Abstract
Platelet derived growth factor B (PDGF-B) and its receptor, PDGFR-β, play a critical role in pericyte maturation; however, the mechanisms by which PDGF-B is up-regulated in the tumor microenvironment remain unclear. We previously demonstrated that up-regulating stromal-derived factor, SDF-1α, in vascular endothelial growth factor (VEGF165)-inhibited Ewing’s sarcoma tumors (TC/siVEGF7-1) induced PDGF-B mRNA expression, increased infiltration and differentiation of bone marrow cells (BMCs) into pericytes and, rescued tumor growth. The purpose of this study was to investigate the mechanism by which SDF-1α increased PDGF-B expression and the role of this pathway in BM-derived pericyte differentiation. We demonstrated that SDF-1α induced expression of PDGF-B mRNA and protein both in vitro and in vivo. In contrast, inhibiting SDF-1α down-regulated PDGF-B. We cloned the 2-kb pdgf-b promoter fragment and showed that SDF-1α activates PDGF-B via a transcriptional mechanism. Chromatin immunoprecipitation demonstrated that the ELK-1 transcription factor binds to the pdgf-b promoter in response to SDF-1α. We confirmed the correlation between the SDF-1α/PDGF-B pathway and the differentiation of PDGFR-β+ BMCs into mature pericytes using an in vitro assay. These findings demonstrate that SDF-1α regulates PDGF-B expression and that this regulation plays a critical role in the differentiation of PDGFR-β+ BMCs into mature pericytes.
Introduction
Platelet derived growth factor B (PDGF-B) is a member of the PDGF family of growth factors. When synthesized and secreted in its homodimer form, PDGF-BB binds to its tyrosine kinase receptor, PDGFR-β1. Both PDGF-B and PDGFR-β are mainly expressed in the developing vasculature2. Pericytes are recruited by PDGF-B-expressing endothelial cells to remodel, stabilize and mature the new vascular tube3. Normally, the expression of PDGF-B is restricted to a limited number of cell types; however, many human tumor cells have been shown to overexpress PDGF-B4, 5. Although the role of PDGF-B and its receptor in the maturation process of pericytes has been described, the mechanisms by which PDGF-B expression can be induced in areas of vessel maturation remain unclear1, 2, 4.
PDGFR-β-expressing pericyte progenitor cells (PPPs) differentiate into NG2+, desmin+, and α-SMA+ pericytes within tumors3. Mature pericytes play a role in vascular stabilization, maturation, and survival and surround endothelial cells, which together form the basement membrane of the microvessels6. Mature pericytes contribute to efficient vascular flow and prevent vascular leakage3. As PDGF-B plays a critical role in the differentiation process for pericytes, understanding the pathways that regulate PDGF-B in the tumor microenvironment will enable the identification and development of agents that interfere with pericyte maturation which results in vascular reduction during tumor vascular development7. Several studies have shown that targeting pericytes in addition to endothelial cells is more effective at inhibiting tumor growth than targeting one or the other alone8, 9.
PDGF-B expression can be regulated by several mechanisms, including transcriptional regulation10. Previous studies by our group demonstrated that treating vascular endothelial growth factor (VEGF165) - inhibited Ewing’s sarcoma tumors (TC/siVEGF7-1) with an adenoviral vector containing the stromal-derived factor gene (Ad-SDF-1α) increased PDGF-B mRNA levels, increased infiltration and differentiation of bone marrow cells (BMCs) into pericytes, and rescued tumor growth11, 12. The purpose of the current study was to investigate the mechanism by which SDF-1α induced the expression of PDGF-B and examine the role of the SDF-1α/PDGF-B pathway in the differentiation of BMCs into pericytes.
SDF-1α or CXCL12 is an alpha chemokine that binds to the 7-transmembrane G-protein coupled receptor CXCR413, 14. Signaling through the SDF-1α/CXCR4 pathway initiates multiple downstream signaling pathways linked to transcription and expression through MEK1/2 and ERK1/2. ERK can phosphorylate and activate other cellular proteins as well as translocate to the nucleus and phosphorylate and /or activate transcription factors13. The binding of SDF-1α to CXCR4 induces phosphorylation of the mitogen-activated protein kinases (MAPKs) p44 ERK-1 and p42 ERK-2, which subsequently initiates the phosphorylation of the nuclear transcription factor ELK-115. The ELK-1 nuclear transcription factor is a member of the 3 ternary complex factors (TCFs), a subfamily of the ETS-domain transcription factors. TCFs are involved in several biological processes; members of this family have been shown to be important in regulating angiogenesis and vasculogenesis16–20. We demonstrate here for the first time that SDF-1α regulates the expression of PDGF-B and that this regulation involves binding of the ELK-1 transcription factor to the pdgf-b promoter. Our data also showed that the SDF-1α/PDGF-B pathway plays a critical role in the differentiation of PDGFR-β+ BMCs into desmin and NG2 expressing mature pericytes.
Materials and methods
Cell lines and culture
We used the human Ewing’s sarcoma cell lines TC71 and VEGF165 -inhibited TC/siVEGF7-1 generated by stable transfection of VEGF165 siRNA into TC7111. TC/siVEGF7-1 cells express all the VEGF isoforms except VEGF165. Human embryonic kidney 293 (HEK293) cells and C3H/10T1/2 murine embryonic mesenchymal cells were purchased from ATCC (Manassas, VA; CRL-1573, CCL-226). TC/siVEGF7-1 and HEK293 cells express low levels of endogenous SDF-1α and PDGF-B compared with C3H/10T1/2 cells. All cells were cultured in complete Dulbecco’s modified Eagle’s medium (DMEM)12.
Proteins and antibodies
Recombinant human SDF-1α was purchased from R&D Systems (Minneapolis, MN; 350-NS-010/CF). The inactive 9SDF-1α recombinant protein used as a negative control was a generous gift from Dr. Qing Ma at The University of Texas MD Anderson Cancer Center. Rabbit polyclonal immunoglobulin (IgG) to human ELK-1 and rabbit polyclonal IgG to mouse desmin were purchased from Abcam (Cambridge, MA; ab28831, ab15-200). ChromPure rabbit control IgG was purchased from Jackson ImmunoResearch Laboratories (West Grove, PA; 011-000-003). Rabbit polyclonal IgG to mouse PDGF-BB (N-30) and rabbit polyclonal IgG to mouse PDGFR-β (P-20) were purchased from Santa Cruz Biotechnology (CA; sc-127, sc-339). Rabbit polyclonal IgG to mouse NG2 chondroitin sulfate proteoglycan was purchased from Millipore (Bedford, MA; AB5320).
q-PCR for PDGF-B
TC/siVEGF7-1 and HEK293 cells were cultured in vitro in the absence of growth factors and supplements for 8 hours and then treated with 100ng/mL of either the recombinant human SDF-1α protein or the inactive 9SDF-α inactive protein for 24 hours. RNA was collected by the TRIzol method (Invitrogen, Grand Island, NY). Briefly, the medium was removed and the cells were treated with 1mL of TRIzol for 5 minutes at room temperature (RT). The cells were collected, and 200µL of chloroform (Fisher Scientific, Pittsburgh, PA) was added for 3 minutes at RT. The mixture was then centrifuged in the Centrifuge 5415-R (Eppendorf, Westbury, NY) for 15 minutes at 4°C. The upper clear phase was collected, and 500µL of isopropanol (Fisher Scientific) was added for 10 minutes at RT. The solution was centrifuged for 10 minutes at 4°C, and the pellet was re-suspended and washed 2 times with 75% ethanol for 5 minutes at 4°C. The pellet was air-dried, re-suspended in 30µL of nuclease-free water (Promega, Sunnyvale, CA), and incubated at 65°C for 5 minutes. cDNA was synthesized using a reverse-transcription system (Promega). Real-time quantitative reverse-transcription polymerase chain reaction (q-PCR) was performed using iQ-SYBR Green Supermix with the iCycler iQ (Bio-Rad Laboratories, Hercules, CA). Primers for human pdgf-b were designed and synthesized using Integrated DNA Technologies (IDT, Coralville, IA). All sequences are available upon request.
PDGF-B immunohistochemical analysis and microscopy
To investigate the effect of SDF-1α on PDGF-B protein levels in vivo, tumor sections obtained from previous published experiments12 were evaluated for PDGF-B by immunohistochemical analysis (IHC) with the primary rabbit polyclonal antibody PDGF-B (N-30) (Santa Cruz Biotechnology, CA; sc-127). Images were captured using a Nikon Microphot-FXA microscope (Nikon Instruments, Melville, NY).
ELISA
TC/siVEGF7-1 and HEK293 cells were cultured in vitro in the absence of growth factors and supplements for 8 hours and then treated with 100ng/mL of either SDF-1α or the inactive 9SDF-1α protein for 36 hours. The supernatant was collected and concentrated using Amicon Ultra-15, PLGC Ultracel-PL Membrane, 10 kDa (Millipore) by centrifuging at 2500rpm for 30 minutes at 4°C in the Centrifuge 5804R (Eppendorf). PDGF-B protein levels were measured by enzyme-linked immunosorbent assay (ELISA) (R&D Systems, Minneapolis, MN; MBB00) according to the manufacturer’s protocol.
ShRNA knockdown of SDF-1α in C3H/10T1/2 cells
SDF-1 shRNA Plasmid (m) (sh-SDF-1) and Control shRNA Plasmid-A (sh-control) were purchased from Santa Cruz Biotechnology (sc-39368-SH, sc-108060). C3H/10T1/2 (10T1/2) cells were cultured in vitro and transfected with 1 µg of either sh-SDF-1 or sh-control using FuGENE 6 (Roche Applied Science, Indianapolis, IN) according to the manufacturer’s directions. At 48 hours after transfection, the medium was replaced with fresh DMEM and the cells were cultured under puromycin selection (2 µg/mL) (InvivoGen, San Diego, CA). RNA was isolated using the TRIzol method, and cDNA was synthesized using a reverse-transcription system (Promega). Down-regulation of SDF-1α was confirmed by q-PCR using primers for mouse SDF-1α. Mouse SDF-1α primers were designed and synthesized using Sigma-Genosys.com (Sigma-Genosys, Spring, TX). All sequences are available upon request.
pdgf-b promoter cloning
DNA was isolated from TC71 Ewing’s sarcoma cells. The 2-kb pdgf-b promoter DNA fragment was isolated by PCR. Briefly, primers were designed and synthesized using Integrated DNA Technologies (IDT). All sequences are available upon request. The PCR product was run on a 1% agarose gel, and the 2-kb pdgf-b promoter DNA fragment was isolated and purified using the QIAquick Gel Extraction Kit (QIAGEN, Valencia, CA; 28704). The purified sequence was ligated into the pCR2.1-TOPO expression vector using the TOPO TA Cloning Kit (Invitrogen, Carlsbad, CA; K4560-01). The vector was isolated and purified with the QIAprep Spin Miniprep Kit (QIAGEN; 27104) according to the manufacturer’s directions, and the DNA sequence was verified by SeqWright DNA Technology Services (Houston, TX). The2 kb pdgf-b promoter DNA fragment was then isolated and cloned into the pGL3-Basic luciferase reporter vector (Promega BioSciences, San Luis Obispo, CA; E1751). The pdgf-b/pGL3 reporter vector sequence was verified by SeqWright DNA Technology Services. To test the functionality of the pdgf-b/pGL3 construct, we used 50ng of the construct to transfect TC71 cells and the pdgf-b promoter activity was tested by measuring the luciferase signal (data not shown).
Dual-luciferase reporter assay
We used 50 ng of either the pdgf-b/pGL3 reporter construct or the pGL3-Control vector (Promega BioSciences; E1761) to transfect TC/siVEGF7-1 and HEK293 cells using FuGENE 6 (Roche Applied Science) according to the manufacturer’s directions. Beginning 48 hours after transfection, the cells were cultured in the absence of growth factors and supplements for 24hours and then treated with 100 ng/mL of either the recombinant SDF-1α or the inactive 9SDF-1α protein for 8hours. The cells were lysed and the luciferase signal measured using the Dual-Luciferase Reporter Assay System (Promega BioSciences; E1910) according to the manufacturer’s instructions.
ChIP assay
Potential ELK-1 binding sites within the 2-kb pdgf-b promoter region were identified using the GeneRegulation.com MATCH software. 2 sites at −600bp and at the transcription start site (TS) which scored > 85% were analyzed by chromatin immunoprecipitation (ChIP) (Millipore) according to the manufacturer’s instructions. Briefly, TC/siVEGF7-1 and HEK293 cells were cultured in the absence of growth factors and supplements for 24 hours and then treated with 100 ng/mL recombinant human SDF-1α for 15, 30, or 60 minutes. The cells were collected and the proteins cross-linked to the DNA by incubating in 36% formaldehyde for 10 minutes at RT. The cells were sonicated and the DNA sheered using the Sonicator 3000 (Misonix, Farmingdale, NY) to produce fragments of 400–800 bp. The lysates were pre-cleared with protein A/G agarose for 2 hours at 4°C and incubated with the anti-ELK-1 rabbit polyclonal antibody (Abcam) O/N at 4°C. The ELK-1/DNA/Ab complex was immunoprecipitated, isolated, and washed. Cros- linking was reversed with 5% NaCl at 65°C O/N. The DNA was isolated using the QIAquick PCR Purification Kit (Qiagen) and q-PCR was performed for the different ELK-1 binding sites. Primers were synthesized using Sigma-Genosys.com. All sequences are available upon request. The data was analyzed using the STATISTICA 6.0 software. The sample values minus the control values were used in one-way analysis of variance, with post hoc analysis using the Fisher least significant difference to determine significant differences. The graph represents data pooled from 3 independent trials +/− SD and is scaled from 0 to 1 with 0 representing no binding and 1 representing complete binding of ELK-1 to the promoter region of interest. P values < 0.05 were considered statistically significant.
In vitro pericyte differentiation model
Whole BMCs were isolated from nude mice by flushing their hind femurs with sterile PBS. Fresh BMCs were analyzed for PDGFR-β, desmin, and NG2 expression. Briefly, the cells were collected and then cytospun onto glass slides by centrifuging at 30 rpm for 2mins at RT in the Shandon Cytospin 2 centrifuge (Shandon Southern Products, Astmore, United Kingdom). To establish an in vitro pericyte differentiation model, BMCs were cultured in a 2-well Culture-Slide (BD Biosciences, Woburn, MA; 354102) either in complete DMEM or in conditioned medium obtained from 10T1/2, sh-SDF-1-10T1/2 or sh-control-10T1/2 cells (generated as described above). Supernatant from the corresponding cells was collected, spun down at 1200 rpm for 5 minutes at 4°C in the Centrifuge 5804R (Eppendorf) to remove any debris or floating cells, and added to the freshly collected BMCs. BMCs were kept in culture for 2 weeks and stained for the expression of PDGFR-β, desmin, and NG2 pericyte markers.
Immunocytochemistry staining
BMCs were collected and cultured as described above. The cells were fixed in cold acetone and blocked with 4% fish gelatin in PBS as previously described12. Primary antibodies for PDGFR-β, NG2 (Santa Cruz Biotechnology, Santa Cruz, CA) and desmin (Abcam, Inc. Cambridge, MA) were used at 1:500 in 4% fish gelatin. The goat-anti rabbit-Cy5 secondary antibody (Jackson ImmunoResearch Laboratories) was used at 1:1000 in 4% fish gelatin. Nuclei were stained with SytoxGreen (Invitrogen, Molecular Probes, Eugene, OR) Images were captured using a Zeiss laser confocal microscope (Carl Zeiss MicroImaging, Thornwood, NY).
Results
SDF-1α regulates the expression of PDGF-B in vitro and in vivo
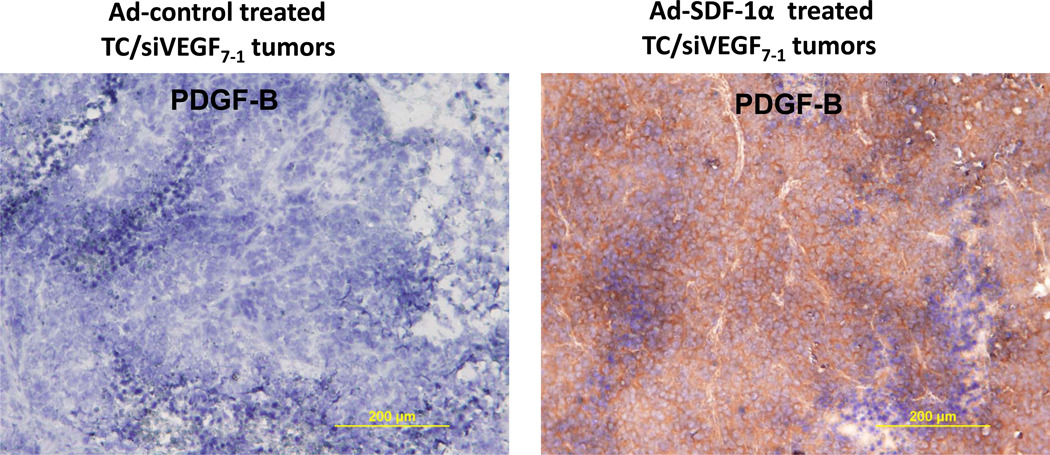
We previously showed that intratumor injections of an adenoviral vector containing SDF-1α (Ad-SDF-1α) into VEGF165 deficient tumors (TC/siVEGF7-1) increased PDGF-B mRNA levels12. To determine whether SDF-1α therapy also altered PDGF-B protein, tumor sections were obtained from the Ad-SDF-1α treated TC/siVEGF7-112 and analyzed by IHC for PDGF-B expression. PDGF-B protein levels were up-regulated in Ad-SDF-1α-treated TC/siVEGF7-1 tumors compared with Ad-control (Figure 1).
Figure 1. SDF-1α up-regulates PDGF-B protein expression in TC/siVEGF7-1 tumors.
Immunohistochemistry analysis for PDGF-B protein. TC/siVEGF7-1 tumors in vivo were injected with either Ad-SDF-1α or Ad-control 2 times weekly for 3 weeks. On day 23 after tumor cell inoculation, tumors were resected, placed in OCT, and snap-frozen in liquid nitrogen. Tumor sections were cut, fixed, and analyzed for expression of PDGF-B by immunohistochemistry (brown staining). Ad-SDF-1α up-regulates PDGF-B protein levels compared with Ad-control.
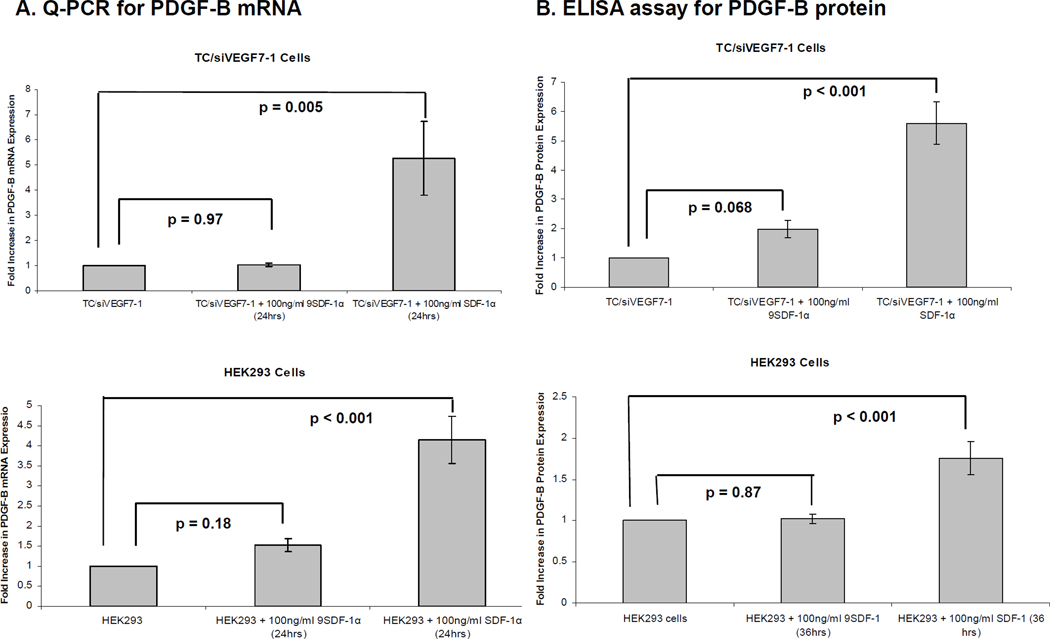
To further elucidate the correlation between SDF-1α and PDGF-B, we examined the effect of recombinant SDF-1α protein on PDGF-B mRNA levels in vitro. TC/siVEGF7-1 and non-tumorigenic HEK293 cells express CXCR4 (supplemental Figure 1). The cells were incubated with either SDF-1α or the inactive 9SDF-1α protein for 24 hours and qPCR was performed. SDF-1α up-regulated PDGF-B mRNA levels in both TC/siVEGF7-1 and HEK293 cells compared with 9SDF-1α (Figure 2A). To determine whether SDF-1α also up-regulated PDGF-B protein levels in vitro, we again treated TC/siVEGF7-1 and HEK293 cells with either SDF-1α or the inactive 9SDF-1α protein for 36hours. As PDGF-BB, the homodimer protein (~ 27 kD), is a secreted protein5, the cultured supernatant was collected and concentrated and the PDGF-B protein levels analyzed by ELISA. SDF-1α up-regulated PDGF-B protein levels compared with 9SDF-1α (Figure 2B).
Figure 2. SDF-1α up-regulates PDGF-B mRNA and protein levels in vitro.
(A) TC/siVEGF7-1 and HEK293 cells were plated, incubated in the absence of growth factors and supplements for 8 hours, and then treated with 100 ng/mL of either SDF-1α or the inactive 9SDF-1α protein for 24 hours. RNA was collected and analyzed by qPCR for the expression of PDGF-B. Each bar graph shows pooled data from 3 independent experiments, each performed in triplicate +/−SD. P < 0.05 was considered significant. (B) ELISA for PDGF-B protein levels. TC/siVEGF7-1 and HEK293 cells were plated, cultured in the absence of growth factors and supplements for 8 hours, and then treated with 100 ng/mL of either SDF-1α or the inactive 9SDF-1α protein for 36 hours. The cultured supernatant was collected and concentrated and the levels of the ~ 27 kDa soluble PDGF-B protein levels quantified by ELISA. Each bar graph shows pooled data from 3 independent experiments, each performed in triplicate, +/− SD. P < 0.05 was considered significant.
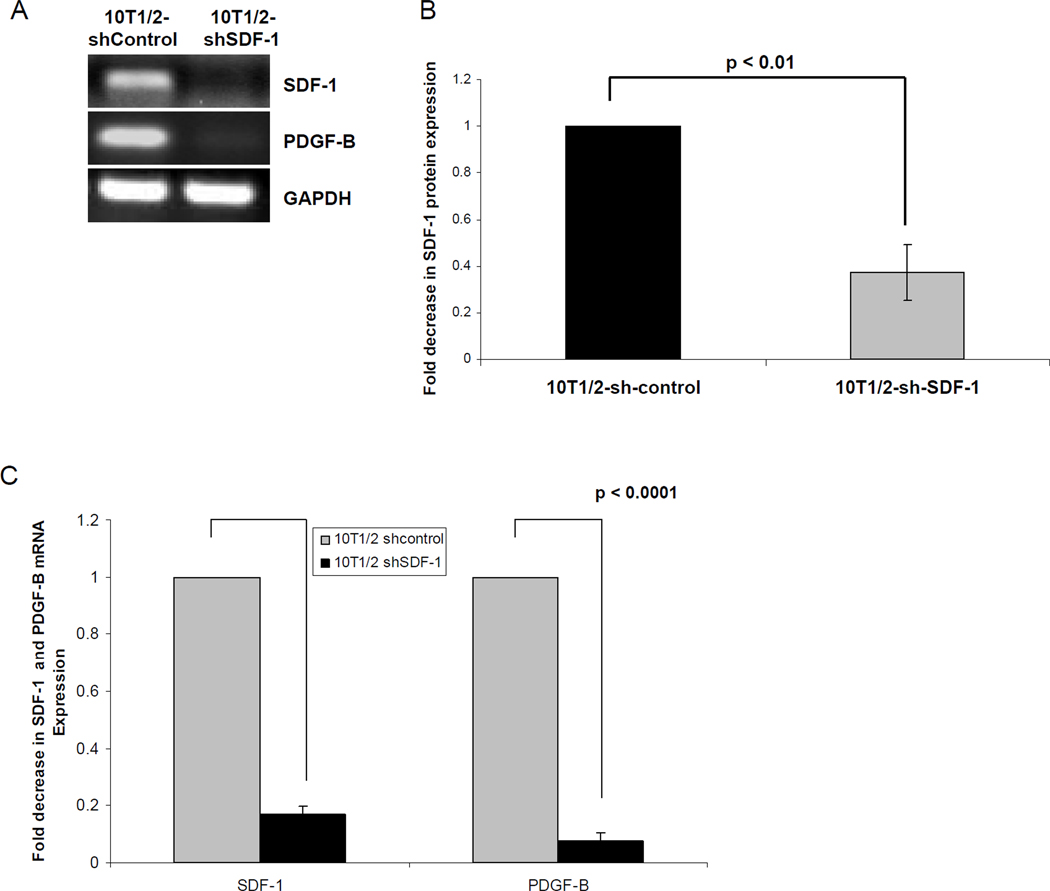
We demonstrated that SDF-1α up-regulates PDGF-B mRNA and protein levels both in vivo in TC/siVEGF7-1 tumors and in vitro in TC/siVEGF7-1 and HEK293 cells. To further confirm that SDF-1α regulates the expression of PDGF-B, we used an shRNA plasmid specific for SDF-1α (sh-SDF-1) to knockdown its expression levels in 10T1/2 cells and determine whether inhibiting SDF-1α also impacts the expression of PDGF-B. 10T1/2 cells, which endogenously express SDF-1α and PDGF-B were transfected with either the sh-SDF-1 (10T1/2-shSDF-1) or sh-control plasmid (10T1/2-shcontrol). As expected, down-regulating SDF-1α resulted in decreased expression of PDGF-B (Figure 3).
Figure 3. Inhibiting the expression of SDF-1α decreases the expression of PDGF-B.
10T1/2 cells were transfected with 1 µg of either sh-SDF-1 (10T1/2-shSDF-1) or sh-control (10T1/2-sh-control) using FuGENE 6. RNA was collected and analyzed by (A) PCR and (C) qPCR for the expression of SDF-1α and PDGF-B. (B) ELISA assay for SDF-1α protein expression. The bar graph shows pooled data from 3 independent experiments, each performed in triplicate, +/− SD. P < 0.05 was considered significant.
SDF-1α regulates the expression of PDGF-B via transcription
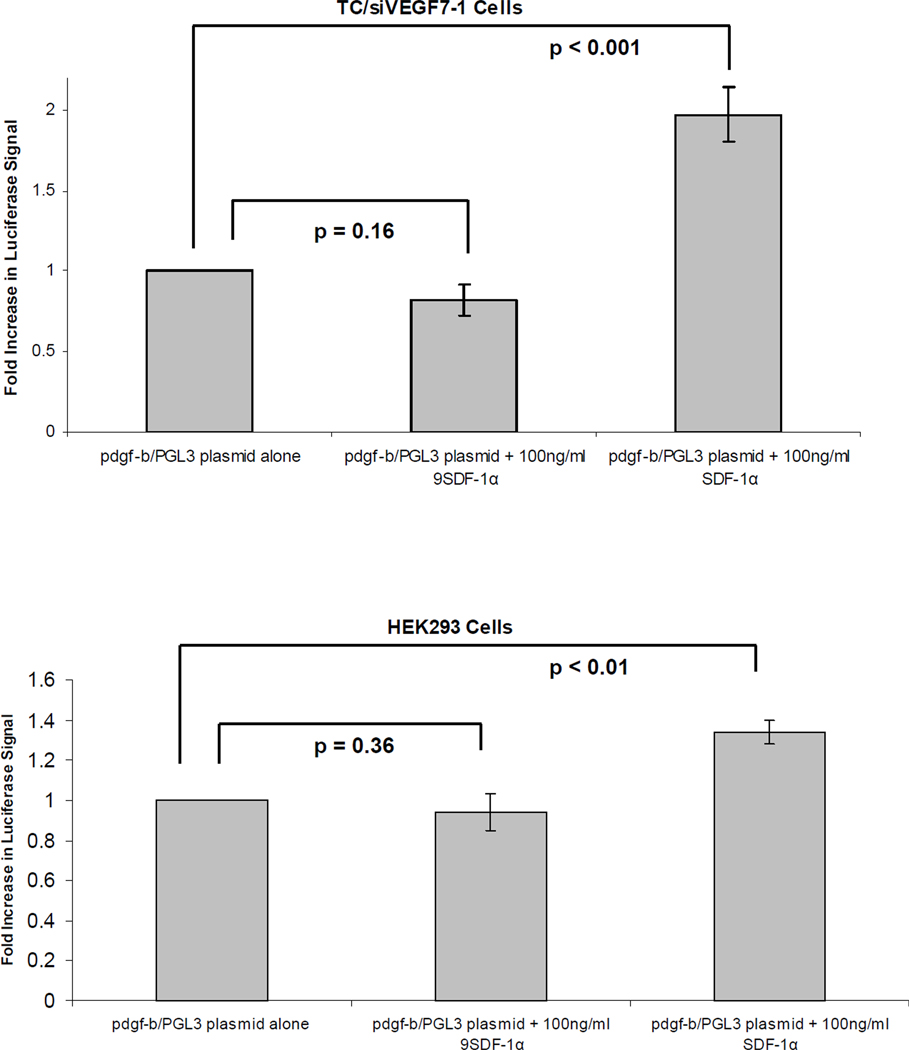
We next investigated whether SDF-1α regulates PDGF-B through a transcriptional mechanism. We cloned the 2-kb human pdgf-b promoter fragment into a luciferase reporter vector. DNA was collected from TC71 cells and used as a template to isolate the 2-kb promoter fragment. The purified fragment was cloned into the PGL3 reporter vector, and the luciferase signal was measured. A luciferase signal indicates promoter activity. The transfected cells were treated with either SDF-1α or the inactive 9SDF-1α protein for 8 hours. SDF-1α activated the pdgf-b promoter in both the TC/siVEGF7-1 and HEK293 cells compared with 9SDF-1α (Figure 4).
Figure 4. SDF-1α regulates the expression of PDGF-B via transcription.
TC/siVEGF7-1 and HEK293 cells were transfected with either the pdgf-b/PGL3 luciferase construct or the control vector. The cells were cultured in the absence of growth factors and supplements for 1 day and then treated with 100 ng/mL of either SDF-1α or the inactive 9SDF-1α protein for 8 hours. The cells were lysed and the luciferase signal quantified. Each bar graph shows pooled data from 3 independent experiments, each performed in triplicate, +/− SD. P< 0.05 was considered significant.
SDF-1α induces binding of the ELK-1 transcription factor to the pdgf-b promoter
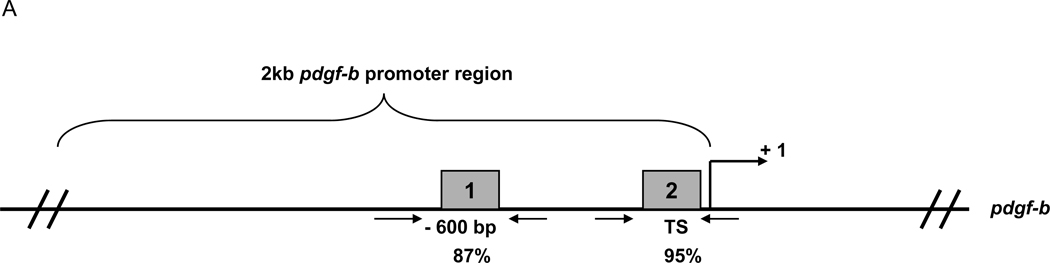
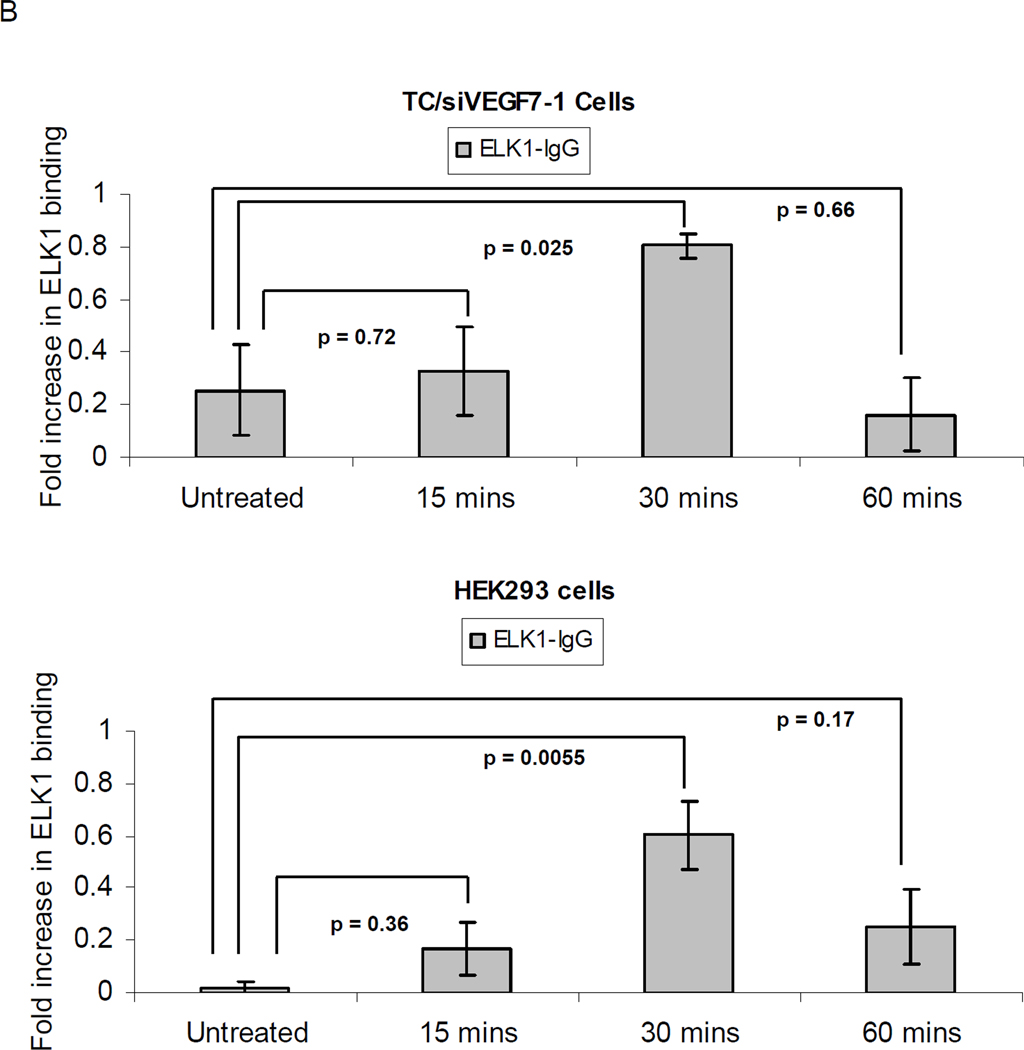
The SDF-1α chemokine binds to its G-protein coupled receptor CXCR4 expressed on the surface of cells21. Once the SDF-1α ligand binds to CXCR4, the signaling cascade activates several MAPKs, including ERK1/2, which phosphorylates and activates the ELK-1 transcription factor15. TC/siVEGF7-1 and HEK293 cells express CXCR4 (supplemental Figure 1). Stimulation with SDF-1α activated the ERK1/2(p44/42)/ELK-1 pathway and induced phosphorylation of both ERK1/2(p44/42) and ELK-1 (S383) in TC/siVEGF7-1 (Supplemental Figure 2). We next investigated whether SDF-1α induced binding of ELK-1 to the pdgf-b promoter. Potential ELK-1 binding sites within the pdgf-b promoter region were identified (Figure 5A). Using ChIP, we investigated 2 of the identified sites that scored above 85% at −600 bp and at the transcription start (TS) site. TC/siVEGF7-1 and HEK293 cells were treated with SDF-1α for 15, 30, or 60 minutes, and ChIP was performed. The ELK-1 transcription factor bound to the pdgf-b promoter with a peak binding at 30 minutes (P < 0.05) and with a higher affinity at the −600 bp site (Figure 5B) than at the TS site (P > 0.05; data not shown).
Figure 5. SDF-1α induces binding of the ELK-1 transcription factor to the pdgf-b promoter.
(A) Schematic of the pdgf-b promoter region with the potential ELK-1 binding sites identified using the GeneRegulation.com MATCH software. Two of the potential ELK-1 binding sites which scored >85% were investigated: at −600 bp (box 1) and at the TS site (box 2). Primer binding sites for the corresponding regions are also illustrated. (B) qPCR for the −600 bp site; the fold increase in ELK-1 binding is scaled from 0 (non-binding) to 1 (complete binding). TC/siVEGF7-1 and HEK293 cells were plated, cultured in the absence of growth factors and supplements for 24 hours, and then treated with 100 ng/mL of SDF-1α for 15, 30, or 60 minutes. The cells were collected and ELK-1 binding quantified by ChIP. Each bar graph shows pooled data from 3 independent experiments, each performed in triplicate, +/− SD. The sample values minus the control values were used in one-way analysis of variance. Negative values were replaced with 0, indicating non-binding. P < 0.05 was considered significant.
PDGFR-β+ BMCs differentiate into pericytes which express PDGFR-β, desmin, and NG2 in response to the SDF-1α/PDGF-B pathway
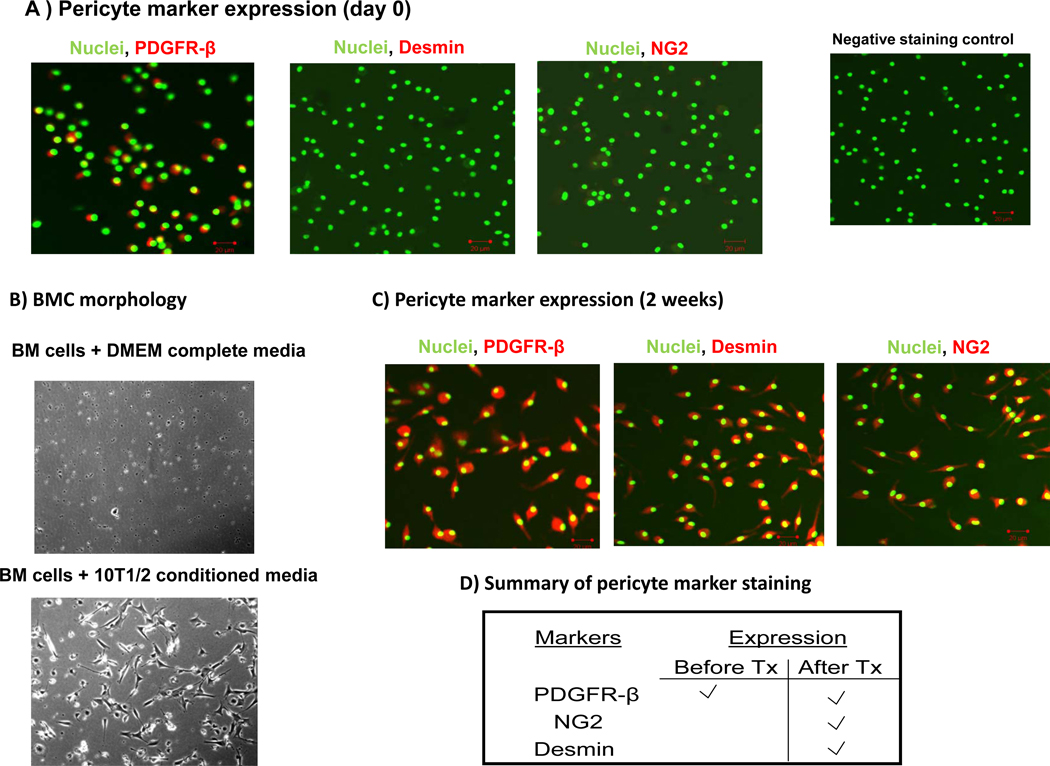
As PDGF-B is required for the recruitment and integration of pericytes in the vascular wall22, we established an in vitro model to study the effect of the SDF-1α/PDGF-B pathway on the differentiation of BMCs into pericytes. Expression of the pericyte markers PDGFR-β, NG2, and desmin were used to indicate pericyte differentiation. While fresh BMCs express PDGFR-β, they do not express the mature pericyte markers desmin and NG2 (Figure 6A). After incubation for 2 weeks in conditioned medium from 10T1/2 cells, PDGFR-β+ BMCs attached to the plate, acquired a pericyte-like morphology (Figure 6B), and expressed desmin and NG2 (Figure 6C, D). The cells cultured in DMEM alone did not attach or acquire a pericyte-like morphology (Figure 6B).
Figure 6. 10T1/2 conditioned medium induces BMCs to differentiate into pericytes as defined by morphology and the expression of NG2 and desmin.
(A) BMCs were flushed from the hind femurs of mice, cytospun onto glass slides, fixed, and stained for PDGFR-β, desmin, and NG2. Fresh BMCs express PDGFR-β but do not express the mature pericyte markers desmin and NG2 (red staining). Green represents SytoxGreen nuclear staining. (B) BMCs were flushed from the hind femurs of mice and cultured in either conditioned medium from 10T1/2 cells or DMEM complete medium alone for 2 weeks. When cultured in 10T1/2 conditioned medium, PDGFR-β+ BMCs attach to the plate and acquire pericyte-like morphology and (C) express the mature pericyte markers desmin and NG2 (red staining). Green represents SytoxGreen nuclear staining. (D) Table summarizing the expression of PDGFR-β, desmin, and NG2 pericyte markers by BMCs before and after culture in conditioned medium from 10T1/2 cells for two weeks.
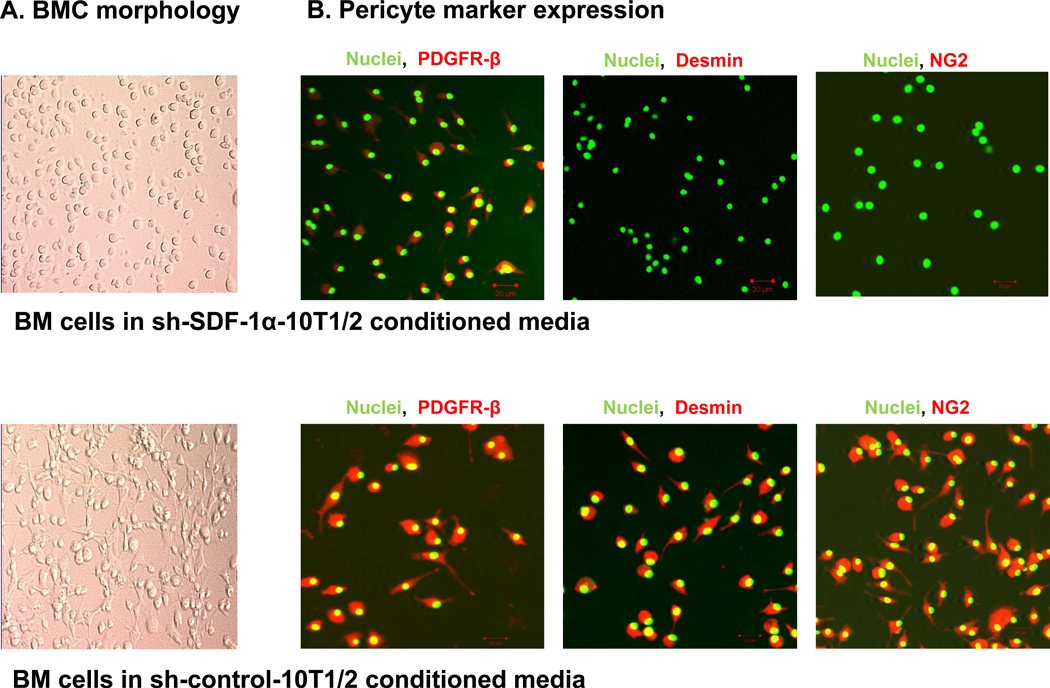
To determine whether SDF-1α in the 10T1/2 conditioned medium played a role in inducing PDGFR-β+ BMCs to express NG2 and desmin, we cultured PDGFR-β+ BMCs in conditioned medium from 10T1/2-shSDF-1 or 10T1/2-shcontrol cells for 2 weeks. We already showed that transfection of 10T1/2 cells with shSDF-1 decreased PDGF-B expression (Figure 3). PDGFR-β+ BMCs cultured in 10T1/2-shSDF-1 conditioned medium behaved in a similar manner to those cultured in DMEM alone: their morphology did not change, and they failed to express NG2 and desmin (Figure 7A). In contrast, PDGFR-β+ BMCs cultured in 10T1/2-shcontrol conditioned medium stained positive for desmin, and NG2 and had a similar pericyte-like morphology to those cultured in conditioned medium from 10T1/2 cells (Figure 7B). These results confirm that the SDF-1α in the conditioned medium was responsible for inducing BMCs to differentiate into pericyte-like cells and that the SDF-1α/PDGF-B pathway participates in the differentiation of PDGFR-β+ BMCs into pericytes.
Figure 7. BMCs cultured in 10T1/2-sh-SDF-1 conditioned medium do not differentiate into pericytes as defined by morphology and expression of NG2 and desmin.
BMCs were flushed from the hind femurs of mice and cultured in conditioned medium from either 10T1/2-sh-SDF-1 cells or 10T1/2-sh-control cells for 2 weeks. (A) PDGFR-β+ BMCs attach to the plate and acquire pericyte-like morphology when cultured in 10T1/2-sh-control conditioned medium but not when cultured in 10T1/2-sh-SDF-1 conditioned medium. (B) PDGFR-β+ BMCs cultured in 10T1/2-sh-control conditioned medium were fixed and stained for the mature pericyte markers desmin and NG2 (red staining). Green represents SytoxGreen nuclear staining.
Discussion
The regulation of PDGF-B expression during vascular remodeling is poorly understood. We previously demonstrated that the inhibition of VEGF165 in TC71 tumor cells (TC/siVEGF7-1) resulted in smaller tumors with decreased perivascular coverage and decreased bone marrow (BM)-derived tumor vascular pericytes12, 23, 24. Intratumoral injections with an adenoviral vector containing the SDF-1α gene (Ad-SDF-1α) up-regulated the expression of SDF-1α (but not of VEGF165), increased BM-derived pericytes surrounding the tumor vessels, and rescued tumor growth12. In the current study, we evaluated tumor sections from our previous study12 for the expression of PDGF-B and found that SDF-1α gene therapy increased PDGF-B protein expression as well as SDF-1α. These findings suggested an association between SDF-1α, PDGF-B, and the differentiation of BMCs into pericytes.
Here, we demonstrated that SDF-1α regulates PDGF-B. Treating two different cell lines (TC/siVEGF7-1 and HEK293) with SDF-1α increased the expression of PDGF-B mRNA and protein levels in vitro. In contrast, an shRNA specific for SDF-1α down-regulated both SDF-1α and PDGF-B. A luciferase assay and chromatin immunoprecipitation indicated that SDF-1α regulates PDGF-B via a transcriptional mechanism and that the ELK-1 transcription factor binds to the pdgf-b promoter in response to SDF-1α stimulation. Furthermore, our findings demonstrated that BMCs cultured in 10T1/2-sh-SDF-1 conditioned medium in which expression of both SDF-1α and PDGF-B is down-regulated, failed to differentiate into pericytes as defined by morphology and the expression of NG2 and desmin mature pericyte markers. Taken together, our data support the concept of an SDF-1α/PDGF-B pathway that plays a critical role in the differentiation of PDGFR-β+ BMCs into mature pericytes.
PDGF-B, a member of the PDGF family of growth factors, is a mitogen for cells of mesenchymal origin such as fibroblasts and smooth muscle cells1, 5. When secreted in its homodimer form, PDGF-BB binds to its tyrosine kinase receptor, PDGFR-β. PDGF-B and PDGFR-β are expressed in the developing vasculature, in which PDGF-B is produced by endothelial cells and PDGFR-β is expressed by pericytes and vascular smooth muscle cells1. Many cancers, including colorectal cancer, pancreatic cancer, glioma, and sarcoma, overexpress PDGF-B4, 5, 25. However, the mechanisms by which PDGF-B is up-regulated are unclear. VEGF-C has been shown to regulate vascular stabilization by controlling PDGF-B expression26. Our data shows that SDF-1α can regulate PDGF-B expression. Our results further show that this regulation is mediated via a transcriptional mechanism which involves the ELK-1 transcription factor.
The ELK-1 transcription factor is a member of the 3 ternary complex factors (TCFs) which is a subfamily of the E twenty-six (Ets) domain transcription factors19. SDF-1α binds to the 7-transmembrane G-protein coupled receptor CXCR4 and induces phosphorylation of the MAPKs p44 ERK-1 and p42 ERK-2, which further leads to the phosphorylation of the nuclear transcription factor ELK-113–15. We identified potential ELK-1 binding sites within the pdgf-b promoter region and found that SDF-1α stimulates binding of ELK-1 to the pdgf-b promoter at the −600 bp site in both TC/siVEGF7-1 and HEK293 cells. We are the first to report the regulation of PDGF-B by SDF-1α and binding of the ELK-1 transcription factor to the pdgf-b promoter in response to SDF-1α stimulation.
We hypothesized that the increased pericyte coverage with BM-derived cells seen in the TC/siVEGF7-1 tumors after SDF-1α gene therapy was a result of the up-regulation of PDGF-B by SDF-1α. PDGF-B and PDGFR-β have been shown to play a critical role in the process of pericyte maturation4. Over expression of PDGF-B in colorectal and pancreatic cancer increased pericyte content4 whereas lack of PDGFR-β signaling led to pericyte loss, which in turn resulted in endothelial cell changes and capillary dilation and rupture2, 5. PDGFR-β+ pericyte progenitor cells (PPPs) have been shown to migrate to sites of tumor growth, where they differentiate into NG2+, desmin+, and α-SMA+ pericytes3. HIF1-α has also been shown to induce recruitment of BM-derived myeloid cells, as well as endothelial and pericyte progenitor cells, and to promote neovascularization in glioblastoma through increases in SDF-1α13, 27. To determine whether SDF-1α played a role in BM-derived pericyte differentiation, we investigated the effect of SDF-1α on PDGFR-β+ BMCs collected from the hind femurs of mice. Whereas fresh BMCs are positive for PDGFR-β, they are negative for the mature pericyte markers NG2 and desmin. Conditioned medium containing SDF-1α induced PDGFR-β+ BMCs to express both NG2 and desmin, with a change in morphology that indicated pericyte differentiation. This effect was abolished when SDF-1α was inhibited. As mentioned above, down-regulating SDF-1α expression also decreased PDGF-B expression. Taken together, these observations suggest that SDF-1α-induced differentiation of BMCs into mature pericytes which express desmin and NG2 is mediated via its regulation of PDGF-B.
Pericytes, also known as smooth muscle cells or mural cells, wrap around endothelial cells which together form the basement membrane of the vessel wall28. Pericytes are recruited by PDGF-B-expressing endothelial cells to the sites of vessel remodeling, where they play a role in vascular stabilization, maturation, and survival3, 6. As pericytes appear sparse and detached from the vessel wall in tumors3, they have been neglected as a potential target for antivascular therapy. However, pericytes are important constituents that support the tumor vasculature3 and studies have demonstrated that targeting pericytes in addition to endothelial cells is more effective in inhibiting tumor growth than targeting one or the other alone8, 9; inhibiting PDGF-B with the AX102 aptamer in combination with bevacizumab in human ovarian carcinoma resulted in increased tumor growth inhibition9. Altogether, our current findings demonstrated that SDF-1α induces the differentiation of PDGFR-β+ BMCs into mature pericytes in vitro and our previous findings demonstrated that up-regulating SDF-1α in the tumor microenvironment led to increased BM-derived pericyte coverage and tumor growth12. Therefore, we anticipate that inhibiting SDF-1α will inhibit BMC differentiation into pericytes, resulting in smaller and less perfused tumor vessels, which, in combination with other anti-angiogenic factors, could subsequently negatively impact tumor growth and metastasis.
In summary, we have shown that SDF-1α regulates PDGF-B via a transcriptional mechanism involving binding of the ELK-1 transcription factor to the pdgf-b promoter. Our findings also suggest that this SDF-1α/PDGF-B pathway plays an important role in the differentiation of PDGFR-β+ BMCs into mature pericytes which express desmin and NG2. All together, our findings demonstrate the importance of SDF-1α not only as a chemotactic factor for BMCs29, 30, but also as a signaling molecule that can regulate the expression of growth factors such as PDGF-B. Our findings emphasize the importance of understanding the different growth factors and signaling pathways that can control the differentiation of BMCs into pericytes which can ultimately affect tumor vascular morphology and function.
Supplementary Material
Acknowledgments
We thank Dr. Pete Taylor, Dr. Gangxiong Huang, and Dr. Ying Cao for their direction, advice, and instruction.
Grant Support
This research is supported by the National Institutes of Health (NIH), through grant R01 CA103986, and MD Anderson’s Cancer Center Support Grant CA016672, and by the Marilyn and Frederick R. Lummis, Jr., M.D., Fellowship in the Biomedical Sciences. The content is the responsibility of the authors and does not represent the official views of NIH.
References
- 1.Fredriksson L, Li H, Eriksson U. The PDGF family: four gene products form five dimeric isoforms. Cytokine Growth Factor Rev. 2004;15(4):197–204. doi: 10.1016/j.cytogfr.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Hellstrom M, Kalén M, Lindahl P, Abramsson A, Betsholtz C. Role of PDGF-B and PDGFR-beta in recruitment of vascular smooth muscle cells and pericytes during embryonic blood vessel formation in the mouse. Development. 1999;126(14):3047–3055. doi: 10.1242/dev.126.14.3047. [DOI] [PubMed] [Google Scholar]
- 3.Song S, Ewald AJ, Stallcup W, Werb Z, Bergers G. PDGFRbeta+ perivascular progenitor cells in tumours regulate pericyte differentiation and vascular survival. Nat Cell Biol. 2005;7(9):870–879. doi: 10.1038/ncb1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCarty MF, Somcio RJ, Stoeltzing O, Wey J, Fan F, Liu W, et al. Overexpression of PDGF-BB decreases colorectal and pancreatic cancer growth by increasing tumor pericyte content. J Clin Invest. 2007;117(8):2114–2122. doi: 10.1172/JCI31334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Westermark B, Heldin CH. Platelet-derived growth factor. Structure, function and implications in normal and malignant cell growth. Acta Oncol. 1993;32(2):101–105. doi: 10.3109/02841869309083897. [DOI] [PubMed] [Google Scholar]
- 6.Hirschi KK, D'Amore PA. Pericytes in the microvasculature. Cardiovasc Res. 1996;32(4):687–698. [PubMed] [Google Scholar]
- 7.Sennino B, Falcón BL, McCauley D, Le T, Kurz JC, Haskell A, et al. Sequential loss of tumor vessel pericytes and endothelial cells after inhibition of platelet-derived growth factor B by selective aptamer AX102. Cancer Res. 2007;67(15):7358–7367. doi: 10.1158/0008-5472.CAN-07-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergers G, Song S, Meyer-Morse N, Bergsland E, Hanahan D. Benefits of targeting both pericytes and endothelial cells in the tumor vasculature with kinase inhibitors. J Clin Invest. 2003;111(9):1287–1295. doi: 10.1172/JCI17929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu C, Shahzad MM, Moreno-Smith M, Lin YG, Jennings NB, Allen JK, et al. Targeting pericytes with a PDGF-B aptamer in human ovarian carcinoma models. Cancer Biol Ther. 9(3):176–182. doi: 10.4161/cbt.9.3.10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kujoth GC, Robinson DF, Fahl WE. Binding of ETS family members is important for the function of the c-sis/platelet-derived growth factor-B TATA neighboring sequence in 12-O-tetradecanoylphorbol-13-acetate-treated K562 cells. Cell Growth Differ. 1998;9(7):523–534. [PubMed] [Google Scholar]
- 11.Guan H, Zhou Z, Wang H, Jia SF, Liu W, Kleinerman ES. A small interfering RNA targeting vascular endothelial growth factor inhibits Ewing's sarcoma growth in a xenograft mouse model. Clin Cancer Res. 2005;11(7):2662–2669. doi: 10.1158/1078-0432.CCR-04-1206. [DOI] [PubMed] [Google Scholar]
- 12.Reddy K, Zhou Z, Jia SF, Lee TH, Morales-Arias J, Cao Y, et al. Stromal cell-derived factor-1 stimulates vasculogenesis and enhances Ewing's sarcoma tumor growth in the absence of vascular endothelial growth factor. Int J Cancer. 2008;123(4):831–837. doi: 10.1002/ijc.23582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teicher BA, Fricker SP. CXCL12 (SDF-1)/CXCR4 Pathway in Cancer. Clin Cancer Res. doi: 10.1158/1078-0432.CCR-09-2329. [DOI] [PubMed] [Google Scholar]
- 14.Horuk R. Chemokine receptors. Cytokine Growth Factor Rev. 2001;12(4):313–335. doi: 10.1016/s1359-6101(01)00014-4. [DOI] [PubMed] [Google Scholar]
- 15.Majka M, Ratajczak J, Kowalska MA, Ratajczak MZ. Binding of stromal derived factor-1alpha (SDF-1alpha) to CXCR4 chemokine receptor in normal human megakaryoblasts but not in platelets induces phosphorylation of mitogen-activated protein kinase p42/44 (MAPK), ELK-1 transcription factor and serine/threonine kinase AKT. Eur J Haematol. 2000;64(3):164–172. doi: 10.1034/j.1600-0609.2000.90112.x. [DOI] [PubMed] [Google Scholar]
- 16.Oettgen P. The role of ets factors in tumor angiogenesis. J Oncol. 2010 doi: 10.1155/2010/767384. 767384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lelièvre E, Lionneton F, Soncin F, Vandenbunder B. The Ets family contains transcriptional activators and repressors involved in angiogenesis. Int J Biochem Cell Biol. 2001;33(4):391–407. doi: 10.1016/s1357-2725(01)00025-5. [DOI] [PubMed] [Google Scholar]
- 18.Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2(11):827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 19.Buchwalter G, Gross C, Wasylyk B. Ets ternary complex transcription factors. Gene. 2004;324:1–14. doi: 10.1016/j.gene.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 20.Kasza A, Wyrzykowska P, Horwacik I, Tymoszuk P, Mizgalska D, Palmer K, et al. Transcription factors Elk-1 and SRF are engaged in IL1-dependent regulation of ZC3H12A expression. BMC Mol Biol. 11:14. doi: 10.1186/1471-2199-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kucia M, Jankowski K, Reca R, Wysoczynski M, Bandura L, Allendorf DJ, et al. CXCR4-SDF-1 signalling, locomotion, chemotaxis and adhesion. J Mol Histol. 2004;35(3):233–245. doi: 10.1023/b:hijo.0000032355.66152.b8. [DOI] [PubMed] [Google Scholar]
- 22.Abramsson A, Lindblom P, Betsholtz C. Endothelial and nonendothelial sources of PDGF-B regulate pericyte recruitment and influence vascular pattern formation in tumors. J Clin Invest. 2003;112(8):1142–1151. doi: 10.1172/JCI18549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee TH, Bolontrade MF, Worth LL, Guan H, Ellis LM, Kleinerman ES. Production of VEGF165 by Ewing's sarcoma cells induces vasculogenesis and the incorporation of CD34+ stem cells into the expanding tumor vasculature. Int J Cancer. 2006;119(4):839–846. doi: 10.1002/ijc.21916. [DOI] [PubMed] [Google Scholar]
- 24.Reddy K, Cao Y, Zhou Z, Yu L, Jia SF, Kleinerman ES. VEGF(165) expression in the tumor microenvironment influences the differentiation of bone marrow-derived pericytes that contribute to the Ewing's sarcoma vasculature. Angiogenesis. 2008 doi: 10.1007/s10456-008-9109-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fomchenko EI, Holland EC. Platelet-derived growth factor-mediated gliomagenesis and brain tumor recruitment. Neurosurg Clin N Am. 2007;18(1):39–58. viii. doi: 10.1016/j.nec.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 26.Onimaru M, Yonemitsu Y, Fuji T, Tanii M, Nakano T, Nakagawa K, et al. VEGF-C regulates lymphangiogenesis and capillary stability by regulation of PDGF-B. Am J Physiol Heart Circ Physiol. 2009;297(5):H1685–H1696. doi: 10.1152/ajpheart.00015.2009. [DOI] [PubMed] [Google Scholar]
- 27.Du R, Lu KV, Petritsch C, Liu P, Ganss R, Passequé E, et al. HIF1alpha induces the recruitment of bone marrow-derived vascular modulatory cells to regulate tumor angiogenesis and invasion. Cancer Cell. 2008;13(3):206–220. doi: 10.1016/j.ccr.2008.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bergers G, Song S. The role of pericytes in blood-vessel formation and maintenance. Neuro Oncol. 2005;7(4):452–464. doi: 10.1215/S1152851705000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lapidot T, Kollet O. The essential roles of the chemokine SDF-1 and its receptor CXCR4 in human stem cell homing and repopulation of transplanted immune-deficient NOD/SCID and NOD/SCID/B2m(null) mice. Leukemia. 2002;16(10):1992–2003. doi: 10.1038/sj.leu.2402684. [DOI] [PubMed] [Google Scholar]
- 30.Kucia M, Reca R, Miekus K, Wanzeck J, Wojakowski W, Janowska-Wieczorek A, et al. Trafficking of normal stem cells and metastasis of cancer stem cells involve similar mechanisms: pivotal role of the SDF-1-CXCR4 axis. Stem Cells. 2005;23(7):879–894. doi: 10.1634/stemcells.2004-0342. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.