulkar A, Durani S. Proteins. 2009;74:155–163. doi: 10.1002/prot.22142. [DOI] [PubMed] [Google Scholar]
- 81.Marsili S, Chelli R, Schettine V, Procacci P. Phys Chem Chem Phys. 2008;10:2673–2685. doi: 10.1039/b718519g. [DOI] [PubMed] [Google Scholar]
- 82.Raudino A, Mauzerall D. Biophys J. 1986;50:441–449. doi: 10.1016/S0006-3495(86)83480-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stern HA, Feller SE. J Chem Phys. 2003;118:3401–3412. [Google Scholar]
- 84.Boudon S, Wipff G, Maigret B. J Phys Chem. 1990;94:6056–6061. [Google Scholar]
- 85.Keasler SJ, Nellas RB, Chen B. J Chem Phys. 2006;125:144520–1–5. doi: 10.1063/1.2338316. [DOI] [PubMed] [Google Scholar]
- 86.Vondrášek J, Mason PE, Heyda J, Collins KD, Jungwirth P. J Phys Chem B. 2009;113:9041–9045. doi: 10.1021/jp902377q. [DOI] [PubMed] [Google Scholar]
- 87.No KT, Nam K-Y, Scheraga HA. J Am Chem Soc. 1997;119:12917–12922. [Google Scholar]
- 88.Magalhaes A, Maigret B, Hoflack J, Gomes JN, Scheraga HA. J Protein Chem. 1994;13:195–215. doi: 10.1007/BF01891978. [DOI] [PubMed] [Google Scholar]
- 89.Garcija-Yolgi I, Miller JS, Novoa JJ. J Phys Chem A. 2009;113:484–492. doi: 10.1021/jp807022h. [DOI] [PubMed] [Google Scholar]
- 90.Carvajal MA, Garcija-Yoldi I, Novoa JJ. J Mol Struct Theochem. 2005;727:181–189. [Google Scholar]
- 91.Bowie JU. Curr Opin Struct Biol. 2011;21:42–49. doi: 10.1016/j.sbi.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Joh NH, Oberai A, Yang D, Whitelegge JP, Bowie JU. J Am Chem Soc. 2009;131:10846–10847. doi: 10.1021/ja904711k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hong H, Park S, Flores-Jiménez RH, Rinehart D, Tamm LK. J Am Chem Soc. 2007;129:8320–8327. doi: 10.1021/ja068849o. [DOI] [PubMed] [Google Scholar]
- 94.Krepkiy D, Mihailescu M, Freites JA, Schow EW, Worcester DL, Gawrisch K, Tobias DJ, White SH, Swartz KJ. Nature. 2009;426:473–479. doi: 10.1038/nature08542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chan DI, Prenner EJ, Vogel HJ. Biochim Biophys Acta. 2006;1758:1184–1202. doi: 10.1016/j.bbamem.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 96.Schmidt N, Mishra A, Hwee Lai G, Wong GCL. FEBS Letters. 2010;584:1806–1813. doi: 10.1016/j.febslet.2009.11.046. [DOI] [PubMed] [Google Scholar]
- 97.Tossi A, Sandri L, Giangaspero A. Biopolimers. 2000;55:4–30. doi: 10.1002/1097-0282(2000)55:1<4::AID-BIP30>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 98.Lynden-Bell RM. Phys Chem Chem Phys. 2010;12:1733–1740. doi: 10.1039/b916987c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.