Abstract
Hepatitis C virus (HCV) replication in primary liver cells is less robust than that in hepatoma cell lines, suggesting that innate antiviral mechanisms in primary cells may limit HCV replication or spread. Here, we analyzed expression of 47 genes associated with interferon (IFN) induction and signaling following HCV infection of primary human fetal liver cell (HFLC) cultures from 18 different donors. We report that cell culture-produced HCV (HCVcc) induced expression of Type III (λ) IFNs and of IFN-stimulated genes (ISGs). Little expression of Type I IFNs was detected. Levels of IFNλ and ISG induction varied among donors and, often, between adapted and non-adapted HCV chimeric constructs. Higher levels of viral replication were associated with greater induction of ISGs and of λ IFNs. Gene induction was dependent on HCV replication, as UV-inactivated virus was not stimulatory and an antiviral drug, 2′-C-methyladenosine, reduced induction of λ IFNs and ISGs. The level of IFNλ protein induced was sufficient to inhibit HCVcc infection of naïve cultures.
Conclusion
Together, these results indicate that despite its reported abilities to blunt the induction of an IFN response, HCV infection is capable of inducing antiviral cytokines and pathways in primary liver cell cultures. Induction of ISGs and λ IFNs may limit the growth and spread of HCV in primary cell cultures and in the infected liver. HCV infection of HFLC may provide a useful model for the study of gene induction by HCV in vivo.
Keywords: Hepacivirus, Type III IFN, Type I IFN, hepatocyte, RT-PCR
Between 130 and 170 million persons worldwide are persistently infected with hepatitis C virus (HCV) (1). Chronic HCV infection can lead to cirrhosis, hepatocellular carcinoma, and liver failure, and is a leading indication for liver transplant. HCV persists in part because of failures in innate and immune-mediated control of viral replication and spread (2, 3). A better understanding of HCV-host interactions is urgently needed in order to develop improved therapies. Our ability to study HCV-host-virus interactions has been hampered by the lack of robust in vitro systems that faithfully replicate viral growth and spread in the liver. Until recently, most studies of HCV infection and replication have been performed in derivatives of a hepatoma cell line (Huh-7) that were selected for their ability to support high-level HCV replication in culture. These transformed cells may not reproduce the more complex environment in the infected liver. Indeed, they may work so well for in vitro studies because of defects in innate antiviral pathways (4, 5). Furthermore, studies based on Huh-7 derivatives may not address the impact of host genetic diversity on the outcome of HCV infection.
Using cell lines, many groups have reported that HCV infection can block innate antiviral mechanisms within infected cells (reviewed in (3)). The NS3-4A protease of HCV cleaves the signaling adapter IPS-1 (6–8), and it is believed that this cleavage prevents IPS-1-dependent signal transduction needed to stimulate IFNα/β expression. Furthermore, NS3-4A can cleave the adapter TRIF and thereby interfere with signaling by TLR3, an endosomal dsRNA receptor (9). In a hepatoma line, HCV infection induced transient IRF-3 activation, but the activation was not sustained (10). In HEK293 kidney cells, ectopic NS3-4A expression reduced virus-stimulated activation of IFN (α, β, λ) and chemokine promoters, suggesting that HCV-infected cells might be deficient in IFN induction and response (11). Much less is known about the degree to which HCV influences innate antiviral mechanisms within the infected liver. In contrast to the robust spread of cell culture-derived HCV in cell lines, primary liver cells support lower levels of HCV infection in vitro (12) and in vivo (13, 14). Furthermore, IFN-stimulated genes (ISGs) are highly expressed in liver biopsies during acute (13, 15) and chronic HCV infection (14, 16, 17), even in tissues lacking detectable IFNα/β expression (13, 16–18). ISG expression may produce an antiviral state restricting HCV spread in vivo (14, 19, 20). The cellular source of IFNs in the HCV-infected liver has not been identified. Also important, many of the hundreds of genes known as ISGs are directly inducible by viral proteins and nucleic acids in the absence of IFN (21).
Genome-wide association studies have revealed important roles for the λ IFNs in control of HCV infection and responses to antiviral therapy (reviewed in (22)). There are three human IFNλ: IL29 (IFNλ1), IL28A (IFNλ2), and IL28B (IFNλ3). IL28A and IL28B are closely related in sequence and promoter structures. Analysis of the IL29 and IL28A/B promoters suggests that these genes are differentially regulated, with IL29 controlled in a manner similar to IFNβ, and IL28A/B control more similar to IFNα (23). All three IFNλ are inducible by viral infection (24). Hepatocytes, including those in the HCV-infected liver, express the IFNλ receptor (25). IFNλ activates a transcriptional program resembling that of IFNα/β in liver cells, and inhibits HCV replication in cell lines as well as primary fetal liver cells (25–29).
Recent advances in the field have included the development of cell culture-adapted HCV (HCVcc) chimeras with useful properties (30–34), and of cell-based reporters permitting real-time visualization of HCV infection (35). New systems permit prolonged culture of untransformed human hepatocytes that retain their differentiated phenotype and HCV susceptibility over weeks in vitro (12) (Andrus et al, submitted). Here we have taken advantage of these advances to evaluate the impact of HCV infection on gene expression in primary human fetal liver cell (HFLC) cultures. Our results demonstrate that HCV infection activates expression of IFNλ and a number of ISGs. Both viral and host factors influence the level of gene induction. IFNλ levels produced following HCV infection are sufficient to inhibit HCV replication in primary liver cells, and an inhibitor of IFN receptor signaling enhances HCV replication even in the absence of added IFNs. These results suggest that endogenous IFNs may limit the spread of HCV in primary liver cells.
Materials and Methods
Human fetal liver cells, HFLC
The preparation of HFLC is detailed in the accompanying paper by Andrus et al. Briefly, de-identified human fetal liver tissue (16–22 weeks gestation) was procured through Advanced Bioscience Resources (Alameda, CA) or the human fetal tissue repository of the Albert Einstein College of Medicine (New York, NY). The Rockefeller University Institutional Review Board exempted this use of fetal liver tissue from review. Fetal gestational ages are listed in Table S1. Liver cells were isolated by collagenase and DNAse I digestion, and hepatoblasts enriched relative to hematopoietic cells and other small cell populations by a series of low-speed spins and sedimentation at 1×g. For some experiments, hepatoblasts were further enriched by centrifugation through lymphocyte separation medium (Cellgro, Manassas, VA) as described (36). Hepatoblasts were plated at 1×105/cm2 in 24- or 48-well collagen I-coated plates (Biocoat-I, BD Biosciences, Bedford, MA), cultured overnight, and washed extensively to remove non-adherent cells. After overnight plating, cultures contained predominantly cells with hepatoblast morphology. Cultures were maintained in Hepatocyte Culture Medium (BD Biosciences) with medium aspiration and replacement every 2 days (or less, as shown for each experiment).
Viruses
We used two chimeric HCVcc constructs, each encoding a fully infectious HCV (genotype 2a) with different advantages. Jc1FLAG2(p7nsGLuc-2A) (Jc1G) (33) is a J6/JFH chimeric genome encoding a Gaussia princeps luciferase (GLuc) reporter between p7 and NS2. Infected cells secrete GLuc, allowing measurement of viral replication by sampling the supernatant. Clone 2 (34) is a J6/JFH chimeric genome selected by serial passage in Huh-7.5 cells. This HCVcc bears twelve mutations that increase infectious titers, and does not include a reporter. Infectious titers of both Jc1G and Clone 2 were determined by titration on Huh-7.5 cells using the TCID50 method (37).
HCV-dependent fluorescent reporter (HDFR)
The construction of a modified HDFR with hepatocyte-specific expression is detailed in the Supplemental Information.
Monitoring infection in HFLC
On the third or fourth day after plating, HFLC (1×105/cm2) were infected with 3×106 TCID50 units/well of HCVcc. Inocula were left in place for 6 hours and then removed with three washes (500 μl) of Williams E media without serum. Jc1G replication was monitored by measurement of secreted GLuc (33), quantitation of HCV RNA using the EraGen MultiCode-RTx method (EraGen Biosciences, Madison, WI) (38), and assessment of HDFR translocation (35). Because HCVcc Clone 2 does not encode a reporter, we used RNA quantitation and HDFR translocation to monitor Clone 2 infection.
Quantitative RT-PCR
RNA was prepared using the Qiagen RNeasy mini kit (Qiagen, Valencia, CA) and yields determined by absorption spectroscopy using a NanoDrop (NanoDrop Products, Wilmington, DE). Five μl of RNA was used as a template for reverse transcription using SuperScript III (Invitrogen, Carlsbad, CA) and random hexamers. Five μl of tenfold-diluted cDNA was used as in a SYBRR Green qPCR assay (Applied Biosystems, Carlsbad, CA) on the LightCyclerR 480 Real-Time PCR System (Roche Applied Sciences, Indianapolis, IN). Primer pairs were selected using the Harvard Medical School Center for Computational and Integrative Biology Primer Bank website (http://pga.mgh.harvard.edu/primerbank/), and are listed in Supplemental Table 2. Expression was normalized to that of the housekeeping gene RPS11 and to expression in uninfected cultures from the same donor at the same time point.
UV inactivation
Five ml of virus, in one well of a six-well plate, was placed on a 1.5-inch platform and exposed to 82 mJoule of UV light (GS Gene Linker, Bio-Rad, Hercules, CA). UV-inactivated virus was non-infectious as determined by titration on naïve Huh-7.5 cells.
Drugs and cytokines
2′C-methyladenosine (2′CMA), an inhibitor of HCV’s NS5B polymerase (39), was generously provided by Drs. D. Olsen and S. Carroll, Merck Research Laboratories (West Point, PA) and used at 2.5 μM. This is >80X the previously reported IC50 for this drug using J6/JFH HCVcc and Huh-7.5 cells (30); however, for unknown reasons, 2′CMA is less effective in HFLC than in Huh-7.5 cells. Pyridone-6 (Calbiochem, San Diego, CA), a pan-JAK inhibitor (40), was used at 1 μM. Pyridone-6 and 2′CMA were replaced at each media change. Recombinant human IFNβ (produced in CHO cells), IFNλ1 and IFNλ2 (produced in E. coli) were purchased from PeproTech (Rocky Hill, NJ). Cytokine endotoxin levels were <1EU/μg. Polyinosinic:polycytidylic acid (p(I:C)) was purchased from GE Healthcare (Piscataway, NJ) and used at 5 μg/ml.
Protein measurement
IL29 was measured by ELISA (eBioscience, San Diego, CA) according to the manufacturer’s instructions. The linear range of this assay was 31–1000 pg/ml. CXCL10 and CXCL11 ELISAs (R&D Systems, Minneapolis, MN) were performed according to the manufacturer’s instructions. The linear ranges of these assays were 15–500 pg/ml (CXCL10) and 125–4000 pg/ml (CXCL11).
IL28B SNP genotyping
The IL28B rs12979860 SNP (41) was genotyped by sequencing following PCR amplification of a 429-bp region using the following primers: forward, 5′-cgcttatcgcatacggcta-3′; reverse, 5′-gggaccgctacgtaagtcac-3′.
Data analysis
Real-time PCR data were acquired on the Roche Light Cycler 480 (software release 1.5.0 SP3) and analyzed using the 2−ΔΔC(T) method (42). HCV RNA was quantitated with MultiCode-RTx Analysis Software-v1.1.5d from EraGen. Graphing and statistical analysis were performed with Prism 5.0d (GraphPad Software, San Diego, CA). All data are presented as the mean±SD of duplicate cultures.
Results
HCVcc infection in HFLC cultures
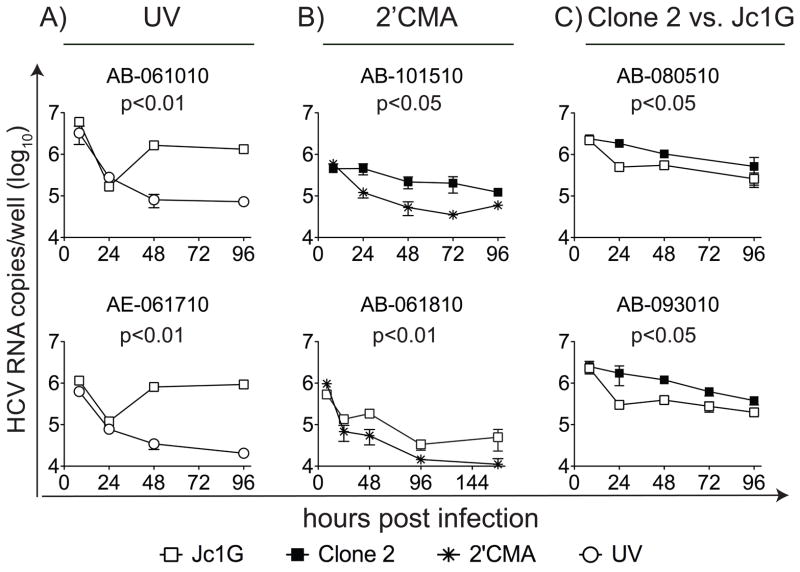
We used two chimeric HCV viruses to monitor HFLC infection: Clone 2 (34) and Jc1G (33). Limited and variable replication of both viruses was detected over 24–96 hours (Figure 1). UV irradiation of the virus inoculum blocked replication without significantly reducing amounts of input viral RNA (Figure 1A). Replication was sensitive to the nucleoside analog 2′CMA, an inhibitor of the HCV RNA-dependent RNA polymerase NS5B (Figure 1B). In side-by-side comparisons, Clone 2 RNA levels were often significantly higher than those of Jc1G (p <0.05, repeated measures ANOVA) (Figure 1C). HCV replication was also demonstrated by nuclear translocation of the HDFR (35) in infected cells (Figure S1A–B). Using the HDFR system, we confirmed that Clone 2 infection of HFLC is more robust than Jc1G, and that inhibiting replication with 2′CMA reduced the signal from the reporter. Replication of Jc1G could also be demonstrated by measurement of secreted GLuc (Figure S1C; also see the accompanying manuscript by Andrus et al.).
Figure 1. HCV infection in HFLC cultures from six donors.
The HFLC source is indicated above each graph; see Table S1 for additional information. At the indicated time points, cultures were harvested for measurement of HCV RNA. HCV RNA is indicated in copies/culture (mean ± SD) on a log10 scale. (A), HFLC cultured with infectious or UV-inactivated Jc1G. (B), HFLC cultured with HCVcc clone 2 (top) or Jc1G (bottom) ± 2′CMA. (C), Comparing HCVcc Jc1G and Clone 2 replication in HFLC from the same donors. Data shown are representative of 18 experiments with similar results. Values of P were determined by repeated measures ANOVA, and are indicated above each graph.
Gene induction in HCVcc-infected HFLC cultures
HCVcc replication in HFLC was less robust than in hepatoma cell lines. Additionally, as reported in the accompanying manuscript by Andrus et al., paramyxovirus V proteins that interfere with IFN induction and signaling enhanced HCV replication in HFLC. Therefore, we hypothesized that innate antiviral mechanisms in primary cells limited HCV replication and/or spread. We used quantitative RT-PCR approaches to measure expression of 47 genes associated with IFN signaling and innate antiviral responses (Supplemental Table 2) in HCVcc-infected HFLC. HCVcc infection stimulated the expression of many of these genes, notably including IL29, certain chemokines, and several ISGs (Table 1). The experiments shown represent the two patterns of gene expression observed when comparing Clone 2-stimulated to Jc1G-stimulated gene expression in cells from the same liver: greater gene induction by Clone 2 than by Jc1G (three of four experiments), and similar gene induction by both viruses (one of four experiments). Time courses for induction of selected genes by both Clone 2 and Jc1G are shown in Figure S2. The experiments shown compare Clone 2 and Jc1G-stimulated gene induction in HFLC from the same livers; mRNA levels are compared to uninfected control cultures for each time point. IL29 mRNA was transiently induced (Figure S2A). IL29 expression was more frequently detected than was IL28B expression, consistent with the differential control of these genes (23, 24). Robust increases in CXCL10, CXCL11 (Figure S2A), and ISGs such as Viperin, IFITM1, IFIT2, MX2, and OAS2 (Figure S2B) were observed within 24–48 hours of infection. Low-level IFNβ induction was observed in response to Clone 2 in two of 18 experiments; no induction of IFNα, IFNγ, or IFNω was observed in any experiment (out of 6 experiments). The synthetic dsRNA analog, p(I:C), stimulated expression of IFNs and ISGs in HFLC; exogenous IFNβ and IL29 each stimulated expression of ISGs (Figure S3). Gradient-enriched hepatoblasts prepared as described (36) demonstrated a further increase in HCV-stimulated gene expression (Figure S4A). After HCVcc infection, gradient-purified HFLC cultures were primed to respond to p(I:C) stimulation with increased expression of IL29, IL28B, IFNβ, and IFITM1 when compared to uninfected cultures (Figure S4B).
Table 1. Genes induced following HCV infection in HFLC.
Results of 4 analyses on two independent livers are shown. Liver AB-080510 and AE-102910 were each infected with both Clone 2 and Jc1G. Values shown represent the peak fold increase in mRNA levels for each gene, relative to uninfected cultures from the same liver at the same time point. See Figure S2 for kinetics of gene induction. Expression of IFNα1, IFNω, IFNγ, IL1β, IL6, IL8, MDK, MOV10, DDIT4, c15orf8, and c8orf4 was not detected at any time point. In three of four comparisons, the relative gene induction in response to Clone 2 was dramatically higher than that in response to Jc1G, as shown for AB-080510. In one of four comparisons (AE-102910, shown here), high levels of gene induction were observed in response to both Jc1G and Clone 2.
| Cytokines and chemokines | ||||
|---|---|---|---|---|
Liver
|
AB-080510 | AE-102910 | ||
| Clone 2 | Jc1G | Clone 2 | Jc1G | |
Gene

|
Mean (SD) | Mean (SD) | Mean (SD) | Mean (SD) |
| CXCL10 | 13127.6 (643.2) | 102.5 (23.9) | 17554.8 (4092.3) | 4482.6 (548.0) |
| CXCL11 | 7666.6 (488.2) | 59.3 (9.8) | 16527.0 (5177.9) | 3594.0 (35.2) |
| IL29 | 49.6 (10.1) | 1.3 (0.6) | 20.0 (5.0) | 14.2 (1.3) |
| IL28B | 45.1 (16.2) | 1.1 (0.2) | 7.8 (5.7) | 11.3 (0.7) |
| IFNβ | 4.7 (1.7) | 0.9 (0.7) | 1.4 (0.9) | 2.5 (1.0) |
| TNFƒα | 5.3 (1.8) | 1.2 (0.3) | 4.2 (2.0) | 1.7 (1.1) |
| Interferon stimulated genes | ||||
Liver
 |
AB-080510 | AE-102910 | ||
| Clone 2 | Jc1G | Clone 2 | Jc1G | |
Gene
 |
Mean (SD) | Mean (SD) | Mean (SD) | Mean (SD) |
| Viperin | 544.6 (100.9) | 15.5 (5.9) | 344.4 (208.5) | 231.1 (9.0) |
| IFITM1 | 468.2 (252.7) | 13.2 (3.1) | 219.1 (18.2) | 85.3 (27.1) |
| IFIT2 | 379.5 (44.5) | 6.2 (0.8) | 422.0 (217.1) | 279.7 (43.7) |
| MX2 | 395.3 (80.8) | 13.9 (1.7) | 131.2 (69.2) | 65.0 (17.6) |
| IFI27 | 276.0 (69.6) | 10.6 (3.3) | 112.4 (53.0) | 87.2 (54.5) |
| IFIT1 | 197.5 (11.6) | 12.8 (0.1) | 229.4 (117.0) | 498.0 (428.8) |
| OASL | 187.2 (53.4) | 4.9 (1.0) | 143.6 (96.1) | 118.3 (12.7) |
| OAS2 | 146.7 (25.8) | 14.2 (1.8) | 136.5 (4.7) | 107.1 (4.2) |
| IFI44L | 146.0 (3.6) | 8.2 (0.2) | 354.0 (162.2) | 308.6 (91.0) |
| IFI44 | 144.3 (18.4) | 11.7 (2.8) | 79.0 (70.6) | 63.8 (11.8) |
| GBP4 | 91.6 (18.3) | 3.5 (0.8) | 506.0 (341.7) | 68.1 (20.4) |
| MX1 | 78.2 (13.0) | 18.9 (4.6) | 27.8 (16.1) | 22.6 (6.9) |
| ISG15 | 67.6 (7.6) | 9.6 (1.3) | 269.5 (152.2) | 139.5 (0.7) |
| USP18 | 44.1 (2.4) | 3.3 (0.5) | 29.8 (26.4) | 24.3 (11.9) |
| ISG20 | 37.0 (7.7) | 1.8 (0.3) | 589.9 (152.5) | 74.3 (0.7) |
| MAC2BP | 27.1 (0.4) | 9.4 (6.5) | 2.3 (2.5) | 15.7 (3.3) |
| OAS1 | 25.1 (2.0) | 4.4 (0.4) | 15.3 (9.9) | 6.7 (0.5) |
| IFI6 | 23.7 (3.4) | 2.7 (1.1) | 82.5 (8.9) | 61.4 (11.9) |
| IFP35 | 20.3 (2.0) | 3.5 (0.4) | 127.4 (29.1) | 71.0 (23.9) |
| DDX60 | 19.5 (3.7) | 4.2 (1.1) | 91.6 (10.3) | 50.9 (31.8) |
| PKR | 12.3 (0.8) | 6.4 (3.3) | 47.4 (4.9) | 16.3 (7.9) |
| HERC6 | 8.4 (2.7) | 1.8 (1.3) | 16.8 (3.1) | 4.7 (1.6) |
| MECL1 | 7.2 (0.1) | 2.8 (0.4) | 19.4 (3.6) | 7.2 (0.8) |
| IFITM3 | 7.8 (6.0) | 1.6 (0.3) | 36.9 (2.5) | 3.6 (0.9) |
| Signal transduction | ||||
Liver
 |
AB-080510 | AE-102910 | ||
| Clone 2 | Jc1G | Clone 2 | Jc1G | |
Gene
 |
Mean (SD) | Mean (SD) | Mean (SD) | Mean (SD) |
| RIGI | 50.5 (4.7) | 3.0 (0.7) | 123.6 (30.0) | 27.2 (0.7) |
| MDA5 | 29.0 (2.8) | 1.9 (0.3) | 60.3 (22.2) | 27.5 (8.0) |
| STAT1 | 13.6 (2.5) | 2.4 (0.9) | 36.9 (2.5) | 18.1 (1.2) |
| IRF1 | 9.0 (0.3) | 1.5 (0.3) | 38.2 (2.6) | 12.4 (0.7) |
| IRF2 | 4.2 (0.8) | 2.8 (0.0) | 2.3 (2.9) | 1.9 (0.7) |
| IRF7 | 10.5 (2.3) | 2.2 (0.0) | 97.4 (1.9) | 141.1 (28.8) |
| IRF9 | 4.7 (1.2) | 2.3 (0.1) | 18.9 (4.2) | 7.2 (0.7) |
Gene induction is influenced by HCV replication and the JAK signaling pathway
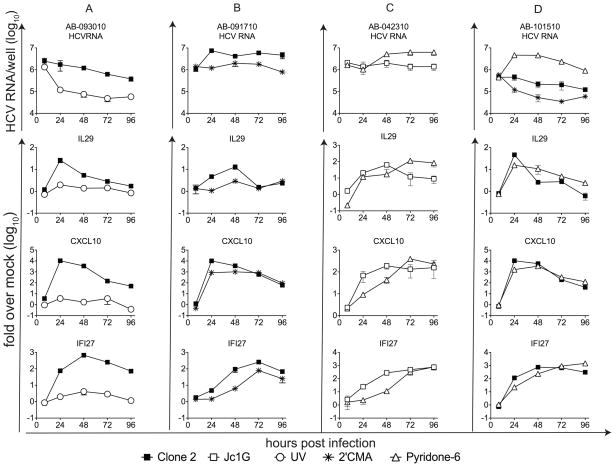
In general, greater gene induction was seen in cultures supporting higher levels of viral replication. In three of four comparisons, gene induction was greater in cultures infected with Clone 2 than with Jc1G (Table 1; Figure S2). IL29 was reproducibly induced at the RNA and/or protein level in HFLC infected with Clone 2 (seven of eight donors); in contrast, Jc1G stimulated detectable IL29 mRNA expression in three of 14 donors (p <0.01, Fisher’s exact test). Notably, Clone 2 typically has higher specific infectivity than does Jc1G, meaning that the same number of infectious units of the latter typically contain more copies of viral RNA. In contrast to replicating HCVcc, UV-irradiated HCV stimulated little if any gene expression (Figure 2A). Gene induction by HCVcc was reduced in the presence of 2′CMA (Figure 2B). IFNs signal through activation of JAK and Tyk kinases (43). The pan-JAK inhibitor, Pyridone-6, enhanced HCVcc replication in HFLC, and reduced or delayed the induction of some ISGs (but not IFNλ) during early time points of infection (Figure 2C–D). At later time points, ISG expression was increased in Pyridone-6-treated cultures that demonstrated enhanced HCV replication.
Figure 2. Gene induction in HCVcc-infected HFLC.
Four independent HFLC cultures are shown. At the indicated times, cultures were harvested and HCV, IL29, CXCL10, and IFI27 RNA levels measured by real-time RT-PCR as detailed in Materials and Methods. HCV RNA is indicated in copies/culture (mean ± SD) on a log10 scale. IL29, CXCL10, and IFI27 mRNA are shown as fold change (mean ± SD) over uninfected cultures at the same time point on a log10 scale. (A), HFLC (AE- 093010) were cultured with infectious or UV-inactivated Clone 2. (B), HFLC (AB-091710) were cultured with HCVcc Clone 2 ± 2.5 μM 2′CMA. (C), HFLC (AB-042310) were cultured with HCVcc Jc1G ± 1 μM Pyridone-6. (D), HFLC (AB-101510) were cultured with HCVcc Clone 2 ± 1 μM Pyridone-6. Four experiments are shown out of six with similar results.
Expression of IL29 and chemokine proteins in HCVcc-infected cultures
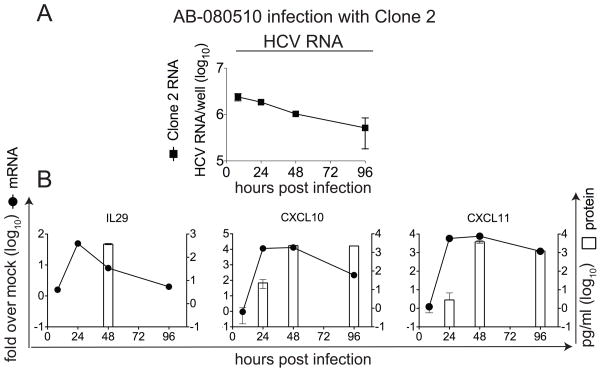
We performed ELISAs to confirm the expression of selected genes following HCVcc infection of HFLC (Figure 3). In the experiment shown, HCV RNA persisted but did not increase during culture (Figure 3A). IL29 mRNA expression was transient, with 45-fold induction at 24 hours, and IL29 protein peaked after the mRNA peak (Figure 3B). Maximal IL29 protein levels in five experiments averaged 211±110 pg/ml (range, 64–388 pg/ml). IL29 protein peaked at day 2 after infection in 3/5 experiments, but peaks at day 1 and day 4 after infection were observed in one experiment each. A >10,000-fold induction of CXCL10 mRNA was observed beginning 24 hours after the start of infection; CXCL10 protein was detectable as early as 24 hours, with maximal levels detected 48–96 hours after infection. In two experiments, CXCL10 protein peaked at an average of 2300 pg/ml (range, 2000–2500 pg/ml). Similarly, CXCL11 mRNA was induced >5000-fold over uninfected cultures beginning 24 hours after infection, and CXCL11 protein was readily detected beginning 48 hours after infection. In two experiments, CXCL11 protein peaked at an average of 3800 pg/ml (range, 300–4000 pg/ml).
Figure 3. HCVcc induces expression of IL29 and chemokine proteins.
HFLC (AB-080510) were infected with HCVcc Clone 2. At the indicated times, supernatants were harvested for ELISA and cells were harvested for RNA preparation. HCV RNA is indicated in copies/culture (mean ± SD) on a log10 scale. IL29, CXCL10, and CXCL11 mRNA are shown as fold change (mean ± SD) over untreated cultures at the same time point, also on a log10 scale. Proteins are indicated in pg/ml (mean ± SD) of culture supernatant on a log10 scale. (A), HCV RNA. (B), mRNA and protein for IL29 (left), CXCL10 (middle) and CXCL11 (right); one experiment is shown out of five (IL29) or two (CXCL10 and CXCL11) with similar results.
IFNs inhibit HCVcc infection in HFLC
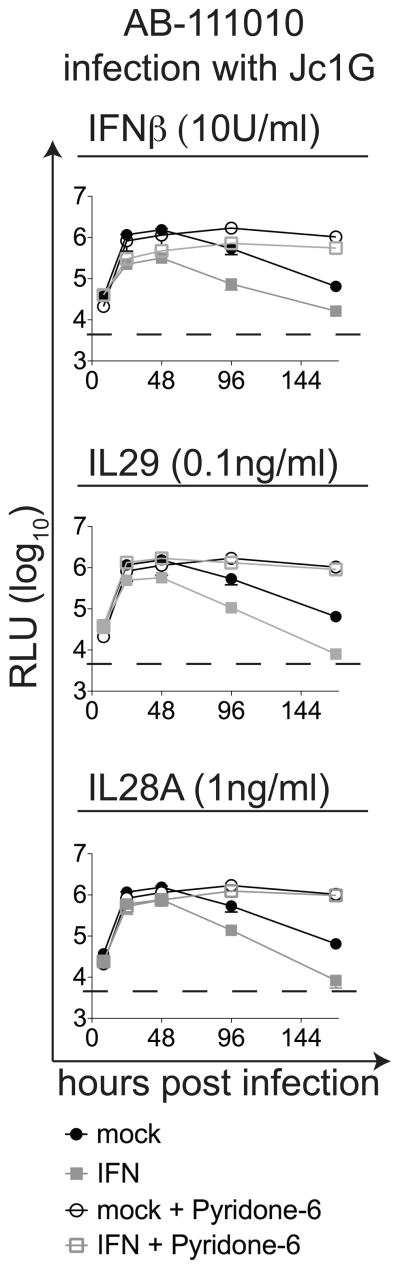
Since IL29 protein was detected in HCV-infected cultures (Figure 3), we evaluated the effects of IL29 on HCV replication in HFLC (Figure 4). We measured the GLuc reporter to monitor Jc1G infection. As a control, IFNβ inhibited HCVcc infection in a dose-dependent fashion, and the inhibitory effect of 10U IFNβ was diminished in the presence of Pyridone-6. Recombinant IL29 inhibited HCV-dependent luciferase expression at 100 pg/ml, a level below that detected in HCV-infected HFLC cultures. This IL29-mediated inhibition of HCV replication was reversed in the presence of Pyridone-6. Recombinant IL28A also inhibited Jc1G replication in HFLC, and the inhibitory effect of 1 ng/ml IL28A was blocked in the presence of Pyridone-6. Pyridone-6 did not reverse inhibition by higher doses of IFNs. These results extend previous reports that Type III IFNs (at higher levels) can inhibit HCV replication in hepatoma cells and primary liver cells (26–29), and suggest that such inhibition is dependent on JAK activity. They also suggest that endogenous IFNs may contribute to the low HCV infection efficiency in HFLC.
Figure 4. Cytokines mediate JAK-dependent inhibition of HCV replication in HFLC.
HFLC (AB-111010) were infected with Jc1G ± the indicated cytokines and ± Pyridone-6, as indicated. At the indicated times, culture supernatants were sampled for GLuc measurement. Top: Jc1G replication in cultures treated with 10 U/ml IFNβ or no cytokine (mock) ± 1 μM Pyridone-6. Middle: Jc1G replication in cultures treated with IL29 (100 pg/ml) or no cytokine (mock) ± 1 μM Pyridone-6. Bottom: Jc1G replication in cultures treated with IL28B (1 ng/ml) or no cytokine (mock) ± 1 μM Pyridone-6. Pyridone-6 enhances HCV replication in cultures without added cytokine and restores HCV replication in HFLC treated with IFNβ, IL29, or IL28A. Dashed lines indicate background RLU from uninfected cultures.
HCV replication, but not IL28B genotype, correlates with IFNλ and ISG induction
In genome-wide association studies, single nucleotide polymorphisms close to and within the IL28B locus have been found to correlate with spontaneous clearance vs. persistence of untreated HCV infection, and with sustained virologic control vs. non-response in HCV (genotype 1) patients treated with pegylated IFNα plus ribavirin (22). In light of the varied outcomes in HCVcc-infected cultures shown here, we performed IL28B SNP rs12979860 genotyping (41) on the HFLC used in these assays (subject to the availability of cells for DNA preparation). The CC genotype at rs12979860 is associated with increased likelihood of spontaneous recovery from HCV infection, while CT and TT genotypes are associated with increased risks of persistent infection and treatment failure (22). As shown in Figure 5, the rs12979860 genotype did not predict the ability of HFLC to support robust HCV replication, nor did it correlate with levels of IL29, CXCL10, or CXCL11 induction. Instead, the levels and kinetics of gene induction correlated most closely with the levels and kinetics of HCV replication.
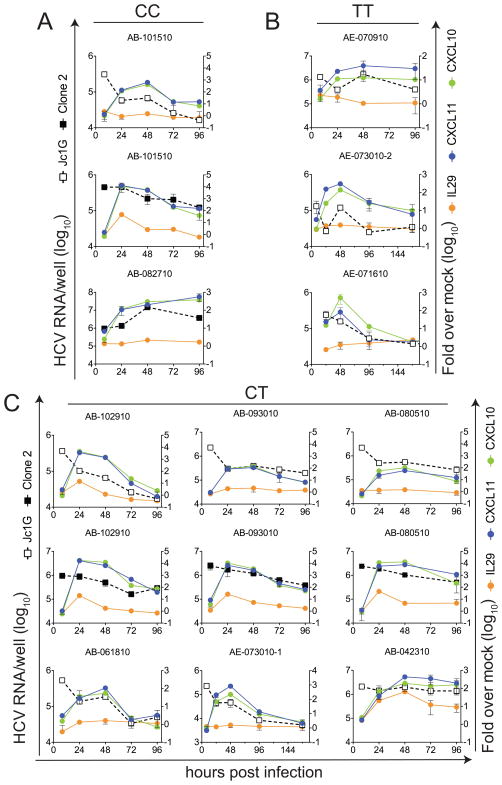
Figure 5. IL28B SNP genotype does not correlate with HCV replication or HCV-stimulated gene expression.
DNA was prepared from the available material (organ identification numbers are indicated above each graph) and subjected to rs12979860 SNP genotyping as described in Materials and Methods. Cultures were infected with Jc1G or Clone 2, as indicated. Data shown represent HCV RNA levels and induction of IL29, CXCL10, and CXCL11 RNA. (A), Samples with CC genotype. Note that IL29 protein (135 ±100 pg/ml) was detected in supernatants from AB-082710 at 96 hours, but no 72-hour RNA sample was available for RT-PCR. (B), Samples with TT genotype. (C), Samples with CT genotype. Data shown represent 15 experiments using 11 organs.
Discussion
In vitro studies of virus-host interactions in HCV infection have until recently been restricted largely to derivatives of a hepatoma cell line selected for their ability to support high-level HCV replication. These cell lines may not accurately reproduce the complex host-virus interactions occurring in vivo. In an accompanying paper (Andrus et al.), we report that HFLC support persistent HCV infection in vitro. Here, we demonstrate that HCV infection of primary liver cells stimulates expression of a number of potential antiviral genes, including IL29 (IFNλ1), but not IFNα or (in 16/18 experiments) IFNβ. IL29 protein was produced in quantities sufficient to inhibit HCV infection of HFLC. Pyridone-6, a JAK inhibitor that blocks the antiviral effect of IL29 (Figure 4), enhanced HCV replication even in the absence of exogenous IFNs (Figures 2 and 4), suggesting that endogenous IFNs may decrease HCV replication in these cultures. The level of gene induction varied between donors, suggesting that host polymorphisms influence the response to HCV infection. For some donors, gene induction also varied depending on the infecting virus. Induction of some ISGs was reduced or delayed by inhibition of JAK signaling. Together, these results indicate that HCV infection of HFLC can serve as a valid in vitro model for studying innate immune responses to hepatotropic viruses.
The precise functions of many ISGs remain to be determined (21). However, recent work demonstrates that specific ISGs including RIG-I, MDA-5, IRF1, and IRF7 each mediate significant antiviral activities against HCV and other viruses (44). A common feature of these genes is their potential to stimulate downstream expression of other ISGs. All four of these ISGs are induced in HCV-infected HFLC. Therefore, we hypothesize that expression of these and other ISGs in HFLC may limit HCV replication in these cultures. Other genes, such as CXCL10 and CXCL11, may serve in vivo to recruit immune cells to the site of infection. These chemokines are highly expressed in HCV patients (reviewed in (45, 46)).
Our observation of transient IFNλ production following HCV infection, and the ability of the JAK inhibitor, Pyridone-6, to increase HCV replication in HFLC even without added IFNs, suggest that IFNλ may contribute to control of HCV replication and gene induction in HFLC and in vivo. Notably, in HCV patient liver biopsies, levels of HCV RNA—but not of IFNβ mRNA—correlated with ISG expression (14). Thus, factors other than IFNα/β may modulate intrahepatic ISG expression.
Although genetic variation at the IL28B locus is associated with outcomes of HCV infection and treatment (22), the mechanisms behind this association are poorly understood. IL28B SNP genotype did not correlate with IL28A/B expression levels in HCV patient liver biopsies (47). Furthermore, the associations between IL28B and HCV infection may be weaker for subjects infected with HCV genotype 2 or 3 than those infected with genotype 1 (22). The JFH strain on which so much in vitro HCV research depends is of genotype 2. As other HCV culture systems are developed, it will be important to learn how HCV isolates representing other HCV genotypes interact with IFN systems.
HCV replicates less efficiently in primary HFLC cultures than in Huh-7.5 cells. Furthermore, the ability of these mixed cell populations to support HCV infection and spread varies between different donors. These observations highlight important differences between primary tissue and highly selected cell lines. HCV may infect only a minor population of hepatocytes in vivo (13, 14, 48). ISG expression is readily detected in the liver during acute and chronic HCV infection, even when type I IFN expression is not (13–18). This study demonstrates that a similar gene expression profile is observed during acute HCV infection in HFLCs. It has been proposed that IFNs induce an antiviral state in most uninfected hepatocytes in vivo, thereby limiting the number of cells capable of supporting infection (19, 20). This may benefit HCV by restricting antigen levels, delaying recognition by the adaptive immune system (2), and preventing immune-mediated liver damage. In the infected liver, transient, localized lessening of ISG expression may provide HCV with a steady supply of susceptible hepatocytes for new infectious events.
Supplementary Material
Acknowledgments
We thank S.M. di Vittorio, C. Murray, and J. Sable for expert administrative assistance and laboratory management; we are grateful to W.T. Barry, M.E. Castillo, B.R. Flatley, M. Holz, S. Pouzol, S. Shirley, and A. Webson for technical assistance.
Funding: This work was supported in part by the National Institutes of Health R01 AI60561 (LBD, SM, EDC), K08 AI075031 (EDC), R01 DK085713 (AP, CMR), R01 AI090055 (AP, CMR), F32 AI084448 (TPS), by the Irma T. Hirschl / Monique Weill-Caulier Trust (LBD), and by the Greenberg Institute for Medical Research.
List of abbreviations
- 2′CMA
2′C-methyladenosine
- dsRNA
double-stranded RNA
- GLuc
Gaussia princeps luciferase
- HCV
hepatitis C virus
- HCVcc
cell culture-adapted HCV
- HDFR
HCV-dependent fluorescent reporter
- HFLC
human fetal liver cells
- JAK
Janus kinase
- IFN
interferon
- IPS
IFNβ promoter stimulator
- ISG
interferon-stimulated gene
- NLS
nuclear localization signal
- p(I
C), polyinosinic:polycytidylic acid
- RFP
red fluorescent protein
- RT-PCR
reverse transcription and polymerase chain reaction
- TCID50
50% tissue culture-infectious dose
- UV
ultraviolet light
Contributor Information
Svetlana Marukian, Email: marukis@rockefeller.edu.
Linda Andrus, Email: landrus@rockefeller.edu.
Timothy P. Sheahan, Email: tsheahan@rockefeller.edu.
Christopher T. Jones, Email: christophert.jones@novartis.com.
Edgar D. Charles, Email: charlee@rockefeller.edu.
Alexander Ploss, Email: aploss@rockefeller.edu.
Charles M. Rice, Email: ricec@rockefeller.edu.
Lynn B. Dustin, Email: dustinl@rockefeller.edu.
Bibliography
- 1.Lavanchy D. The global burden of hepatitis C. Liver Int. 2009;29 (Suppl 1):74–81. doi: 10.1111/j.1478-3231.2008.01934.x. [DOI] [PubMed] [Google Scholar]
- 2.Dustin LB, Rice CM. Flying under the radar: The immunobiology of hepatitis C. Annu Rev Immunol. 2007;25:71–99. doi: 10.1146/annurev.immunol.25.022106.141602. [DOI] [PubMed] [Google Scholar]
- 3.Rehermann B. Hepatitis C virus versus innate and adaptive immune responses: a tale of coevolution and coexistence. J Clin Invest. 2009;119:1745–1754. doi: 10.1172/JCI39133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sumpter R, Jr, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, et al. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Binder M, Kochs G, Bartenschlager R, Lohmann V. Hepatitis C virus escape from the interferon regulatory factor 3 pathway by a passive and active evasion strategy. Hepatology. 2007;46:1365–1374. doi: 10.1002/hep.21829. [DOI] [PubMed] [Google Scholar]
- 6.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 7.Foy E, Li K, Sumpter R, Jr, Loo YM, Johnson CL, Wang C, Fish PM, et al. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc Natl Acad Sci U S A. 2005;102:2986–2991. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, et al. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A. 2005;102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM, Carney DS, et al. Viral and therapeutic control of IFN-β promoter stimulator 1 during hepatitis C virus infection. Proc Natl Acad Sci U S A. 2006;103:6001–6006. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaukinen P, Sillanpaa M, Kotenko S, Lin R, Hiscott J, Melen K, Julkunen I. Hepatitis C virus NS2 and NS3/4A proteins are potent inhibitors of host cell cytokine/chemokine gene expression. Virol J. 2006;3:66. doi: 10.1186/1743-422X-3-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ploss A, Khetani SR, Jones CT, Syder AJ, Trehan K, Gaysinskaya VA, Mu K, et al. Persistent hepatitis C virus infection in microscale primary human hepatocyte cultures. Proc Natl Acad Sci U S A. 2010;107:3141–3145. doi: 10.1073/pnas.0915130107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bigger CB, Brasky KM, Lanford RE. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. J Virol. 2001;75:7059–7066. doi: 10.1128/JVI.75.15.7059-7066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stiffler JD, Nguyen M, Sohn JA, Liu C, Kaplan D, Seeger C. Focal distribution of hepatitis C virus RNA in infected livers. PLoS One. 2009;4:e6661. doi: 10.1371/journal.pone.0006661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thimme R, Bukh J, Spangenberg HC, Wieland S, Pemberton J, Steiger C, Govindarajan S, et al. Viral and immunological determinants of hepatitis C virus clearance, persistence, and disease. Proc Natl Acad Sci U S A. 2002;99:15661–15668. doi: 10.1073/pnas.202608299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bigger CB, Guerra B, Brasky KM, Hubbard G, Beard MR, Luxon BA, Lemon SM, et al. Intrahepatic gene expression during chronic hepatitis C virus infection in chimpanzees. J Virol. 2004;78:13779–13792. doi: 10.1128/JVI.78.24.13779-13792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarasin-Filipowicz M, Oakeley EJ, Duong FH, Christen V, Terracciano L, Filipowicz W, Heim MH. Interferon signaling and treatment outcome in chronic hepatitis C. Proc Natl Acad Sci U S A. 2008;105:7034–7039. doi: 10.1073/pnas.0707882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su AI, Pezacki JP, Wodicka L, Brideau AD, Supekova L, Thimme R, Wieland S, et al. Genomic analysis of the host response to hepatitis C virus infection. Proc Natl Acad Sci U S A. 2002;99:15669–15674. doi: 10.1073/pnas.202608199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Major ME, Dahari H, Mihalik K, Puig M, Rice CM, Neumann AU, Feinstone SM. Hepatitis C virus kinetics and host responses associated with disease and outcome of infection in chimpanzees. Hepatology. 2004;39:1709–1720. doi: 10.1002/hep.20239. [DOI] [PubMed] [Google Scholar]
- 20.Dahari H, Major M, Zhang X, Mihalik K, Rice CM, Perelson AS, Feinstone SM, et al. Mathematical modeling of primary hepatitis C infection: noncytolytic clearance and early blockage of virion production. Gastroenterology. 2005;128:1056–1066. doi: 10.1053/j.gastro.2005.01.049. [DOI] [PubMed] [Google Scholar]
- 21.Sen GC, Sarkar SN. The interferon-stimulated genes: targets of direct signaling by interferons, double-stranded RNA, and viruses. Curr Top Microbiol Immunol. 2007;316:233–250. doi: 10.1007/978-3-540-71329-6_12. [DOI] [PubMed] [Google Scholar]
- 22.Balagopal A, Thomas DL, Thio CL. IL28B and the control of hepatitis C virus infection. Gastroenterology. 2010;139:1865–1876. doi: 10.1053/j.gastro.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Österlund PI, Pietilä TE, Veckman V, Kotenko SV, Julkunen I. IFN regulatory factor family members differentially regulate the expression of type III IFN (IFN-λ) genes. J Immunol. 2007;179:3434–3442. doi: 10.4049/jimmunol.179.6.3434. [DOI] [PubMed] [Google Scholar]
- 24.Onoguchi K, Yoneyama M, Takemura A, Akira S, Taniguchi T, Namiki H, Fujita T. Viral infections activate types I and III interferon genes through a common mechanism. J Biol Chem. 2007;282:7576–7581. doi: 10.1074/jbc.M608618200. [DOI] [PubMed] [Google Scholar]
- 25.Doyle SE, Schreckhise H, Khuu-Duong K, Henderson K, Rosler R, Storey H, Yao L, et al. Interleukin-29 uses a type 1 interferon-like program to promote antiviral responses in human hepatocytes. Hepatology. 2006;44:896–906. doi: 10.1002/hep.21312. [DOI] [PubMed] [Google Scholar]
- 26.Robek MD, Boyd BS, Chisari FV. Lambda interferon inhibits hepatitis B and C virus replication. J Virol. 2005;79:3851–3854. doi: 10.1128/JVI.79.6.3851-3854.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcello T, Grakoui A, Barba-Spaeth G, Machlin ES, Kotenko SV, MacDonald MR, Rice CM. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006;131:1887–1898. doi: 10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]
- 28.Lazaro CA, Chang M, Tang W, Campbell J, Sullivan DG, Gretch DR, Corey L, et al. Hepatitis C virus replication in transfected and serum-infected cultured human fetal hepatocytes. Am J Pathol. 2007;170:478–489. doi: 10.2353/ajpath.2007.060789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diegelmann J, Beigel F, Zitzmann K, Kaul A, Goke B, Auernhammer CJ, Bartenschlager R, et al. Comparative analysis of the lambda-interferons IL-28A and IL-29 regarding their transcriptome and their antiviral properties against hepatitis C virus. PloS ONE. 2010;5:e15200. doi: 10.1371/journal.pone.0015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 31.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, et al. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marukian S, Jones CT, Andrus L, Evans MJ, Ritola KD, Charles ED, Rice CM, et al. Cell culture-produced hepatitis C virus does not infect peripheral blood mononuclear cells. Hepatology. 2008;48:1843–1850. doi: 10.1002/hep.22550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diamond DL, Syder AJ, Jacobs JM, Sorensen CM, Walters KA, Proll SC, McDermott JE, et al. Temporal proteome and lipidome profiles reveal hepatitis C virus-associated reprogramming of hepatocellular metabolism and bioenergetics. PLoS Pathog. 2010;6:e1000719. doi: 10.1371/journal.ppat.1000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones CT, Catanese MT, Law LM, Khetani SR, Syder AJ, Ploss A, Oh TS, et al. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat Biotechnol. 2010;28:167–171. doi: 10.1038/nbt.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmelzer E, Zhang L, Bruce A, Wauthier E, Ludlow J, Yao HL, Moss N, et al. Human hepatic stem cells from fetal and postnatal donors. J Exp Med. 2007;204:1973–1987. doi: 10.1084/jem.20061603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindenbach BD. Measuring HCV infectivity produced in cell culture and in vivo. Methods Mol Biol. 2009;510:329–336. doi: 10.1007/978-1-59745-394-3_24. [DOI] [PubMed] [Google Scholar]
- 38.Mulligan EK, Germer JJ, Arens MQ, D'Amore KL, Di Bisceglie A, Ledeboer NA, Moser MJ, et al. Detection and quantification of hepatitis C virus (HCV) by MultiCode-RTx real-time PCR targeting the HCV 3' untranslated region. J Clin Microbiol. 2009;47:2635–2638. doi: 10.1128/JCM.02170-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carroll SS, Tomassini JE, Bosserman M, Getty K, Stahlhut MW, Eldrup AB, Bhat B, et al. Inhibition of hepatitis C virus RNA replication by 2'-modified nucleoside analogs. J Biol Chem. 2003;278:11979–11984. doi: 10.1074/jbc.M210914200. [DOI] [PubMed] [Google Scholar]
- 40.Pedranzini L, Dechow T, Berishaj M, Comenzo R, Zhou P, Azare J, Bornmann W, et al. Pyridone 6, a pan-Janus-activated kinase inhibitor, induces growth inhibition of multiple myeloma cells. Cancer Res. 2006;66:9714–9721. doi: 10.1158/0008-5472.CAN-05-4280. [DOI] [PubMed] [Google Scholar]
- 41.Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O'hUigin C, Kidd J, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–802. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC(T) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 43.Schindler C, Plumlee C. Inteferons pen the JAK-STAT pathway. Semin Cell Dev Biol. 2008;19:311–318. doi: 10.1016/j.semcdb.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Charles ED, Dustin LB. Chemokine antagonism in chronic hepatitis C virus infection. J Clin Invest. 2011;121:25–27. doi: 10.1172/JCI45610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeremski M, Petrovic LM, Talal AH. The role of chemokines as inflammatory mediators in chronic hepatitis C virus infection. J Viral Hepat. 2007;14:675–687. doi: 10.1111/j.1365-2893.2006.00838.x. [DOI] [PubMed] [Google Scholar]
- 47.Urban TJ, Thompson AJ, Bradrick SS, Fellay J, Schuppan D, Cronin KD, Hong L, et al. IL28B genotype is associated with differential expression of intrahepatic interferon-stimulated genes in patients with chronic hepatitis C. Hepatology. 2010;52:1888–1896. doi: 10.1002/hep.23912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang Y, Shilagard T, Xiao SY, Snyder N, Lau D, Cicalese L, Weiss H, et al. Visualizing hepatitis C virus infections in human liver by two-photon microscopy. Gastroenterology. 2009;137:1448–1458. doi: 10.1053/j.gastro.2009.07.050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.