Abstract
Integrins are adhesion receptors involved in bidirectional signaling that are crucial for various cellular responses during normal homeostasis and pathological conditions such as cancer progression and metastasis. Aberrant expression of noncoding microRNAs has been implicated in the deregulation of integrin expression and activity, leading to the development and progression of cancer tumors, including their acquisition of the metastatic phenotype. miR-31 is a key regulator of several critical genes involved in the invasion-metastasis cascade in cancer. Using diverse cell based, genetic, biochemical, flow cytometry and functional analyses, we report that miR-31 is a master regulator of integrins as it targets multiple α subunit partners (α2, α5 and αV) of β1 integrins and also β3 integrins. We found that expression of miR-31 in cancer cells resulted in a significant repression of these integrin subunits both at the mRNA and protein levels. Loss of expression of α2, α5, αV and β3 was a direct consequence of miR-31 targeting conserved seed sequences in the 3’UTR of these integrin subunits leading to their posttranscriptional repression, which was reflected in their diminished surface expression in live cells. The biological consequence of decreased the cell surface of these integrins was a significant inhibition of cell spreading in a ligand-dependent manner. While different reports have shown that a single integrin can be regulated by several microRNAs, here we show that a single microRNA, miR-31, is able to specifically target several integrin subunits to regulate key aspects of cancer cell invasion and metastasis.
Keywords: miR-31, Integrins, cell adhesion and spreading, cancer cell invasion
INTRODUCTION
As a large family of cell-cell and cell-extracellular matrix (ECM) receptors, integrin α/β heterodimers play prominent roles during embryonic development and postnatal physiology and pathology 1. In addition to their adhesive functions, integrins mediate bidirectional signaling across the cell membrane that regulates numerous cellular responses, including motility, differentiation, growth and gene expression 2,3, 4,5-7. In this context, alterations in integrin expression and function impart phenotypic properties to transformed cells that have significant impact on cancer progression and metastasis 8, 9.
microRNAs, short non-coding, single-stranded RNAs that regulate protein translation and messenger RNA (mRNA) degradation of their target genes 10, have been shown to be involved in a large number of biological processes. It is estimated that about one-third of all human mRNAs are regulated by microRNAs 11; and, thus far, more than 1000 microRNAs have been identified in humans with the potential to control more than 30,000 target genes. The broad spectrum of genes that can be targeted by a single microRNA is attributed to the high level of conserved target motifs, seed sequences, within the 3’untranslated regions (UTR) of the affected genes, thus making them powerful regulators of gene expression in numerous and complex cellular responses, including cancer cell invasion and metastasis 12-15. microRNAs have also been found to regulate integrin activity in such complex biological processes, such as angiogenesis, cell migration and invasion (reviewed in 16). Recently, we and others have identified and characterized miR-31 as a master regulator of different steps in the invasion-metastasis-cascade by virtue of its capacity to modulate key metastasis-promoter genes, including WAVE3 and integrin α5 subunit 17, 18. In this study, we show that in addition to α5, miR-31 directly targets seed sequences in the 3’UTRs of two other integrin alpha subunits, α2 and αV, and the β3 subunit. We also find that the miR-31-mediated suppression of the integrin alpha subunits indirectly suppresses cell surface expression of their β1 subunit partner and show that suppressed expression of these integrins reduces cell spreading on specific target ECM ligands of these affected integrins.
MATERIALS AND METHODS
Reagents
All monoclonal antibodies (mAbs) to integrins were from Millipore (Bedford, MA) except for anti-β5 integrin, which was from eBioscience (San Diego, CA) and β8 integrin, which was from R&D Systems (Minneapolis, MN). UV Live Dead dye and all secondary antibodies were from Molecular Probes (Eugene, OR). Other reagents were from Sigma (St. Louis, MO).
Flow cytometry
MDA-MB-231 cells expressing GFP-pBABE vector or GFP-miR-31, a kind gift from Dr. Robert Weinberg 18, were propagated in DMEM culture medium supplemented with 10% FBS and puromycin (5 μg/ml). Sub-confluent cells were detached by trypsinization and washed with Hank's Balanced Salt Solution with Ca2+, Mg2+ and 0.1% BSA. Cells were processed for flow cytometry as described 19. Alexa Fluor 647 tagged goat anti-mouse antibody was used to detect all integrin mouse mAbs, including the β1 activation specific HUTS-4, except for rat anti-α6 integrin mAb, which was detected with Alexa Fluor 647 tagged anti-rat antibody. All primary and secondary antibodies were used at 10 μg/ml. As a positive control for integrin activation, 3.5 mM MnCl2 was used to treat the cells for 5 min at 37°C in a humidified tissue culture incubator. All data were acquired in a BD LSRII instrument and analyzed with FlowJo 7.6.3 (Treestar, Ashland, OR) software. Binding of only live, GFP+ cells to different antibodies was used in the analyses. Non-specific binding due to secondary antibody alone to empty vector transfected cells was subtracted from all flow cytometric data. The resultant fluorescence intensity values were then plotted graphically.
Cell Culture, transfections and immunoblotting
The established human cancer cell lines were obtained from American Type Culture Collection (ATCC) and cultured as previously described 20. Growth and morphology of each cell line were routinely monitored and key features were found to be consistent with prior description of each cell line. No further genetic analyses or fingerprinting were performed. Transient transfections and immunoblotting analyses were performed as described previously 20-23.
Plasmid Construction, Site-directed Mutagenesis and 3’UTR luciferase reporter analysis
The nucleotide sequence (~300 bp) flanking the miR-31 seed sequence within the 3’ untranslated region (3’UTR) of each integrin subunit under study was PCR-amplified from MDA-MB-231 genomic DNA and subcloned into the pmiRGlo vector downstream of the firefly luciferase expression cassette (Promega, Madison, WI). The correct sequence and orientation were verified by sequencing. QuikChange site-directed mutagenesis kit (Stratagene, Cedar Creek, TX) was used to generate the 3’UTR mutants where the seed sequence recognized by miR-31 was deleted. pmirGlo reporter plasmids (1 μg total plasmid) were transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) into the specified cancer cells and then seeded in 12-well plates (3 × 104 cells per well). Cells were collected after 48 h for assay using the Dual-Luciferase reporter assay system (Promega, Madison, WI), as described previously 17. For co-transfection experiments, synthetic microRNA mimics or miR Negative Control (5 nM), were added to the transfection mix. All experiments were performed in triplicate with data averaged from at least three independent experiments.
Semi-quantitative and real-time quantitative–RT-PCR
Total RNA was extracted from established cancer cell lines using TRIzol reagent (Invitrogen, Carlsbad, CA), following the manufacturer's instructions. cDNA was generated and used as a template for semi-quantitative RT-PCR as previously described 20, 22-24. Expression levels of microRNAs were quantified by real-time quantitative RT-PCR using the human TaqMan MicroRNA Assays Kits (Applied Biosystems, Carlsbad, CA). The reverse transcription reaction was carried out with TaqMan microRNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA) following the manufacturer's instructions. For each gene, quantification of expression levels was performed using the respective gene-specific primers (Table S1) and the RT2 SYBR Green/Fluorescein qPCR Master Mix (SABiosciences, Frederick, MD) following the manufacturer's instructions. Quantitative RT-PCR was performed on the BioRad (Hercules, CA) iCycler PCR system where the reaction mixtures were incubated at 95°C for 10 min, followed by 40 cycles of 95°C for 15s and 60°C for 1 min. The cycle threshold (Ct) values were calculated with SDS 1.4 software (Bio-Rad). The expression levels of each transcript were normalized using the 2−ΔΔCt method 25 relative to GAPDH. The ΔCt was calculated by subtracting the Ct values of GAPDH from the Ct values of the transcript of interest. The ΔΔCt was then calculated by subtracting ΔCt of the parental cells from the control vector- or miR-31-expressing cells. Fold change in the gene expression was calculated according to the equation 2-ΔΔCt.
Immunofluorescence
MDA-MB-231 cells expressing miR-31 or the control vector were seeded onto 6-well plates coated with 10 μg/ml laminin, 10 μg/ml fibronectin, 10 μg/ml collagen or 5 times diluted growth factor-reduced Matrigel for 1 hour, fixed, permeabilized with 0.1% Triton X-100, blocked in horse serum, and stained for 30 min with Alexa 568-phalloidin to visualize actin filaments 26.
Cell spreading and integrin blocking
Cell spreading assays were carried out on round coverslips (Fisher Scientific, Pittsburg, PA) in 12-well plates (Falcon, Becton Dickinson&Comp., NY). Coverslips were coated 1 h with Matrigel Matrix (BD Biosciences, Bedford, MA) diluted 1:6 in PBS, 10 μg/ml collagen I, 10 μg/ml laminin or 2 μg/ml vitronectin (all from Sigma, St. Louis, MO) at 37°C. Following three washes with PBS coverslips were blocked for 1 h with 1% BSA in PBS at 37°C. BSA (1 mg/ml) coated coverslips were used as negative controls.
MDA-MB-231-vector and MDA-MB-231-miR-31 breast cancer cells grown to ~80% confluency were detached with trypsin, washed with serum-free-DMEM, resuspended in the same medium and then plated in triplicate at a cell density of 6×104 cells/well. For α2β1 integrin blocking experiments, cells were re-suspended in media containing 10 μg/ml of the α2β1 integrin blocking mAb, LS-C247646, (LS-Bio, Seattle, WA) and incubated 30 min at 37°C before they were seeded on coverslips coated with collagen I or Matrigel. Control cells were re-suspended with non-blocking IgG control under the same conditions. Cells were allowed to adhere and spread for 30 min and 1 h in a 37°C CO2 incubator. At the end of the incubation period, unattached cells were removed by careful aspiration and washing two times with PBS. Adherent cells were fixed with 4% PFA for 10 min and washed twice with PBS. Fixed cells were permeabilized with 0.1% Triton X-100 in PBS for 5 min and blocked with 4 % bovine serum overnight at 4°C and then stained for 1 h with Alexa Fluor 568-conjugated phalloidin (Invitrogen, Molecular Probes, Eugene, OR). After washing, the coverslips were transferred on slides and mounted in immune-fluor mounting medium (MP Biomedicals, Solon, OH). Images were acquired with a 20x/0.7 or 40x/1.25 numerical aperture oil objective on a fluorescent microscope (Leica) equipped with digital camera using Image-Pro Plus version 5.1.2.59. Cells were quantified using the ImageJ software version 1.43u and the results are presented as the total surface area covered by cells, in square micrometer, averaged to 300 cells.
Oligonucleotide sequences
Sequences of the oligonucleotide primers used for genomic PCR, RT-PCR, to amplify the entire WAVE3 3’UTR or the individual seed sequences, as well as the primers used for mutagenesis were from IDT (San Diego, CA) and are listed in supplementary Table S1.
Statistical analyses
The data are presented as the means ± standard errors of at least three independent experiments. The results were tested for significance using an unpaired Student's t test and p values of <0.05 were considered statistically significant.
RESULTS
miR-31 targets several integrins subunits
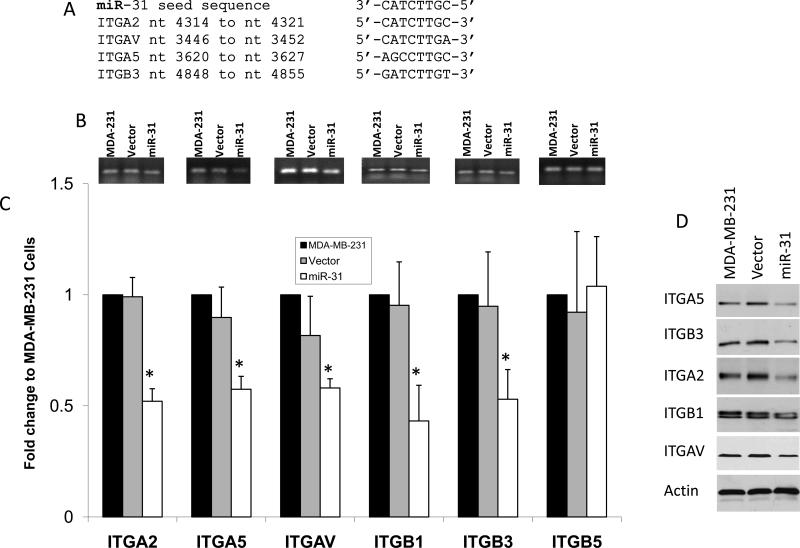
Previous reports have shown that microRNA miR-31 targets the integrin α5 subunit (ITGA5) by interacting with a perfectly matched seed sequence in the 3’UTR of the ITGA5 transcript 17, 18. We sought to determine whether miR-31 could also target other integrin subunits. By in silico analyses, we found that the seed sequence of miR-31 was conserved in the 3’UTR of three other integrin subunits; α2, αV and β3 (ITGA2, ITGAV and ITGB3, respectively, Figure 1A). Since binding of microRNAs to their specific seed sequences often represses gene expression, we examined the effect of miR-31 on the expression levels of these candidate target integrin subunits. The analysis was performed in MDA-MB-231 cells because these breast cancer cells have very low levels of endogenous miR-31 17, 18. Stable expression of miR-31 in these cells had no effect on proliferation compared to control vector transfected cells (Fig. S1). Overexpression of miR-31 in MDA-MB-231 cells resulted in ~50% decrease in the levels of α2, α5, αV and β3. This suppression was found both at the mRNA as observed by semi-quantitative RT-PCR (Fig. 1B) and qRT-PCR (Fig. 1C) and the cellular protein levels as determined by Western blots (Fig. 1D and Fig. S2). All these four integrin subunits contain target sites for miR-31 in their respective 3’UTRs (Fig. 1A), suggesting that the silencing of their expression is consequent to miR-31 targeting of the 3’UTR in their transcripts. Integrin subunit β5, which lacks a miR-31 seed sequence, was not affected by miR-31 expression in these cells. We also found that miR-31 overexpression resulted in ~2-fold decrease in β1 subunit (ITGB1) mRNA and protein expression levels compared to the parental cells or the vector-transfected cells (Fig. 1, all panels), even though β1 does not contain a target sequence for miR-31 in its 3’UTR. This result could be a secondary to effect of miR-31 on the expression levels of α2, α5 and αV, alpha subunits partners of β1; which could lead to degradation of surplus β1 subunit. The effect of miR-31 on the expression of these specific integrin subunits, α2, α5 and αV, and β1, was also confirmed in LNCaP prostate cancer cells, and β5, which lacks the miR-31 target sequence, was again unaffected (Fig. S3).
Fig. 1. miR-31 inhibits expression of specific α and β integrin subunits.
(A) Alignment of miR-31 seed sequence with candidate target sequences within the 3’UTRs of the indicated integrin subunits. (B) semi-quantitative RT-PCR, (C) Real-time qt RT-PCR and (D) immunoblot of the indicated integrin subunits from MDA-MB-231 cells or MDA-MB-231 cells with stable expression of either control or miR-31 vector. Densitometry and statistical analyses of the Western Blot data is shown in Fig. S2.
Surface expression of α-subunits partners of β1 integrins are reduced due to miR-31
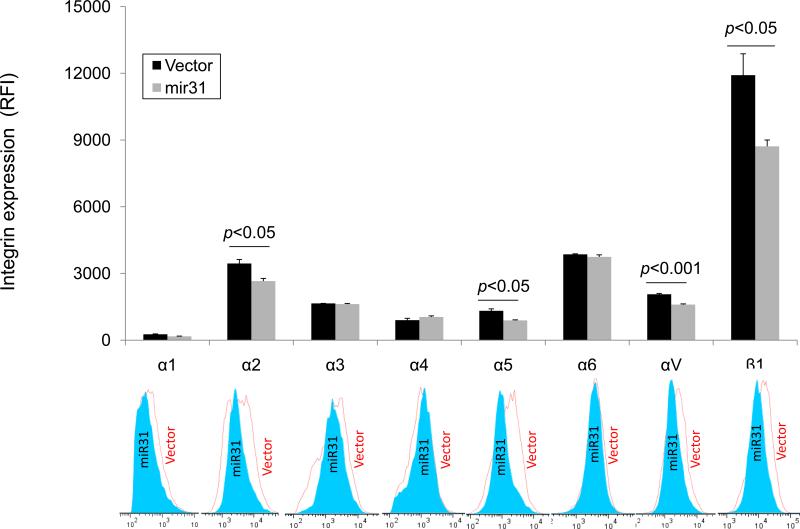
Since integrin mediated cellular responses depend upon the expression at the cell surface, as a next step, we monitored the surface expression patterns of the different α-subunits that commonly form heterodimers with β1 subunit in cells stably expressing either the empty control vector or miR-31 by flow cytometry. In accord with our qRT-PCR, semi-quantitative RT-PCR and western blot results (Fig. 1), flow cytometry analyses revealed that miR-31 significantly reduced the surface expression of α2 (p<0.05), α5 (p<0.05) and αV (p<0.001) subunits (Fig. 2). Consistent with the observed reduction in expression of β1 integrin subunit detected at the mRNA and protein levels, there was also a reduction in surface expression of the β1 subunit partner of these α subunits on the surface of the miR-31 expressing cells.
Fig. 2. Surface expression of integrin α-subunits and their β1 subunit partner is suppressed by miR-31.
Expression profiles of different integrin α subunits in MDA-MB-231 cells overexpressing GFP-pBABE vector control or GFP-miR-31. Surface expression of different α subunit partners of the β1 integrin subunit is quantitated as the relative fluorescence intensity (RFI) with subunit specific mAbs by flow cytometry. Values are means± S.E. and are representative of 3 independent experiments. Representative histograms are provided below in which the empty histograms are for the vector control and the filled histograms are for miR-31 overexpressing cells.
β3 integrin is a target of miR-31
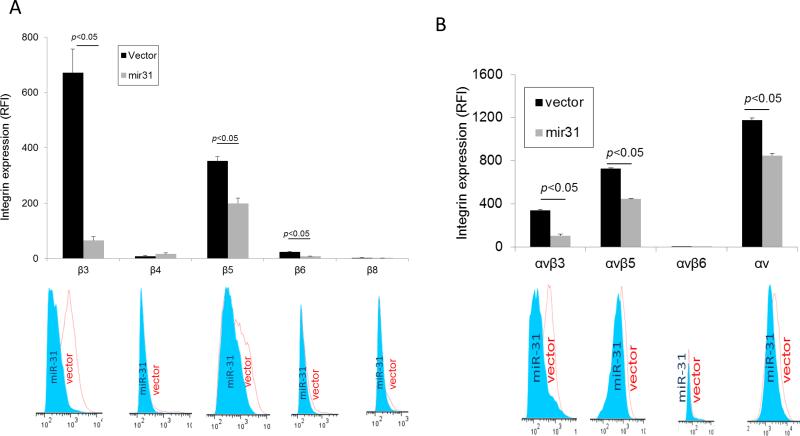
αV integrin subunit is known to complex with several β integrins, including β1, β3, β5 and β6 integrins. Since our results show miR-31 can affect β1 indirectly, we evaluated whether other common partners of αV, β3, β5, β6 and β8, were also targeted by miR-31. Only one of these 4 integrin β subunits, the 3’ UTR of β3, was found to harbor the seed sequence that could be targeted by miR-31 (Fig. 1). Consistent with this prediction, we found β3 surface levels were significantly reduced (p<0.05) in cells expressing high levels of miR-31 compared to vector control (Fig. 3A), which is concordant with the RT-PCR and western blotting analyses (Fig. 1). Surface expression of β5 subunit was also reduced in the presence of miR-31, probably as an indirect effect since there is no seed sequence of miR-31 in the 3’UTR of β5 (Fig. 3A). Reduced expression levels of either αV or β3 subunits resulted in a significant reduction of cell surface expression of the αVβ3 and the αVβ5 heterodimers using antibodies that specifically recognize these integrins heterodimers (Fig. 3B). Thus, miR-31 can simultaneously regulate the expression of multiple α and β integrin subunits in a cellular environment.
Fig. 3. Expression profile of different integrin β subunits in MDA-MB-231 cells expressing and GFP-miR-31 or control GFP-pBABE.
(A) Surface expression of different β integrin subunits (other than β1) as determined by flow cytometry with representative histograms given below. Empty histogram is for vector and filled histogram is for miR-31 overexpressing cells. Values are means ± S.E. and are representative of 3 independent experiments. (B). Surface levels of αVβ3, αVβ5 and αVβ6 reveal differences between vector and miR-31 cells for αVβ3 and αVβ5 integrins. The integrin αV level has been used as the control. Empty histogram is for vector and filled histogram is for miR-31 overexpressing cells. Values are means± S.E. and are representative of 3 independent experiments.
Reduced expression of α2, α5 and αV subunits by miR-31 results in decreased expression of activated β1 integrin on the cell surface
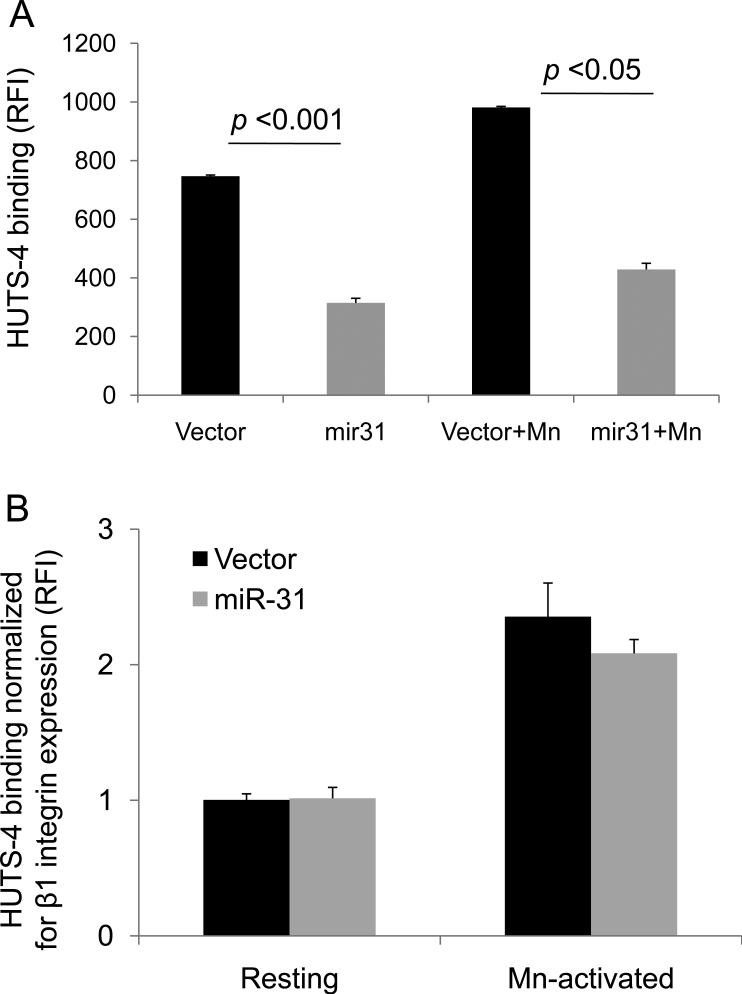
Based on the above results, we assessed whether miR-31-mediated suppression also affected the activation state of integrins expressed on the surface of the MDA-MB-231 cells. This analysis was performed using HUTS-4, a mAb that recognizes the active conformation of β1 integrins upon dimerization with one of its α subunit partner 27. miR-31 transfectants showed a significantly reduction in their capacity to spontaneously bind HUTS-4 compared to vector control (p<0.001) , and this difference remained significant in the presence of Mn2+ (p<0.05), which activates integrins upon binding to their extracellular domain (Fig 4A). However, this decrease in HUTS-4 binding was proportional to the decrease in the surface expression of β1 integrins in these cells (Fig. 4B). Thus, when the activation status of these integrins is expressed as a ratio of the total β1 surface expression, measured in HUTS-4 binding, no significant differences were found between the vector or miR-31 expressants in either unstimulated (resting) cells or cells in which integrins were activated with Mn2+ (Fig. 4B).
Fig. 4. Targeting of α2, α5 and αV subunits by miR-31 decreases β1 integrin activation.
MDA-MB-231 cells stably overexpressing GFP-miR-31 show reduced β1 integrin activation due to a quantitative reduction in β1 integrin surface expression. (A) Activation of β1 integrins was monitored with HUTS-4 antibody, specific for activated β1 integrins, by flow cytometry. Spontaneous HUTS-4 binding or that induced by MnCl2 are shown. Cells were either left untreated or treated with 3.5mM MnCl2 and incubated with HUTS-4 antibody followed by antimouse Alexa Fluor 647 IgG to monitor β1 integrin activation status. (B) HUTS-4 binding normalized to total β1 integrin expression measured with mAb MAB2000 (Millipore) on control vector- or the miR-31-expressing cells in the resting and Mn2+-activated states. HUTS-4 binding was done as in (A) and integrin expression was monitored using the same protocol as for HUTS-4 staining, using the expression-specific β1 antibody. Values in both panels are means±S.E. and representative of 3 independent experiments.
miR-31 directly targets the 3’UTR of ITGA2, ITGA5, ITGAV and ITGB3 integrins and represses their expression
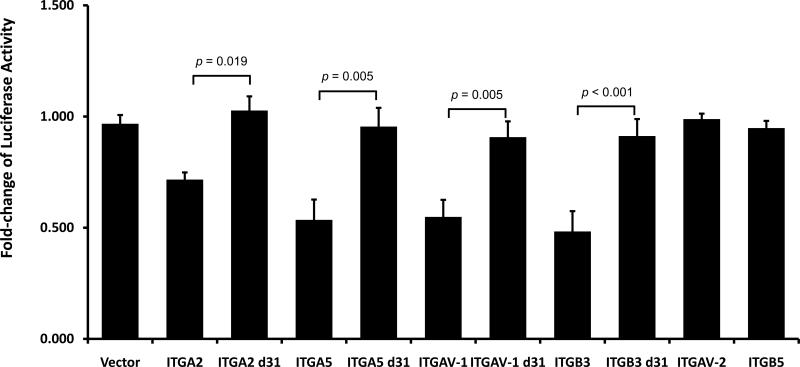
We used the Firefly-Renilla dual luciferase reporter gene assay to demonstrate that the posttranscriptional repression of the α2, α5, αV and β3 integrin subunits is a consequence of direct binding of miR-31 to target seed sequences within the 3’UTR of the transcripts for these integrin subunits. A DNA fragment, ~350 bp-long from the 3’UTR sequence of these integrins encompassing the miR-31 seed sequence, was subcloned within the 3’UTR of the firefly luciferase gene into the pmirGlo vector, and luciferase activity was measured in MDA-MB-231 breast cancer cells, which express very low or no levels of miR-31. For each construct, firefly luciferase activity was normalized to renilla luciferase activity and the ratio of miR-31 compared to control miR treatment was calculated. We found no difference in luciferase activity between MDA-MB-231 cells transfected with control vector (Glo) alone or with control vector cotransfected with either miR-31 or control microRNA (Fig. S4), demonstrating the lack of a miR-31 effect in the absence of its specific target sequence. In contrast, in cells transfected with the individual integrin 3’UTR-containing vector, overexpression of miR-31 resulted in a significant reduction in luciferase activity, ranging from 30 to 55% depending on the integrin subunit compared to the cells transfected with the same vector in the absence of miR-31 or cotransfected with control microRNA in MDA-MB-231 (Fig. 5 and S4). As a control to confirm that miR-31 is specifically targeting and binding seed sequences within these 3’UTR, we deleted the seed sequence of miR-31 in the 3’UTR of α2, α5, αV and β3 integrin subunits. This maneuver abrogated the effect of the exogenous miR-31 on these integrin subunit reporters in the MDA-MB-231 cells (Fig. 5 and Fig. S4). Our in silico analyses also found a second potential seed sequence of miR-31 in the 3’UTR of ITGAV (ITGAV-2, Fig. 5), which only partially aligned to the miR-31 target sequence. However, this second seed sequence was insufficient to be regulated by miR-31; its presence had no effect on luciferase activity. As a negative control for the effect of miR-31 on the luciferase activity, the 3’UTR of ITGB5 which does not contain a seed sequence for miR-31, showed no different in renilla luciferase activity in the low or high miR-31 expressing cells. The luciferase assay for ITGB1 could not be performed because of the large size (more than 1200 bp) of its 3’UTR. Similar results were obtained when these experiments were repeated in the LNCaP prostate cancer cells (Fig. S5); namely, miR-31 repressed expression of the genes encoding for α2, α5, αV and β3 integrin subunits but did not affect β5 expression. These results demonstrate that the effect of miR-31 is not restricted to a single cell line but appears to extend to other cancers and show that miR-31 specifically targets and represses several integrin subunits simultaneously.
Fig. 5. miR-31 directly targets the 3’UTR of ITGA2, ITGA5, ITGAV and ITGB3 integrins and represses their expression in MDA-MB-231 cells.
MDA-MB-231 cells were transfected with the luciferase reporter plasmids containing the 3’UTR of the indicated integrin subunit or with deletion of the miR31 seed sequences, and co-transfected with or without miR31. Luciferase activities were measured after 24 h. For all luciferase activity assays, renilla luciferase activity was used for normalization. The normalized firefly luciferase activity of each construct was plotted as a ratio of the control microRNA over the miR-31 treatment. The data are the mean ± s. d. of at least 3 independent transfections.
miR-31 affects cell spreading in a ligand-dependent manner
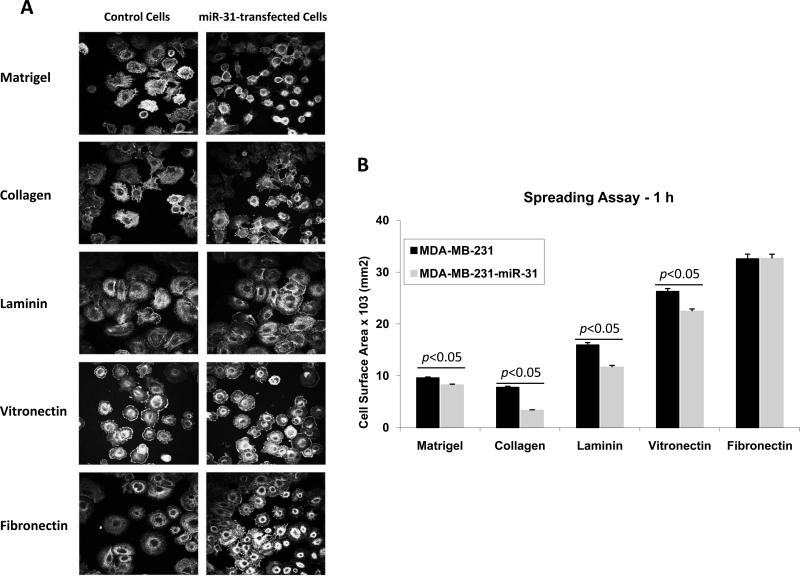
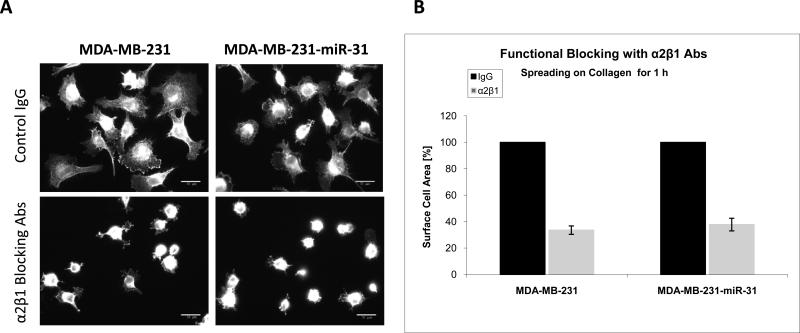
Since miR-31 overexpression leads to diminished expression of β1 integrin and several of its α subunits partners on the cell surface (Fig. 2), we anticipated that specific integrin-mediated cellular responses might be suppressed. Accordingly, we plated control or miR-31-MDA-MB-231 cells on different ECM ligands that are recognized by various β1-contaning integrins and examined cell spreading. The cells were allowed to spread for 30 or 60 min on collagen (an α2β1 ligand), fibronectin (an α5β1 ligand) or laminin (an α6β1 ligand) or Matrigel (containing mostly laminin and collagen IV, ligands for α1β1, α2β1, α3β1 and α6β1 integrins 28). At the two selected time points, cells were fixed and stained with phalloidin to visualize actin (Fig. 6A). We found a striking difference in cell size and actin organization between control cells and miR-31-expressing cells on collagen and Matrigel after 1 hour (Fig. 6, upper two panels). The effect of miR-31 on the ability of cells to spread on these two substrates was also observed after 30 min (Fig S6) but became more pronounced after 1 hour. While the control cells became large and fully spread, and developed extensive stress fibers and lamellipodia at the cell periphery, the miR-31 expressing cells remained small and round on these substrata. In control experiments, a blocking mAb (LS-C247646) to α2β1 completely inhibited spreading of both the parental and the miR-31 expressing MDA-MB-231 cells on collagen (Fig. 7), but not on Matrigel (not shown), suggesting the roles of not only α2β1 but also other integrins that were suppressed by miR-31 expression. The difference in cell spreading between the control cells and the miR-31 expressing cells was also evident on laminin and vitronectin, but no difference was observed when cells were plated on fibronectin (Fig. 6, bottom panels).
Fig. 6. miR-31 affects cell spreading in a ligand-dependent manner.
Wild-type and miR-31-expressing MDA-MB-231 cells spread on collagen, Matrigel, fibronectin and laminin for 1 h, were fixed, permeabilized and stained for 30 min with Alexa 568-phalloidin to visualize actin filaments. (A) In comparison to vector control cells, miR-31-transfected cells showed significant inhibition of spreading and actin organization on collagen and Matrigel; lesser perturbation on laminin, and no difference on fibronectin. The images shown are representative micrographs of 12-15 independent scans for each integrin ligand. Bar = 40 μm. (B) Quantitation of cell spreading presented as the total surface area covered by cells, in square micrometer, averaged to 300 cells.
Fig. 7. A function blocking mAb to α2β1 completely inhibits cell spreading in both the control cells and the miR31-expressing MDA-MB-231 cells.
Wild-type and miR-31-expressing MDA-MB-231 cells where incubated with either an α2β1 monoclonal blocking antibody (LS-C247646) or control IgG and spread on collagen for 1 h. Cells were then fixed, permeabilized and stained for 30 min with Alexa 568-phalloidin to visualize actin filaments. A. The images shown are representative micrographs of 12-15 independent scans for each treatment. Bar = 40 μm. B. Quantitation of cell spreading presented as the total surface area covered by cells, in square micrometer, from 300 cells.
DISCUSSION
Cell adhesion, spreading and migration on ECM proteins are critical processes for both physiological responses, such as organ development, cell-mediated immunity and wound healing, and pathological responses, such as cancer progression and metastasis. Cell adhesion and spreading are triggered by cell contact with ECM proteins such as collagen, laminin, fibronectin, vitronectin and fibrinogen. Physiological regulation of cell-matrix interactions is mediated by cell-to-ECM (inside-out) and ECM-to-cell (outside-in) signals that are themselves transmitted by various adhesion receptors on the cell surface, including integrins28-31. The β1 integrin subunit, in association with several different α integrin subunits, serve as the principal receptors for type I collagen, the most abundant extracellular matrix protein in mammals 28. Binding of β1 integrin to collagen results in downstream rearrangements of the actin cytoskeleton by providing a scaffold for cytoskeletal proteins and multiple signaling molecules that are involved in the regulation of cell adhesion and spreading 32. In this report we identify the regulation of expression and activity of multiple integrins by miR-31 as a mechanism for the modulation of cancer cell spreading. We and others have previously reported the targeting of integrin α5 subunit by miR-31 17, 18. In this report, we show that, in addition to α5, miR-31 directly targets and represses the mRNAs and proteins of two other integrin alpha subunits (α2 and αV). This repression is mediated by recognition of a conserved seed sequence of miR-31 in the 3’UTRs of the respective target transcripts. The suppression of these integrin alpha subunits by miR-31 was observed in a series of in vitro molecular and cellular assays, as well as independent experiments to verify that miR-31 regulates the expression and activity of these integrins subunits. First, we showed that overexpression of miR-31 in MDA-MB-231 breast cancer cells, which has low endogenous levels of miR-31, resulted in a significant decrease in the expression levels of multiple alpha integrins subunits along with the β1 subunit, using both real time qt-RT-PCR and western blotting analyses. Second, we used a luciferase reporter gene assay to demonstrate that the miR-31 mediated suppression of these integrins subunits arises from direct and specific targeting of a conserved miR-31 seed sequence in their 3’UTRs, and that mutation of the seed sequence abrogates the effect of miR-31. Targeting of these α subunits by miR-31 is consistent with indirect repression of the β1 integrin subunit which associates with α2, α5 and αV partners to form functional integrin heterodimers. The expression of α1, α3, α4 and α6 integrins remained similar between the vector and miR-31 expressing cells, showing that the effect of miR-31 is selective for specific α integrins. Interestingly, although analysis of β1 integrin 3’UTR revealed it lacks a seed sequence for miR-31, it was significantly (p<0.05) reduced at the mRNA and protein levels, which further resulted in a significant reduction in its cell surface expression. Since there was no compensatory increase in the levels of the α1, α3, α4 and α6 integrins in the miR-31 transfectants, it can be expected that there is a net decrease in the number of available α integrin subunits for β1 to complex within these cells. Third, we showed that loss of mRNA and protein expression of these integrins downstream of miR-31 resulted in loss of both surface expression as well as activation as demonstrated by flow cytometric analyses with integrin subunit specific mAbs and the β1 activation-specific mAb, HUTS4. Fourth, and importantly, we provided evidence that the miR-31 mediated loss of α2, α5 and αV integrins, and their β1 partner, and β3 integrins resulted in a significant inhibition of cell spreading in a ligand-dependent manner. The inhibition in cell spreading could reflect reduction in surface expression of total or activated integrins.
This study, together with the previously published reports from both our group 17and others 16, 18 establish the important role of miR-31 in regulating the different aspect of cell adhesion, spreading and cancer cell invasion and metastasis and solidifies its role a master metastasis suppressor gene 17, 18. Natural regulation of cancer metastasis is mediated by cancer metastasis suppressor genes which either prevent the progression of tumor cells to a metastatic phenotype or reverse the metastatic phenotype. miR-31 is one of these metastasis suppressor genes, whose pleiotropic functions regulate multiple steps of the invasion-metastasis cascade by targeting a cohort of pro-metastatic targets genes all of which are known to play an important role in metastasis, including integrins. The pleiotropic effect of miR-31 is of potential interest from a translational perspective since modulation of a single miRNA can affect the function of several genes involved in cancer metastasis. Thus, these data reinforce the important role of miR-31 in the regulation of the cancer invasion-metastasis cascade and identify miR-31 as an attractive therapeutic target for blunting cancer metastasis.
Supplementary Material
Supplemental Figure legends:
Fig. S1. miR31 overexpression did not affect cell proliferation. Equal numbers of cells were seeded in 6-well plates. At each day, the number of viable cells was counted. Each experiment was performed in triplicates, and the data are representative of three independent experiments.
Fig. S2. Densitometry and statistical analyses of western blot data.
Semi-quantitative digital image analysis was performed with ImageJ software to detect the intensity of integral density of particular bands. The pixel density of each individual protein was calculated on the basis of the pixel density for the β-actin as a housekeeping control protein, the values of which were equal to 1. The plotted data are the mean ± s. d. of at least 3 independent western blots. * Student TTest determined that the p < 0.05 when compared to either the control or the GFP cells.
Fig. S3. miR-31 inhibits expression of several alpha and beta integrin subunits in a prostate cancer cell line. Real-time qt RT-PCR of the indicated integrin subunits from untransfeted LNCaP cells or LNCaP cells transfected of either control microRNA or miR-31.
Fig. S4. miR-31 directly targets the 3’UTR of several integrins subunits and represses their expression. (A) Firefly luciferase reporter plasmid, pmirGlo was transiently co-transfected into MDA-MB-231 cells with either miR-31 of the control miR, and luciferase activities were measured after 24 h. (B) Firefly luciferase reporter plasmids, pmirGlo control or pmirGlo containing the 3’UTR of each indicated integrin subunit, were transfected into MDA-MB-231 cells that were transiently co-transfected with either miR-31 or control miR, and luciferase activities were measured after 24 h. The values were plotted as a ratio of the control microRNA over the miR31 treatment. (C-F) Mutation of miR-31 seed sequences in the 3’UTR of the indicated integrins subunits abrogated the miR-31 effect. MDA-MB-231 cells were transfected with the luciferase reporter plasmids containing the 3’UTR of the indicated integrin subunit with deletion of the miR31 seed sequences, and co-transfected with or without miR31. Luciferase activities were measured after 24 h. For all luciferase activity assays, Renilla luciferase activity was used for normalization. The data are the mean ± s. d. of at least 3 independent transfections.
Fig. S5. miR-31 directly targets the 3’UTR of ITGA2, ITGA5, ITGAV and ITGB3 integrins and represses their expression in LNCaP cells. The firefly luciferase activity of each construct was normalized to the internal control renilla luciferase activity and plotted a ratio of the control microRNA over the miR31 treatment.
Fig. S6. miR-31 affects cell spreading in a ligand-dependent manner. Wild-type and miR-31-expressing MDA-MB-231 cells spread on collagen, Matrigel, fibronectin and laminin for 30 min, were fixed, permeabilized and stained for 30 min with Alexa 568-phalloidin to visualize actin filaments. miR-31-transfected cells showed significant inhibition of spreading on collagen and Matrigel, less extensive inhibition on laminin and no inhibition on fibronectin, compared to control cells expressing the empty vector. Quantitation of cell spreading presented as the total surface area covered by cells, in square micrometer, from an average of 300 cells.
Acknowledgments
We thank Tim Burke for proofreading the manuscript. We also thank members of the flow cytometry core with assistance with flow cytometry analyses.
Grant Support.
DOD Breast Cancer Idea Award grant W81XWH0810236 (K. Sossey-Alaoui), NIH grants P01HL073311 and P50HL077107 (E.F. Plow). K. Augoff is funded in part by a research fellowship “Development program of Wroclaw Medical University” from the European Social Fund, Human Capital, national Cohesion Strategy” (contract # UDA-POKL.04.01.01-00-010/08-00)”.
REFERENCES
- 1.Johnson MS, Lu N, Denessiouk K, Heino J, Gullberg D. Integrins during evolution: evolutionary trees and model organisms. Biochim Biophys Acta. 2009 April;1788(4):779–89. doi: 10.1016/j.bbamem.2008.12.013. [DOI] [PubMed] [Google Scholar]
- 2.Humphries JD, Byron A, Humphries MJ. Integrin ligands at a glance. J Cell Sci. 2006 October 1;119(Pt 19):3901–3. doi: 10.1242/jcs.03098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavallaro U, Dejana E. Adhesion molecule signalling: not always a sticky business. Nat Rev Mol Cell Biol. 2011 March;12(3):189–97. doi: 10.1038/nrm3068. [DOI] [PubMed] [Google Scholar]
- 4.Schmidt S, Friedl P. Interstitial cell migration: integrin-dependent and alternative adhesion mechanisms. Cell Tissue Res. 2010 January;339(1):83–92. doi: 10.1007/s00441-009-0892-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer. 2010 January;10(1):9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danen EH, van RJ, Franken W, et al. Integrins control motile strategy through a Rho-cofilin pathway. J Cell Biol. 2005 May 9;169(3):515–26. doi: 10.1083/jcb.200412081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mierke CT, Frey B, Fellner M, Herrmann M, Fabry B. Integrin alpha5beta1 facilitates cancer cell invasion through enhanced contractile forces. J Cell Sci. 2011 February 1;124(Pt 3):369–83. doi: 10.1242/jcs.071985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabodi S, del PC- L, Di SP, Defilippi P. Integrin signalling adaptors: not only figurants in the cancer story. Nat Rev Cancer. 2010 December;10(12):858–70. doi: 10.1038/nrc2967. [DOI] [PubMed] [Google Scholar]
- 9.Tabatabai G, Weller M, Nabors B, et al. Targeting integrins in malignant glioma. Target Oncol. 2010 September;5(3):175–81. doi: 10.1007/s11523-010-0156-3. [DOI] [PubMed] [Google Scholar]
- 10.Arsanious A, Bjarnason GA, Yousef GM. From bench to bedside: current and future applications of molecular profiling in renal cell carcinoma. Mol Cancer. 2009;8:20. doi: 10.1186/1476-4598-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005 June 9;435(7043):834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 12.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006 April;6(4):259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 13.Korpal M, Kang Y. The emerging role of miR-200 family of microRNAs in epithelial mesenchymal transition and cancer metastasis. RNA Biol. 2008 July;5(3):115–9. doi: 10.4161/rna.5.3.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peter ME. Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle. 2009 March 15;8(6):843–52. doi: 10.4161/cc.8.6.7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spaderna S, Brabletz T, Opitz OG. The miR-200 family: central player for gain and loss of the epithelial phenotype. Gastroenterology. 2009 May;136(5):1835–7. doi: 10.1053/j.gastro.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 16.Valastyan S, Weinberg RA. Roles for microRNAs in the regulation of cell adhesion molecules. J Cell Sci. 2011 April 1;124(Pt 7):999–1006. doi: 10.1242/jcs.081513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sossey-Alaoui K, Downs-Kelly E, Das M, Izem L, Tubbs R, Plow EF. WAVE3, an actin remodeling protein, is regulated by the metastasis suppressor microRNA, miR-31, during the invasion-metastasis cascade. Int J Cancer. 2011;129(6):1331–43. doi: 10.1002/ijc.25793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valastyan S, Reinhardt F, Benaich N, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009 June 12;137(6):1032–46. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Ithychanda SS, Das M, Ma YQ, et al. Migfilin, a molecular switch in regulation of integrin activation. J Biol Chem. 2009 February 13;284(7):4713–22. doi: 10.1074/jbc.M807719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sossey-Alaoui K, Bialkowska K, Plow EF. The miR200 family of microRNAs regulates WAVE3-dependent cancer cell invasion. J Biol Chem. 2009 November 27;284(48):33019–29. doi: 10.1074/jbc.M109.034553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sossey-Alaoui K, Li X, Ranalli TA, Cowell JK. WAVE3-mediated cell migration and lamellipodia formation are regulated downstream of phosphatidylinositol 3-kinase. J Biol Chem. 2005 June 10;280(23):21748–55. doi: 10.1074/jbc.M500503200. [DOI] [PubMed] [Google Scholar]
- 22.Sossey-Alaoui K, Ranalli TA, Li X, Bakin AV, Cowell JK. WAVE3 promotes cell motility and invasion through the regulation of MMP-1, MMP-3, and MMP-9 expression. Exp Cell Res. 2005 August 1;308(1):135–45. doi: 10.1016/j.yexcr.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 23.Sossey-Alaoui K, Safina A, Li X, et al. Down-Regulation of WAVE3, a Metastasis Promoter Gene, Inhibits Invasion and Metastasis of Breast Cancer Cells. Am J Pathol. 2007 Jun;170(6):2112–21. doi: 10.2353/ajpath.2007.060975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sossey-Alaoui K, Li X, Cowell JK. c-Abl-mediated phosphorylation of WAVE3 is required for lamellipodia formation and cell migration. J Biol Chem. 2007 September 7;282(36):26257–65. doi: 10.1074/jbc.M701484200. [DOI] [PubMed] [Google Scholar]
- 25.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 26.Bialkowska K, Ma YQ, Bledzka K, et al. The integrin co-activator Kindlin-3 is expressed and functional in a non-hematopoietic cell, the endothelial cell. J Biol Chem. 2010 June 11;285(24):18640–9. doi: 10.1074/jbc.M109.085746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luque A, Gomez M, Puzon W, Takada Y, Sanchez-Madrid F, Cabanas C. Activated conformations of very late activation integrins detected by a group of antibodies (HUTS) specific for a novel regulatory region (355-425) of the common beta 1 chain. J Biol Chem. 1996 May 10;271(19):11067–75. doi: 10.1074/jbc.271.19.11067. [DOI] [PubMed] [Google Scholar]
- 28.Barczyk M, Carracedo S, Gullberg D. Integrins. Cell Tissue Res. 2010 January;339(1):269–80. doi: 10.1007/s00441-009-0834-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo BH, Springer TA. Integrin structures and conformational signaling. Curr Opin Cell Biol. 2006 October;18(5):579–86. doi: 10.1016/j.ceb.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plow EF, Haas TA, Zhang L, Loftus J, Smith JW. Ligand binding to integrins. J Biol Chem. 2000 July 21;275(29):21785–8. doi: 10.1074/jbc.R000003200. [DOI] [PubMed] [Google Scholar]
- 31.Qin J, Vinogradova O, Plow EF. Integrin bidirectional signaling: a molecular view. PLoS Biol. 2004 June;2(6):e169. doi: 10.1371/journal.pbio.0020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stossel TP, Condeelis J, Cooley L, et al. Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol. 2001 February;2(2):138–45. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure legends:
Fig. S1. miR31 overexpression did not affect cell proliferation. Equal numbers of cells were seeded in 6-well plates. At each day, the number of viable cells was counted. Each experiment was performed in triplicates, and the data are representative of three independent experiments.
Fig. S2. Densitometry and statistical analyses of western blot data.
Semi-quantitative digital image analysis was performed with ImageJ software to detect the intensity of integral density of particular bands. The pixel density of each individual protein was calculated on the basis of the pixel density for the β-actin as a housekeeping control protein, the values of which were equal to 1. The plotted data are the mean ± s. d. of at least 3 independent western blots. * Student TTest determined that the p < 0.05 when compared to either the control or the GFP cells.
Fig. S3. miR-31 inhibits expression of several alpha and beta integrin subunits in a prostate cancer cell line. Real-time qt RT-PCR of the indicated integrin subunits from untransfeted LNCaP cells or LNCaP cells transfected of either control microRNA or miR-31.
Fig. S4. miR-31 directly targets the 3’UTR of several integrins subunits and represses their expression. (A) Firefly luciferase reporter plasmid, pmirGlo was transiently co-transfected into MDA-MB-231 cells with either miR-31 of the control miR, and luciferase activities were measured after 24 h. (B) Firefly luciferase reporter plasmids, pmirGlo control or pmirGlo containing the 3’UTR of each indicated integrin subunit, were transfected into MDA-MB-231 cells that were transiently co-transfected with either miR-31 or control miR, and luciferase activities were measured after 24 h. The values were plotted as a ratio of the control microRNA over the miR31 treatment. (C-F) Mutation of miR-31 seed sequences in the 3’UTR of the indicated integrins subunits abrogated the miR-31 effect. MDA-MB-231 cells were transfected with the luciferase reporter plasmids containing the 3’UTR of the indicated integrin subunit with deletion of the miR31 seed sequences, and co-transfected with or without miR31. Luciferase activities were measured after 24 h. For all luciferase activity assays, Renilla luciferase activity was used for normalization. The data are the mean ± s. d. of at least 3 independent transfections.
Fig. S5. miR-31 directly targets the 3’UTR of ITGA2, ITGA5, ITGAV and ITGB3 integrins and represses their expression in LNCaP cells. The firefly luciferase activity of each construct was normalized to the internal control renilla luciferase activity and plotted a ratio of the control microRNA over the miR31 treatment.
Fig. S6. miR-31 affects cell spreading in a ligand-dependent manner. Wild-type and miR-31-expressing MDA-MB-231 cells spread on collagen, Matrigel, fibronectin and laminin for 30 min, were fixed, permeabilized and stained for 30 min with Alexa 568-phalloidin to visualize actin filaments. miR-31-transfected cells showed significant inhibition of spreading on collagen and Matrigel, less extensive inhibition on laminin and no inhibition on fibronectin, compared to control cells expressing the empty vector. Quantitation of cell spreading presented as the total surface area covered by cells, in square micrometer, from an average of 300 cells.