Abstract
Prostate cancer (PCa) is the most prevalent cancer, a significant contributor to morbidity and a leading cause of cancer-related death in men in Western industrialized countries. In contrast to genetic changes that vary among individual cases, somatic epigenetic alterations are early and highly consistent events. Epigenetics encompasses several different phenomena, such as DNA methylation, histone modifications, RNA interference, and genomic imprinting. Epigenetic processes regulate gene expression and can change malignancy-associated phenotypes such as growth, migration, invasion, or angiogenesis. Methylations of certain genes are associated with PCa progression. Compared to normal prostate tissues, several hypermethylated genes have also been identified in benign prostate hyperplasia, which suggests a role for aberrant methylation in this growth dysfunction. Global and gene-specific DNA methylation could be affected by environmental and dietary factors. Among other epigenetic changes, aberrant DNA methylation might have a great potential as diagnostic or prognostic marker for PCa and could be tested in tumor tissues and various body fluids (e.g., serum, urine). The DNA methylation markers are simple in nature, have high sensitivity, and could be detected either quantitatively or qualitatively. Availability of genome-wide screening methodologies also allows the identification of epigenetic signatures in high throughput population studies. Unlike irreversible genetic changes, epigenetic alterations are reversible and could be used for PCa targeted therapies.
Keywords: Epigenetics, Genome, Methylation, Prostate cancer.
1. INTRODUCTION
Prostate cancer (PCa) is the most frequently diagnosed non-skin malignancy and the third leading cause of cancer-related death in men in the Western world [1]. PCa is one of the most complicated human tumors and, like many other malignancies, arises from progressive genetic and epigenetic alterations. The field of epigenetics has rapidly evolved and influenced research in different biological phenotypes such as aging, memory formation, and embryological development. Overall, epigenetic defects reported in cancers include reactivation of embryonic genes, loss of imprinted genes changing inactive and active alleles, dysregulated expression of micro-RNAs, increased gene recombination, and transcriptional silencing of tumor-suppressor and housekeeping genes. Epigenetics encompasses several different phenomena, such as DNA methylation, histone modifications, RNA interference, and genomic imprinting [2]. Epigenetic processes regulate gene expression and can change malignancy-associated phenotypes such as growth, migration, invasion, or angiogenesis.
Unlike many other genetic changes, epigenetic processes are reversible and do not affect DNA sequence or quantity. However, they promote genomic instability that might lead to oncogenic activation and / or inactivation of tumor suppressors. Normal methylation levels of various genes preserve cellular homeostasis. Among all recognized epigenetic alterations, aberrant DNA methylation (hypo- and hypermethylation) is the most important and the best characterized change in PCa [3]. Hypermethylated genes in PCa include DNA damage repair genes (e.g., glutathione S-transferase Pi (GSTP1) and the DNA alkyl-repair gene O6-methylguanine DNA methyltransferase (MGMT)), hormonal response genes (e.g., androgen receptor (AR), estrogen receptor (ER)), cell-cycle control genes (e.g., CDKN2A), tumor-suppressor genes (e.g., VHL, RB1, APC), apoptosis genes (e.g., death-associated protein kinase (DAPK)), and invasion and metastasis genes (e.g., Cadherins, CD44, TIMPs). In addition, global and gene-specific hypomethylation have also been associated with PCa. Epigenetic events may also play a role in benign prostatic hyperplasia (BPH). Epigenetic regulatory mechanisms appear very sensitive to external stimuli or influences such as diet and oxidative stress. A comprehensive review on epigenetic alterations in PCa is beyond the scope of this review. Our intention is to provide a synopsis of widely known DNA methylated genes and their biological activity and pathways, interaction between DNA methylation and histone modification, epigenetic biomarkers for PCa diagnosis and prognosis, and epigenetic targets for PCa therapy. Finally, we review a number of methods for detection of DNA methylation and available data bases and computational analytical tools.
2. DNA METHYLATION AND REGULATION OF GENE EXPRESSION
Epigenetic mechanisms such as DNA methylation and histone modification play an essential role in many molecular and cellular alterations associated with the development and progression of prostate cancer [4, 5]. In mammalian cells, most of the chromatin exists in a condensed, transcriptionally silenced form called hetero-chromatin. Euchromatin is less condensed and contains most of the actively transcribed genes. Histones and DNA are chemically modified with epigenetic markers that influence the chromatin structure by altering the electrostatic nature of the chromatin or by altering the affinity of chromatin binding proteins. DNA methylation is usually associated with histone deacetylation, chromatin condensation, and gene silencing [5-7].
DNA methylation leads to gene-silencing either by inhibiting the access of target binding sites to the transcriptional activators [8] or by promoting the binding of methyl-binding domain proteins, which can mediate repression through interaction with histone deacetylases (HDACs) that promote chromatin condensation into transcriptionally repressive conformations [9, 10].
DNA methylation involves the addition of a methyl group to the fifth carbon position of the cytosine pyrimidine ring via a methyltransferase. DNA methylation refers to the covalent bonding of a methyl group specifically to the dinucleotide CpG, which is catalyzed by the family of enzymes known as DNA methyltransferases (DNMTs). It is thought that DNA methylation alters chromosome structure and defines regions for transcriptional regulation. This covalent modification of multiple DNA sites by methylation is a heritable and reversible epigenetic process, which is involved in the regulation of a diverse range of biological processes [11-13]. Clusters of CpG sites are dispersed around the genome and are referred to as CpG islands, stretches of DNA ranging from 0.5 to 5 kb with a guanine-cytosine (GC) content of at least 50%. These islands are found in the promoter region of about 60% of genes, in exons and introns, and in repetitive elements. Most CpG islands in the promoter regions are unmethylated, whereas CpG islands in intronic regions and repetitive sequences are heavily methylated, perhaps to help the cell identify regions for gene transcription [14].
There are two distinct classes of DNMTs. The first class consists of de novo methyltransferases (DNMT3a and DNMT3b) that methylate DNA irrespective of whether the template is hemi-methylated or not. These enzymes are involved in the establishment of new DNA methylation patterns during development. The maintenance DNA methyltransferase DNMT1 belongs to the second class of enzymes. Disruption of the mouse Dnmt1 gene results in genome-wide demethylation and developmental arrest. Thus, the role of DNMT1 in propagating parental DNA methylation during replication cannot be substituted by other DNMTs [15]. Without the DNMTs, the replication machinery itself would produce daughter strands that are unmethylated and, over time, would lead to passive demethylation.
Outside of CpG islands, CpG methylation is thought to suppress the transcription of transposable elements and spurious initiation of transcription elsewhere. DNA methylation abnormalities, such as the gain of methylation in normally unmethylated promoters or other regulatory regions (hypermethylation), contribute to tumorigenesis by decreasing the activity of tumor-suppressor genes. Loss of methylation in normally methylated repetitive sequences that lead to activation of proto-oncogenes and genomic instability is evident in almost all human tumor types [16,17]. DNA methylation is the best established epigenetic mark that is critical for the allele-specific expression of imprinted genes [18]. Hypomethylation of specific chromosomal domains has been linked to chromosomal instability [19]. Chromosomal abnormalities associated with hypomethylation include isochromosomes, unbalanced juxtacentromeric translocations, and whole-arm deletions. DNA hypomethylation of repetitive elements, retrotransposons, and CpG poor promoter regions plays an important role in tumorigenesis [20]. Hypomethylation of repetitive sequences and retrotransposons are associated with chromosomal rearrangements and translocation to other genomic regions, thereby promoting genomic instability [21, 22].
CpGs are underrepresented in mammalian genomes and occur in only 1% of the genome, lower than the expected statistical fraction of 6%. The methyl donor for this reaction is supplied by S-Adenosyl Methionine (SAM). This substrate is recycled through a folate- and cobalamin-dependent pathway. Hypomethylation or loss of methylation can be accelerated by altering this regenerative process through a deficiency of Vitamin B9 (folate), vitamin B12, or other substrates [23]. There are multiple roles for DNA methylation in mammals and disruption of this process during early development by the inactivating DNMTs is lethal [24].
DNA methylation also has a putative role in genome defense. All cells in culture or organisms in environment challenged life are subject to stringent conditions of selection, even the cell which has been forced by its innate recombination mechanisms to tolerate the genomic insertion of foreign DNA can avail itself of an ancient defense mechanism against the genetic activity of foreign DNA that could carry active genes. Since promoter methylation has been identified as part of a mechanism for the long-term silencing of genes and DNA segments, the de novo methylation of integrated foreign DNA can be contemplated as a defense mechanism or at least as an integral part of it [25].
Methylation is also involved with genomic organization and silencing unneeded genes in differentiated cells. During this process, DNA methylation acts as a stable tag on the promoter of a gene that recruits methyl-binding proteins (MBPs) and other proteins such as HDACs, to form large-scale heterochromatic structures that silence the associated genes. Changes in DNA methylation during cancer formation have seemingly divergent effects on the cell.
Three types of altered DNA methylation patterns have been observed in human tumors: hypomethylation, hypermethylation, and loss of imprinting [26]. DNA hypomethylation occurs in many tumors, particularly in advanced stages, and is generally assumed to be a genome-wide event [27, 28]. By comparison, DNA hypermethylation occurs at specific regulatory sites, such as in the promoter regions of tumor-suppressor genes, and thereby decreases the expression of individual genes [29-31]. As a consequence, hypermethylation may be functionally equivalent to an inactivating gene mutation. Loss of imprinting, which refers to the loss of differential expression of the parental alleles, is often seen in embryonal tumors [32, 33]. While genome-wide hypomethylation, which could lead to activation of previously silenced genes, is seen in some advanced, metastatic PCas [28], most studies have emphasized DNA hypermethylation as an important mechanism for inactivation of key regulatory genes in PCas [27, 34]. For example, decreased expression of E-Cadherin, a component of the E-Cadherin/catenin cell adhesion complex, is associated with poorly differentiated and late-stage PCa and is closely linked to the progression of the disease [35]. Since the promoter of E-Cadherin is frequently found to be hypermethylated in prostate tumors and PCa cell lines and can be reversed by demethylation, it is likely that hypermethylation accounts for the reduced expression [31]. In contrast, hypermethylation is not responsible for the observed reduction in PCa of P-Cadherin expression, another member of the Cadherin family [36], illustrating the gene selectivity of DNA hypermethylation (Fig. 1).
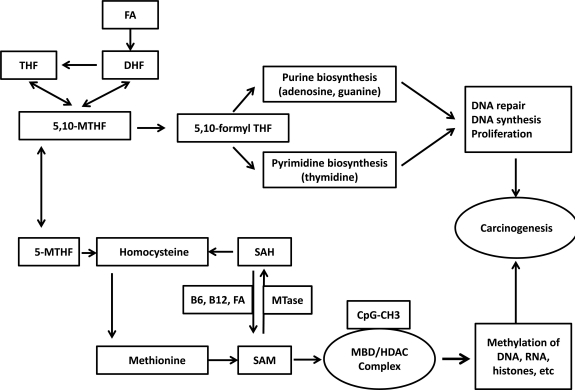
Fig. (1).
Schematic representation of dietary factors, one-carbon metabolism, methionine cycle in DNA methylation. One-carbon metabolism is the best understood pathway of dietary regulation of DNA methylation. Folic acid is converted to dihydroflate (DHF) and then to tetrahydrofolate and finally to methylene tetrahydrofolate (MTHF). 5, 10-MTHF is required for the synthesis of nucelic acids, and 5- MTHF is required for the formation of methionine from homocysteine via folate- and B12-dependent methionine synthase reaction. Methionine adenosyl transferase transfers adenosine to methionine and generates S-adenosylmethionine (SAM), which is the main methyl donor. Subsequently, SAM is converted to S-adenosyl homocysteine (SAH), which has high binding affinity to methyltransferases (MTase). A dietary supply of vitamins B12, B6, and folic acid via several steps regenerate SAH to SAM. Methylene tetrahydrofolate (MTHF) can direct folic acid (FA) to nucleotide synthesis as an important path for DNA synthesis, cell growth, and DNA repair or for conversion of homocysteine to methionine. DNA-methyl transferases (MTase) methylates the CpG island which recruits the methyl binding domain (MBD) and histone deacetylases (HDACs) to the methylated DNA and leads to histone deacetylation, condensation of chromatin, loss of transcription factor binding, and silencing of the gene expression in cancer and other premalignant conditions.
3. HYPERMETHYLATED GENES IN PROSTATE CANCER
DNA hypermethylation is the most common and best-characterized epigenetic abnormality in human malignancies, including PCa. Significantly, many of the affected genes encode proteins that are involved in critical cellular processes and/or have tumor-suppressor activity (Table 1). Pathways frequently disrupted by CpG island hypermethylation include DNA damage repair, hormonal responses, tumor-cell invasion/metastasis, and cell cycle control (Fig. 2). For many of these genes, promoter hypermethylation is often the primary or only mechanism underlying functional loss in PCa. Inappropriate silencing of these genes can contribute to cancer initiation, progression, and metastasis. Some of the hypermethylation occurs in the early stages of PCa progression; along the multistep process of prostate carcinogenesis; some correlate with pathological grade or clinical stages of PCa; some contribute to invasiveness, metastasis and androgen independence of PCa [37].
Table 1.
Genes Frequently Hypermethylated in Prostate Cancer
| Gene | Chra | Role/Function | Hyper methylation | Prostate cancer | Cell linesd | Ref. | |
|---|---|---|---|---|---|---|---|
| Primaryb | Metc | ||||||
| Glutathione S-transferase Pi (GSTP1) | 11q13 | Intracellular detoxification | 70% - 100% | + | + | [50] | |
| Glutathione-S-transferase (GSTM1) | 1p13 | Intracellular detoxification | 58% | + | [178] | ||
| O-6-Methylguanine DNA-Methyltransferase (MGMT) | 10q26 | Remove alkyl adducts from O6-guanine | 76% | + | + | [44] | |
| Retinoic Acid Receptor beta (RAR β) | 3p24 | Tumor suppressor | 84% | + | [68] | ||
| Androgen Receptor (AR) | Xq12 | Hormone regulation | 39% | + | [216] | ||
| Estrogen Receptor Alpha (ERα) | 6q25 | Hormone regulation | 90-95% | + | [56] | ||
| Estrogen Receptor beta (ERβ) | 14q23 | Hormone regulation | 79-100% | + | [56] | ||
| Ras association domain family 1A (RASSF1A) | 3p21 | Tumor suppressor:cell growth | 53-79% | + | [81] | ||
| Death Associated Protein Kinase 1 (DAPK1) | 9q34 | Regulator of cell growth | 36% | + | [46] | ||
| Endothelin Receptor B (ENDRB) | 13q22 | Tumor suppressor | 45-72% | + | + | + | [81] |
| E-Cadherin (CDH1) | 16q22 | Tumor suppressor: invasion and metastasis | 61-72% | + | [76] | ||
| Cyclin-dependent kinase inhibitor2A/p16 (CDKN2A/p16) | 9p21 | Tumor suppressor | 66% | + | + | [217] | |
| Cyclin-dependent kinase inhibitor1C/p57 (CDKN1C/p57) | 11p15 | Tumor suppressor | 56% | + | [218] | ||
| Cyclin-dependent kinase inhibitor1B/p27 (CDKN1B/p27) | 12p13 | Tumor suppressor | 6% | + | [45] | ||
| Cyclin-dependent kinase inhibitor1A/p21 (CDKN1A/p21) | 6p21 | Tumor suppressor | 6% | + | [45] | ||
| Tissue Inhibitors of Metalloproteinase-2 (TIMP-2) | 17q25 | Tumor suppressor | 78.5% | + | [219] | ||
Chr: Chromosomal location.
Primary prostate cancers.
Mets: Metastatic prostate cancers.
Prostate cancer cell lines.
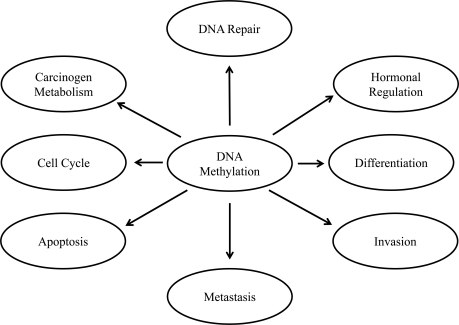
Fig. (2).
Schematic diagram of involvement of DNA methylation in different cellular processes. DNA methylation is the best known epigenetics alteration in prostate cancer. DNA methylation can regulate gene expression and can function in favor of malignancy-associated phenotypes such as cellular differentiation, growth, migration and invasion, metastasis, apoptosis, hormonal regulation of steroid receptors, and DNA repair. DNA methylation appears to be very sensitive to external stimuli or influences such as diet and oxidative stress.
3.1. DNA Damage Repair Genes
DNA repair is a correcting mechanism that maintains genomic integrity during replication or after DNA damage. Cells that are defective in the components of DNA repair pathways exhibit higher rates of spontaneous DNA mutations, which can lead to cancer [38]. The hypermethylation of two genes involved in DNA damage repair, the detoxifier gene glutathione S-transferase Pi (GSTP1) and the DNA alkyl-repair gene O6-methylguanine DNA methyltransferase (MGMT), has been reported in PCa.
Glutathione S-transferase (GST) is a family of enzymes involved in the detoxification of xenobiotics and oxygen radicals [39]. Human GSTs are classified into distinct families; five of which encode cytosolic GSTs, α, µ, π, σ, θ forms. The product of π gene (GSTP1), which is located on 11q13, can detoxify environmental electrophilic carcinogens and oxidants and might have a genomic care taker role by preventing oxidant and electrophilic DNA damage and the resulting mutations [40]. GSTP1 expression is detectable in normal tissues at varying levels in different cell types. Notably, a loss or low level of GSTP1 expression has been reported in high-grade prostatic intraepithelial neoplasia (PIN) and PCa. Methylation of the GSTP1 gene promoter has been the most frequently detected epigenetic alteration, and occurs in over 90% of cancerous samples and about 70% of prostatic intraepithelial neoplasia (PIN) samples [41, 42], but is rarely detected in normal prostate or benign prostatic hyperplasia (BPH) tissues. It is also detected in a subset of proliferative inflammatory atrophy lesions (PIA). At the present time, decreased GSTP1 expression is the most common epigenetic alteration in PCa.
MGMT is a DNA repair protein that removes mutagenic and cytotoxic alkyl adducts from genomic DNA. Tumors that lack MGMT expression have a higher incidence of point mutations in the genes encoding p53 and K-ras, which may influence cancer progression. The MGMT promoter contains a CpG island that is methylated in many human cancers [43]. Over- expression of MGMT reduces the risk of mutations after exposure to methylating agents while loss is associated with increased sensitivity to methylating agents and carcinogenicity. Results from studies evaluating the MGMT promoter methylation in PCa have been unequivocal, with moderate to high levels of methylation detected in some studies [44, 45], but not others [46, 47]. Further work will be necessary to resolve this discrepancy.
3.2. Hormonal Response Genes
The prostate is an endocrine gland that responds to sex hormones such as androgens, estrogens, and progesterones through their specific receptors. These agents bind to specific cellular receptors to mediate their physiological effects.
3.2.1. Androgen Receptor
Testosterone and 5α-dihydrotestosterone are the most important male hormones, with actions mediated through the androgen receptor (AR), resulting in the development and maintenance of the prostate [48]. The AR gene is located on chromosome Xq11–12. The 5’ region of the AR gene contains a CpG island according to the criteria established by Gardiner-Garden and Frommer [49] indicating that it might be regulated by DNA methylation. Jarrard et al. [50] analyzed the influence of AR promoter methylation on AR gene expression in PCa cell lines. Genetic alterations that alter the sensitivity of the AR to androgens, including AR gene mutations [51] and amplifications [52] without loss of AR expression, are thought to play a key role in the development of hormone-independent advanced PCa. Epigenetic changes have been accounted for alterations in the AR expression in 28% of hormone-independent PCa. Epigenetic regulation, including CpG methylation and histone acetylation, play important roles in the regulation of AR [53]; however, the frequency of AR methylation appears to be low in PCa [54-56].
3.2.2. Estrogen Receptors
Estrogens, which have been used for the treatment of PCa for decades [57], require estrogen receptors (ERs) to mediate their activity [58-60]. The prostate expresses two types of estrogen receptors: ERα (ER1) and ERβ (ER2) [61]. Lost or decreased expression of ER1 and ER2 in PCa has been documented [62-65]. The ER1 gene is frequently methylated in PCa and methylation status is associated with tumor progression [66]. The ER2 promoter contains a typical CpG island [67]. Hypermethylation of the ER2 proximal promoter in the PCa cell lines and high frequency in human PCa has been documented [56, 68, 69]. Methylation of the ER1 and ER2 gene promoters is detectable in BPH; however, the extent of ER1 and ER2 promoter methylation is significantly less in the BPH samples than in prostatic tumors, indicating that prostate carcinogenesis induces ER gene hypermethylation [66, 69].
3.2.3. Retinoic Acid Receptors
Retinoids, a group of natural and synthetic vitamin A analogs, are important factors in modulating cell growth, differentiation and apoptosis which have been shown to suppress carcinogenesis in vitro and in vivo. The retinoids are among the most investigated classes of chemo-preventive drugs for PCa. Their effects are primarily mediated through two classes of nuclear receptors – retinoic acid receptors (RARs) and retinoid X receptors (RXRs). These classes belong to the steroid/thyroid hormone – receptor superfamily and are composed of three subtypes (α, β, γ) [70]. The RARβ gene has two different promoters and expresses RNA transcripts that undergo alternative splicing [71]. The RARβ2 promoter contains a CpG island that is aberrantly methylated in PCa [72]. In PCa, RARβ2 expression is also controlled epigenetically, and a high frequency of aberrant methylation has been noted in clinical samples [47,54, 68 70, 73-77]. RAR β2 gene methylation is a frequent event in PCa. In addition, the RAR β2 promoter is methylated in 20% of PIN samples, a putative PCa precursor. Therefore, RAR β2 gene methylation appears to occur early in PCa etiology, and is implicated in cancer initiation [46, 72].
3.3. Cell Cycle Control Genes
A distinguishing characteristic of tumor cells is uncontrolled growth, which is regulated by the cell cycle pathway. Many genes act as checkpoints that regulate the cell cycle and if these genes become defective, may lead to carcinogenesis and progression of PCa [78].
The tumor-suppressor gene p16/CDKN2 is one of the cyclin-dependent kinase inhibitors (CDKIs). CDKN2A, a key protein in the signaling pathway, can be affected by a variety of genetic and epigenetic changes, including hypermethylation in PCa. Aberrant CDKI expression is observed in many tumor tissues, including the prostate [45, 79, 80]. Results regarding the frequency of CDKN2A promoter methylation are inconsistent in prostate tumors, ranging from 3% to 77%; other studies have investigated the role of hypermethylated CDKN2A in the carcinogenesis and progression of PCa [45, 47, 79-85]. Herman et al. first reported that inactivation of CDKN2A by DNA methylation in PCa [86]. The CDKN2A gene was frequently methylated in tumor tissue (77%). These data support p16 methylation as a potential biomarker for early detection of PCa. Other CDKIs such as p14, p21, p27 and RB1 are rarely methylated in prostate tumors, and thus, are probably not good candidates as biomarkers [45].
The RAS family of proto-oncogenes plays a key role in the signal transduction pathways involved in cellular proliferation and survival, interacting with other regulatory circuits of cell growth and death. RAS association domain family protein 1 isoform A (RASSF1A) is known as a tumor suppressor gene. The RASSF1A protein was found to be associated with the DNA repair protein and to mediate the apoptotic effect of oncogenic Ras [87, 88]. Inactivation of RASSF1A may result in dysregulation of DNA repair system and the Ras-dependent growth control in cancer cells. The RASSF1A gene is silenced by aberrant methylation of the promoter in a large fraction of various cancers including prostate [89]. In prostate tumors, RASSF1A promoter methylation is a common event, occurring in 49% to 99% of tumor tissues and it has been shown to be associated with aggressive PCa [44, 47, 89].
3.4. Tumor-Suppressor Genes
Function loss of classic tumor-suppressor genes through DNA hypermethylation is not a common event in PCa. For instance, methylation of RB1 [90], hMLH1 [91], and VHL [30] has been frequently detected in other types of cancer, but not in PCa. Methylation inactivation in CDKN2A and CDH1 has shown moderate to high prevalence in PCa [82, 86, 92].
The adenomous polyposis coli (APC) gene, located on chromosome 5q21-22, is known as tumor-suppressor gene that is responsible for Familial Adenomatous Polyposis (FAP). FAP is frequently identified in colorectal cancers, but not in PCa [93, 94]; however, involvement of APC hypermethylation has been described in different studies [44, 47, 75, 79, 81, 95, 96]. β-catenin mutations have been detected in various tumors and are relatively rare in PCa [94]. However involvement of γ - catenin methylation in prostatic carcinogenesis has been recently documented [97].
Additional genes with putative tumor-suppressor function undergoing epigenetic inactivation by hypermethylation in PCa include KAI1 (a prostate-specific tumor metastasis suppressor gene) [98], inhibin-α (a member of the transforming growth factor–β family of growth and differentiation factors) [99], and DAB2IP, a novel GTPase-activating protein for modulating the Ras-mediated signaling pathway [100]. It is unknown, however, whether hypermethylation of these genes plays a role in prostate carcinogenesis or has a role as a biomarker for PCa diagnosis.
3.5. Apoptosis Genes
Programmed cell death (apoptosis) is a critical process for carcinogenesis in human. Typical morphological characteristics of apoptosis include damage to the plasma membrane, condensation and fragmentation of the nucleus, and DNA fragmentation [101]. A major component of the apoptosis pathway is the caspase family. However, other genes, including death-associated protein kinase (DAPK), fragile histidine triad (FHIT), solute carrier family 5A8 (SLC5A8), vesicular monoamine transporter 2 (SLC18A2), and tumor necrosis factor receptor superfamily, member 10C (TNFRSF10C) are also involved in this pathway. A repressed expression of these genes by hypermethylation in the promoter region has been shown for PCa [46, 47, 81, 102-106]. However, DAPK and FHIT may have a limited value due to a persistently low frequency of methylation in tumors and normal tissues [46, 47, 81, 101]. SLC5A8, SLC18A2, and TNFRSF10C were found to be hyper-methylated in 50% to 88% of PCa and were significantly downregulated in the tumor when compared to normal prostate tissues [103-105, 107-109]. It is noteworthy that, the expression of SLC18A2 and TNFRSF10C is inversely associated with biochemical recurrence after radical prostatectomy [110].
3.6. Invasion and Metastasis Genes
Metastasis is an extremely complicated process that occurs through a series of sequential steps involving invasion, transport, adhesion to distant sites, and outgrowth into a secondary organ. Although metastases are the cause of 90% of human cancer deaths, little is known about the genetic and biochemical determinants of metastasis. Cell–cell adhesion and cell–substrate adhesion are critical to the preservation of the normal tissue architecture. These phenotypes are regulated by a group of proteins known as cell adhesion molecules (CAMs). Cadherins are a large family of CAMs that are involved in cell–cell adhesion. Disruption of the cell adhesion system can lead to tumor infiltration and metastasis [111,112].
E-Cadherin (CDH1), an important member of the Cadherin family of cell adhesion molecules, is a transmembrane glycoprotein whose extracellular domain mediates cell-cell adhesion through calcium-dependent homophilic interaction [113]. Decreased CDH1 expression is observed in many cancers, a significant correlation between loss of CDH1 expression and invasion and metastasis has been documented. With regard to PCa, correlations between decreased CDH1 expression and prognostic factors including tumor grade, stage and ploidy, have been reported [114, 115]. Methylation of the CDH1 promoter region has been detected in PCa cell lines [92]. In human prostate tumors, expression of CDH1 is strongly reduced and its promoter is methylated to varying degrees [44, 74, 92, 116]. The 5’ CpG island of CDH1 is densely methylated in PCa cell lines [31].
Matrix metalloproteinases (MMPs) are proteolytic enzymes that degrade the extracellular matrix and the basement membrane. High expressions of these enzymes have been associated with tumor growth, invasion, and tumor-induced angiogenesis [117]. These pathways are controlled by the balance between the levels of the MMPs and tissue inhibitors of metalloproteinases (TIMPs) [118]. TIMP-1, TIMP-2, and TIMP-3 are widely investigated members of this family involved in tumor progression and metastasis in a variety of human cancers. TIMP-2 expression appears to have a tumor-promoting role in PCa [119]. The promoter region of TIMP-3 was found to be methylated in 97% of prostate tumors [79] and two studies found TIMP-3 promoter methylation in 37% and 41% of urine sediments from PCa patients [84, 102].
CD44 is an integral membrane protein that is involved in matrix adhesion and signal transduction. CD44 is classified as a metastasis suppressor because decreased CD44 expression is associated with the progression of PCa to a metastatic state [120]. It has been reported that loss of standard CD44 expression in PCa predicts a poor prognosis [121, 122]. The methylation of the 5’ CpG island of CD44 is associated with transcriptional inactivation in PCa cell lines, as detected in metastasis, indicating an important role in the progression of PCa [73-76, 123-126].
Others tumor metastasis genes — Caveolin-1 (CAV1), H-Cadherin (CDH13), EPHA7, and S100A2 are often downregulated in prostate tumor tissues when compared to adjacent normal tissues due to methylation [47, 74, 76, 81, 85, 92, 127-134]. Gene-silencing of CAV1 and CDH13 is associated with clinical features of PCa progression [132, 135, 136]. S100A2 methylation was seen in 75% of cases of nonmalignant tissues and in 100% of cases of BPH [133].
4. DNA HYPOMETHYLATION
DNA methylation in mammalian genomes is a DNA preservation mechanism by which repetitive DNA, (~50% of genome’s content), is transcriptionally silenced to prevent their expression and function [137]. Demethylation of normally methylated DNA, also known as hypomethylation, can disrupt such a defense mechanism, leading to structural and functional alterations of the genome.
There are two types of hypomethylation, global or genomic, which refer to an overall decrease of 5-methylcytosine content in the genome, and localized or gene-specific hypomethylation, which refers to a decrease in cytosine methylation relative to the “normal” methylation level. The latter process affects specific regions of the genome, such as the promoter regions of proto-oncogenes or normally highly methylated sequences, such as repetitive sequences and oncogenes [138]. Both global hypomethylation and gene-specific hypomethylation have been implicated in human cancers (Table 2).
Table 2.
Genes Frequently Hypomethylated in Prostate Cancer
| Gene | Chra | Role/Function | Hypo methylation | Prostate cancer | Cell linesd | Ref. # | |
|---|---|---|---|---|---|---|---|
| Primaryb | Metsc | ||||||
| Urokinase Plasminogen Activator (uPA) | 10q24 | Tumor invasion and metastasis | 75-96.9% | + | + | [220] | |
| Heparanase (HPSE) | 4q21 | Tumor invasion and metastasis | 8.5 -30% | + | + | [142] | |
| Cancer/testis Antigen Gene (CAGE) | 6p24 | Cell cycle control: cellular proliferation | 34% | + | [151] | ||
| Cytochrome P4501B1(CYP1B1) | 2p21 | Hydroxylation of estrogens and activation of carcinogens | 5 17% | + | + | [143] | |
Chr: Chromosomal location.
Primary prostate cancers.
Mets: Metastatic prostate cancer.
Prostate cancer cell lines.
4.1. Global Hypomethylation
Global hypomethylation, relative to the normal situation in PCa, has been reported in a few cases [28, 85, 139-143]. Subsequently, more sensitive and precise analysis of genome-wide hypomethylation has been conducted. LINE-1 retrotransposon sequences constitute between 5-10% of human genome and are strongly methylated in somatic tissues [144]. With LINE-1 hypomethylation analysis, the frequencies of DNA hypomethylation in PCa are reported to be 7-53%, leading to the hypothesis of a relationship between LINE- 1 hypomethylation and GSTP1 hypermethylation, or alterations on chromosome 8 [85, 139, 140].
Genome-wide hypomethylation, which could lead to activation of previously silenced genes, is seen in some advanced, metastatic PCa [28]. Heparanase degrades heparin sulfate and has been implicated in tumor progression. Increased heparanase expression in PCa tissue has been reported to be caused by promoter hypomethylation with an up-regulation of the transcriptional factor, Early Growth Factor Response1 (EGFR1) [142]. Cytochrome P450 1B1 (CYP1B1), a member of the CYP superfamily that is overexpressed in PCa, is also regulated by a hypomethylation of its promoter/enhancer region [143].
4.2. Gene-Specific Hypomethylation
Genes from cancer cells but not from normal cells are substantially hypomethylated [145]. Moreover, when compared to adjacent normal tissues, most cancer tissues contain two hypomethylated ras oncogenes, c-Ha-ras and c-Ki-ras [146]. In the prostate, the Plasminogen Activator Urokinase (PLAU) gene is highly expressed in most PCa tissues [147] and invasive PCa cell lines [148]. The PLAU gene encodes urokinase plasminogen activator, a multifunctional protein that can promote tumor invasion and metastasis in several malignancies, including PCa. DNA methylation may also play a role in the regulation of the PLAU gene in PCa, with hypomethylation of the PLAU promoter being associated with its increased expression in hormone-independent PCa cells, higher invasive capacity in vitro, and increased tumorigenesis in vivo. However, in normal prostate epithelial cells and in hormone-dependent LNCaP cells, the PLAU promoter is methylated, resulting in lower expression of the gene [149]. Another hypomethylated PCa gene is the Cancer Associated Gene (CAGE), a novel cancer/testis antigen gene [150]. Hypomethylation of CAGE, which occurs in about 40% of PCa cases, is responsible for its exclusive expression in cancer tissues [151].
5. RACIAL DIFFERENCES AND DNA METHYLATION IN PROSTATE CANCER
PCa is associated with racial disparities. In 2007, PCa was responsible for 37% of all malignancies in black men in the US. The PCa incidence and mortality rate is approximately 60% and up to threefold higher, respectively, in African American (AA) men than in the Caucasians (CAs). This tendency has been validated for more than 20 years before or after the PSA era [152]. Similar incidence and mortality rates have been reported in black men of West African ancestry from the Caribbean and South America [153]. The underlying reasons for such disproportionate ethnic differences in PCa prognosis and mortality are unclear. In part, genuine racial differences in cancer genetics and biology, sociocultural differences and/or access to health care systems are responsible, but these factors do not totally explain the higher mortality rate in African Americans with PCa. Limited knowledge is available for inter-racial differences on gene-specific or genome-wide methylation or other epigenetic processes in normal individuals or patients with PCa. Kwabi-Addo et al. [154] examined the methylation levels of six genes (GSTP1, AR, RARβ2, SPARC, TIMP3, and NKX2-540), which have been previously shown to be hypermethylated in Caucasians with PCa or cell lines. They compared matched and PCa tissues from AAs and CAs who had comparable Gleason scores. They observed significant differences in the methylation levels in all genes, except GSTP1, in the AA samples in comparison with CA samples [154].
This observation is in agreement with work by Woodson et al. [155], which also demonstrated an increase but no significant difference in GSTP1 methylation in PCa tissues and benign prostatic hyperplasia samples from AAs compared to CAs. Also, it was demonstrated that NKX2-5 and TIMPC genes were highly methylated in normal prostate tissue samples from AAs compared to CAs and that methylation of NKX2-5 increased with age in AAs. Thus, it was suggested that NKX2-5 may serve as a marker for PCa detection and increased sensitivity for detection PCa in AAs since the incidence of PCa increases dramatically with age. Although Kwabi-Addo et al. [154] observed the relationship between DNA methylation and PCa risk, they did not detect any significant association between DNA methylation based on race. Therefore, the utilization of these genes as “ethnic-sensitive” biomarkers for PCa is promising and can be further assessed with a larger PCa population size, as noted by the authors.
In addition to the aforementioned genes, the adhesion and signal transduction membrane protein CD44, which is associated with the progression of localized cancer to metastatic disease, has also been shown to be hypermethylated in AAs. Woodson et al. [155] examined the methylation of three genes involved in the progression of PCa, GSTP1, CD44, and E-Cadherin. As noted earlier, there was an increase but no significant difference in GSTP1 methylation, as well as no difference in the frequency between AAs and CAs. While E-Cadherin was not hypermethylated at all, hypermethylation of CD44 was observed among AAs with higher frequencies compared to CAs and correlated with tumor grade but not disease stage. However, Kito et al. reported a correlation between CD44 hypermethylation and disease stage in a Japanese men [156]. Although it is speculated that CD44 hypermethylation may have prognostic implications, its methylation and role in racial differences in PCa should continue to be explored in larger studies.
6. HISTONE MODIFICATIONS
In eukaryotes, genomic DNA is packaged with histone proteins into chromatin, compacting DNA some 10,000-fold. Such condensation of DNA provides a considerable obstacle to the nuclear machinery that drives processes such as replication, transcription or DNA repair. Importantly, the structure of chromatin dynamically changes, permitting localized decondensation and remodeling that facilitates the progress of nuclear machinery.
The chromatin structure is regulated by a variety of post-translational modifications including DNA methylation, modification of histones, and ATP-dependent chromatin remodeling. Histones can be modified by several post-translational mechanisms including acetylation, methylation, phosphorylation, ubiquitination, sumoylation, or ribosylation of distinct amino acids, resulting in either the activation or suppression of gene expression [157-161].
A complete understanding of the precise molecular mechanisms by which the histone tail alterations influence DNA-histone interactions remains elusive. There are two main hypotheses on how histone modifications can affect chromosome function: 1) they may alter the electrostatic charge of the histone, resulting in a structural change in the histones or their binding to DNA; or 2) these modifications are binding sites for protein recognition motifs, such as the bromodomains or chromodomains, that recognize acetylated lysines or methylated lysines, respectively [162].
Enzymes that tightly control the balance of covalent histone modifications are histone acetyl transferases (HATs) and HDACs as well as histone methyltransferases (HMTs) and demethylases (HDM). These enzymes alter the configuration of the chromatin and regulate gene expression. Acetylation of lysine residues in histone tails by HATs unpacks the chromatin structure and renders the DNA accessible to transcription factors, thereby facilitating gene expression. The effects of HATs are counteracted by HDACs, which pack chromatin and repress gene transcription. In mammals, there are 18 HDACs which are subdivided into four distinct classes based on sequence homology to yeast HDACs and functional similarities. Class I HDACs (1, 2, 3, and 8) are primarily located in the nucleus and are ubiquitously expressed. Class II HDACs are divided into two subgroups: IIa HDACs (4, 5, 7, and 9) and IIb HDACs (6 and 10), which shuttle between the nucleus and cytoplasm and show a tissue-specific expression pattern [163, 164]. The NAD+-dependent enzymes of Class III HDACs (also named SIRTUINS) comprise seven members (SirT1– 7) and are ubiquitously expressed. Although most of these enzymes were shown to regulate histone acetylation, their distinct biological functions are largely due to the deacetylation of non-histone proteins such as transcription factors.
Overall, post-translational modifications of histones create an epigenetic mechanism for the regulation of a variety of normal and disease-related processes, including cancers. Drugs affecting histone modifications already have been developed and have shown promising results in the treatment of different tumor types.
7. INTERACTION BETWEEN DNA METHYLATION AND HISTONE MODIFICATION IN PROSTATE CANCER
The silencing mechanism of DNA methylation is only part of a complex set of epigenetic regulatory processes. Another epigenetic change that is distinct from and yet linked to DNA methylation is the modification state of the surrounding histones in which the DNA is packaged. DNA and histones are linked functionally to control transcription and repair. It has been shown that methylated DNA recruits HDAC through methyl-DNA binding proteins (MBPs); consequently, DNA methylation/ histone deacetylation cross talk has been suggested to influence gene silencing [10, 12, 165-169].
The histone modifications exist in relative balance, maintained by competing enzymes that constantly work to place and remove the appropriate modifications. The signals that control these modifications are complex and can include DNA methylation and the binding of other protein co-factors [170]. In PCa, a number of in vitro studies provide evidence that promoter hypermethylation and histone deacetylation interact to maintain chromatin in its inactive state. These studies have shown that combined treatment with the histone deacetylase inhibitor, Trichostatin A, and demethylating agents 5-aza-cytidine or 5-aza-2’- deoxycytidine led to reversing epigenetic silencing of several genes. A loss of hypermethylation in the promoter and concomitant gene activation has been observed for a number of tumor-suppressor genes in various PCa cell lines.
Transcriptional activation of a gene is facilitated by the addition of acetyl groups on the N-terminal lysines of histones to create an open or noncompact chromatin (euchromatin) conformation. The addition of acetyl groups on histones, primarily histones H2a, H3 and H4, is catalyzed by HATs. Increased HAT activities have myriad effects that may alter PCa growth in positive and negative ways. They can regulate transcriptional co-regulator proteins that bind to steroid receptors. For example, the HATs p300, PCAF and Tip60 up-regulate the expression or activation state of the AR which leads to an increase in the AR signaling in the absence of androgen [171, 172]. HATs also increase IGFBP-2 and p21 activation in PCa, slowing cell-cycle progression [4]; thus, HATs affect a range of cellular processes and as such they represent a putative target for therapy.
8. EPIGENETIC CHANGES AND PROSTATE CANCER DIAGNOSIS, PROGNOSIS, AND TREATMENT
Prostate-specific antigen (PSA) is a less than-optimal tumor marker and cannot effectively differentiate between PCa and other conditions such as prostatitis or BPH. The false positive results lead to expensive and invasive critical investigations such as transrectal prostate biopsies. This provides the opportunity for researchers to identify more reliable potential epigenetic markers for PCa diagnosis, prognosis, and follow up treatment.
8.1. Epigenetic Diagnostic Markers
Epigenetic markers, particularly aberrant DNA methylation, have the potential to be useful diagnostic tumor markers. Historically, there have been two general strategies for detecting DNA methylation changes at specific DNA sequences in cancer cells. The older approach exploits the use of methylation-sensitive restriction endonucleases, which cut recognition sites differently depending on whether 5-meCpG is present [173, 174]. The second major strategy for selective detection of genome sequences carrying methylation marks features DNA modification using sodium bisulfite, a procedure that promotes cytosine deamination to uracil but spares 5meC [175, 176]. Methylation markers have several advantages over the mutation-based genetic markers. Aberrant DNA methylations are more frequent than mutations and can be identified by genome-wide screening methodologies. DNA methylation markers potentially could be tested in tumor tissues and body fluids (e.g., serum, urine). The mthylation markers are simple in nature; with high sensitivity, these markers can be detected, either quantitatively or qualitatively, by available well-established techniques (e.g., PCR). Furthermore, the incidences of aberrant DNA methylation are higher than those of mutations and can be discovered by genome wide screening procedures [177].
One of the best characterized epigenetic markers in PCa is GSTpi, a gene that is hypermethylated and silenced in more than 90% of all PCa [178]. It is specific for PCa, although methylation of this gene is also found in proliferative inflammatory atrophy in the prostate, a histological entity that has been linked to PCa development [42]. It has also been reported that a methylation assays combined with histological analysis improves the diagnostic specificity [179]. Several groups examined body fluids containing prostate cells, including serum, urine and semen, for the presence of GSTpi methylation in patients with cancer. The GSTpi protein is clearly detectable in the urine of patients with PCa [180], especially after prostate biopsy. Analysis of prostatic secretions obtained from 100 patients at radical prostatectomy demonstrated 74% sensitivity, and a positive association between the degree of methylation at GSTpi and the extent of the cancer [181]. GSTpi CpG island hypermethylation was found in 72% of serum samples from patients with PCa [182], and it was found to be a significant predictor of PSA recurrence in the cell-free serum of men with PCa [183].
Methylations of several other genes have been studied in PCa diagnosis including, RARβ, CD44, E-Cadherin (ECAD), RASSF1A, APC and tazarotene-induced gene 1 (T1G1) [75, 76]. Others studies have reported that the use of a panel of methylation markers including GSTP1 improves the diagnosis of PCa both in body fluids and tissues. Further studies are needed before these markers can be used as diagnostic markers in routine clinical practice.
8.2. Epigenetic Prognostic Markers
One study demonstrated that GSTP1 hypermethylation is seen in 40% of pre-operative bone marrow aspirate in patients with advanced PCa [184]. Caveolin-1 (CAV1), E-Cadherin (CDH1), H-Cadherin (CDH13), EPHA7, and S100A2 are the tumor metastatic genes that are often more down-regulated in prostate tumor tissues than in adjacent normal tissues due to methylation [47, 74, 76, 81, 84, 85, 127-134]. Silencing of Gene CAV1, CDH1, and CDH13 is associated with clinical features of PCa [132, 135, 136]. These data suggest that the methylation status of CAV1 and CDH1 is not only a potential biomarker for PCa, but may also be a predictive marker of disease outcome [135]. Genes such as CD44 and T1G1 may exhibit specific methylation in high-risk and metastatic tumors and could be used in molecular staging and as predictors of disease progression [78]. Prostate cancers with a high Gleason score are correlated with a higher degree of methylation of many genes, such as RARβ, RASSF1A, and GSTP1 [185].
8.3. Epigenetic Changes as Therapeutic Targets
Epigenetic information is heritable, but has plasticity. Dynamic erasure and writing of epigenetic information take place in specific genes during embryonic development [186]. This makes it possible to modify unwanted epigenetic changes. So far, several classes of drugs, including inhibitors of DNMTs and HDACs, are known to modify epigenetic information in a manner that is not specific to genes. Currently, there are several drugs that are at varying different stages of development. They can be broadly classified in two groups: (i) DNMT inhibitors and (ii) HDAC inhibitors. Some of the drugs are in both groups.
8.3.1. DNMT Inhibitors
DNMTs, especially DNMT1, play important roles in maintaining CpG methylation [182], and their inhibitors are known to induce hypomethylation. In particular, an inhibitor of DNMT1, 5 aza-C, and 5-aza-20-deoxycytidine (5-azadC; Decitabine), are two closely related drugs used experimentally to inhibit DNA methylation in vitro and have been shown to re-activate numerous methylation-silenced genes, such as GSTP1 [183] and RARβ2 [72]. 5-aza-C and 5-aza-dC are cytosine analogues that become incorporated into DNA and trap DNMT1 during replication, leading to the synthesis of nascent DNA in the absence of DNMT activity, resulting in DNA demethylation [187-190]. Myelo-suppression is a major adverse effect, but the treatment is well tolerated. 5-aza-dC has been recently approved by FDA for clinical use in certain hematological conditions. Another drug in the same group, Zebularine can be administered orally or intraperitoneally, but has to be given in high doses; however, it is chemically stable and has low toxicity [191].
Among the chemicals already in clinical use or in food, procainamide, procaine and epigallocatechin-3-gallate (EGCG) have also shown demethylating activity [192-194]. Considering that some aberrant DNA methylation is present in the early stages of carcinogenesis, it is possible that such demethylating agents may be useful for cancer prevention. MG98 is a phosphorothioate antisense oligodeoxynucleotide, and is a specific inhibitor of DNA methyltransferase mRNA; this drug is also used for demethylation [177].
8.3.2. Histone Deacetylase Inhibitors
In recent years, it became evident that HDACs are promising therapeutic targets with the potential to reverse aberrant epigenetic states associated with cancer. Various studies in cancer cell lines and tumor tissues revealed changes in the acetylation levels and the expression of the HDAC enzymes [195]. Increased HDAC activity in prostate tumors provides another avenue for therapeutic inhibition. A variety of natural products exhibit HDAC-inhibitory activity. Commonly used HDAC inhibitors that are being tested include trichostatin A (TSA), Suberoylanilide hydroxamic acid (SAHA) and valproic acid [5]. These drugs appear in vitro to also increase the efficacy of radiation therapy [196] and they have anti-angiogenesis function [197]. SAHA, a class I HDAC inhibitor, significantly represses the growth of LNCaP hormone sensitive prostate tumors in nude mice (97% inhibition vs untreated controls) [198]. Trichostatin A (TSA), leads to hyperacetylation of histones and cell cycle inhibitors, such as p21WAF1 [199]. The class I inhibitor and benzamide derivative, MS-275 increases xenograft radiosensitivity and inhibits prostate tumor growth .The HDACi, LBH589 targets class IIa HDACs and decreases angiogenesis in PC-3 xenograft tumors [200]. Various HDAC inhibitors have been developed for therapeutic purposes, and tumor cells are known to show higher sensitivity than normal cells for unidentified mechanisms. HDAC inhibitors are also reported to be effective even in non-proliferating tumor cells in vitro [201].
The combination of HDAC and DNMT inhibitors has synergistic effect in the reactivation of silenced gene [5]. Another interesting possibility is the combination of epigenetic drugs and conventional anti androgens and chemotherapeutic agents. It should be cautioned that the epigenetic drugs currently lack gene specificity and some of them are associated with significant toxicity. Hence, efforts are being made to develop gene specific epigenetic drugs [177].
9. METHODS FOR DETECTION OF DNA METHYLATION
A wide range of methods are available to either quantitatively or qualitatively to find methylated changes in genomic DNA. Based on the type of technique used, several major groups of detection methods are described here (Fig. 3 and Table 3).
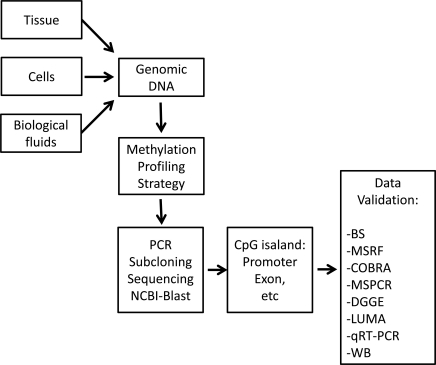
Fig. (3).
Step-by-step approach from the discovery to validation of the CpG methylation. Genomic DNA could be extracted from different types of biological specimens. Based on the investigators need methylation study could begin using a variety of methylation profiling tools and could be followed by PCR and subcloning into a vector, sequencing, aligning into NCBI-Blast data base. After proper identification of the gene or sequence of interest or candidate CpG islands, and designing specific primers, final data validation assays is necessary to confirm the relationship between methylation level and gene or protein expression. This will be performed by different methods listed in the flow chart. BS, Bisulfite sequencing; MSRF, methylation-sensitive restriction fingerprinting; COBRA, Combined bisulfite restriction analysis; MSPCR, methylation-sensitive PCR; DGGE, denaturing gel electrophoresis; LUMA, luminometric methylation assay; qRT-PCR, quantitative real-time PCR; WB, western blotting.
Table 3.
Available Techniques for Detection of DNA Methylation
| Technique | Gene-Specific or Genome-wide | Sample (µg) | Advantage and limitations | Ref. |
|---|---|---|---|---|
| Bisulfite Sequencing | Gene-specific Applicable to genome-wide | 0.2-0.5 | Gene-specific sequence, Simple set up, Most economic, Prone to false positive data, Needs additional confirmatory step, Difficulty in primer design, DNA degradation during bisulfite treatment | [202] |
| MSRF | Genome-wide | 0.1-1.0 | Simple set up suitable for novel genes screening, Special set up for gel electrophoresis | [203] |
| ChIP-chip | Genome-wide | 1.0-10 | High-throughput, Not common, Platform- specific | [204] |
| LUMA | Both | 0.2-0.5 | High-throughput, Relatively expensive, Limited sequence size | [205-209] |
| MALDI-TOF-MS | Gene-specific | 0.01-1.0 | High-throughput, Mostly for specific genes of interest | [210, 211] |
| Illumina Methylation Beadchip Array | Genome-wide | 0.2-0.5 | High-throughput, High accuracy, Relatively expensive, Platform-specific | [12, 213] |
| DGGE | Gene-specific | 0.5-1.0 | Very sensitive to variation in DNA sequence, Simultaneous analysis of multiple samples possible, Time-consuming, No method for automated analyses currently available | [214, 215] |
MSRF: Methylation-Sensitive Restriction Fingerprinting.
ChIP-ChIP: Chromatin Immunoprecipitation on DNA Microarray.
LUMA: Luminometric methylation assayMALDI-TOF-MS.
MALDI-TOF-MS: Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry.
DGGE: Denaturing Gradient Gel Electrophoresis.
9.1. Bisulfite Sequencing
This is the most widely used method to examine the methylation status of individual cytosines within any amplified portion of a gene [202]. It is based on a simple principle; the ability of sodium bisulfite to deaminate un-methylated cytosine residues into uracil in genomic DNA. Following PCR amplification, the uracils will be amplified as thymines. Cloning and sequencing of the DNA fragments will identify the methylated cytosine in CG-islands.
9.2. Methylation-Sensitive Restriction Fingerprinting (MSRF)
In this technique, methylated DNA will be subjected to restriction enzymes MseI and/or BstUI digestion followed by PCR amplification of DNA [203]. Extracted genomic DNA will be digested first by MseI enzyme which recognizes the TTAA restriction site, which rarely exists in CG-rich areas. Therefore, DNA is cleaved into small pieces with the intact CG-regions. Subsequently, a fraction of the MseI-digested DNA will undergo a second digestion by a methylation-resistant restriction enzyme, BstUI. This enzyme is able to cut un-methylated CGCG segments of the DNA which exists in >80% of CG-rich area. The remaining small undigested fragments of DNA of BstUI digested and MseI-only digested DNAs will be amplified using random-primer PCR and compared.
9.3. Chromatin Immunoprecipitation on DNA Microarray (ChIP-chip)
In this method, live cells will be fixed by formaldehyde crosslinking. After isolating intact genomic DNA with transcription factors, antibodies specific to transcription factors of interest will be used to immuoprecipitate the chromatin-protein complex of interest. Next, DNA will be extracted by reversing crosslinking, purified, and chromatin amplicons will be generated by PCR. Amplicons will be labeled with Cy5, and Cy3will be used to label the input reference amplicons. These labeled probes will be applied on a CG array for hybridization and data will be analyzed for the specific interaction between a unique gene-specific CpG site and a specific transcription factor [204].
9.4. Luminometric Methylation Assay for Global DNA Methylation Analysis
Luminometric methylation assay (LUMA) is based on DNA cleavage by a combinations of restriction enzymes, EcoRI/MspI or EcoRI/HpaII, which leaves the TTAA (EcoRI) and methylation-sensitive CG (HpaII) or methylation-resistant MspI overhangs. Bioluminometric polymerase extension will be used to quantify the extent of restriction cleavage by Pyrosequencing. Following the DNA cleavage, the methylation status will be determined by PCR or Southern blotting. The extent of CpG sites methylation could be accurately and quickly determined by Pyrosequencing, which dispenses labeled nucleotides and primers and gel-electrophoresis. LUMA has been used to evaluate the level of methylated cytosine of the whole genome, more frequently to study specific CG islands, to determine the association between changes in methylation status, and diseases, and as diagnostic or prognostic markers of cancers [205-209].
9.5. Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF-MS)
This is a powerful technique to analyze DNA methyla-tion [210, 211]. A variety of mass spectrometry approaches have been developed to measure DNA methylation levels such as rapid screening of single nucleotide polymorphisms and quantitative allele studies. In addition, it has been used to assess nucleotide digestion and DNA sequencing. It involves bisulfite conversion of genomic DNA followed by DNA sequencing, which leads to accurate determination of the levels of genomic DNA methylation. MALDI-TOF-MS can measure the gene-specific or genome-wide content of methylated cytosine in a high-throughput manner with the advantage of high speed, accuracy, and automation.
9.6. Illumina Human Methylation 27/450 Beadchip Array
Genome-wide promoter methylation could be evaluated by Illumina Infinium Human Methylation 27/450 Bead array. This analysis was made at the Bioinformatics and Expression Analysis core facility at the Karolinska Institute. The EZ DNA methylation kit (Zymo Research, Orange, CA) could be used for bisulfite conversion of genomic DNA, and the remaining assay steps could be performed using Illumina-supplied reagents and conditions. The Illumina Infinium II Bead Array uses allele-specific annealing to either methylation-specific probes or non-methylation probes to detect the methylation grade of 27,578 individual CpG sites spread across the promoter regions of 14,495 genes [212, 213]. The HumanMethylation450K BeadChip includes 90% of the content contained on the HumanMethylation27 BeadChip, is designed in the same 12-sample per array format. Its unique feature combines unprecedented quality of coverage and study design flexibility in running genome-scale methylation screens.
9.7. Methylation Analysis by Denaturing Gradient Gel Electrophoresis (DGGE)
PCR products of bisulfate reacted different gene promoter regions could be analyzed using denaturing gradient gel electrophoresis (DGGE). In brief, the PCR products are loaded on a 10–70% denaturant gradient gel together with a fully methylated control (e.g., in vitro methylated DNA) and an unmethylated control (e.g., peripheral blood lymphocytes). After electrophoresis, gels will be stained in Tris/EDTA buffer containing ethidium bromide and photographed under ultraviolet transillumi-nation. Samples are scored as methylated when bands or smears are present on the gels in the area below the band corresponding to unmethylated DNA [214, 215].
10. DNA METHYLATION DATA BASE AND COMPUTATIONAL ANALYTIC TOOLS
During the last decade, epigenetics emerged as a vibrating field in biological sciences and specifically cancer research that led to significant amount of experimental data. Among all others, DNA methylation is the most investigated epigenetic alterations. These developments necessitate the creation of multi-task or subject-specific databases and more and more high-throughput computational methodologies capable of handling the enormous experimental data that could be tailored for discovering the most relevant biological information. Based on the epigenetic profile and in translational settings, this knowledge could be used for risk assessment, screening, prognostic or diagnostic classification, and the development of novel therapeutic strategies toward cancers.
10.1. Databases for CpG Detection Sites
Detection of CG dinucleotides and CpG islands are central to DNA-methylation research. Based on cytosine and guanine frequencies, the CG dinucleotide occurrence in the human genome is considerably less than what is expected for all the methylated cytosine in the entire genome. The CpG islands are defined based on a few default or most common parameters that includes a minimal length of approximately 200 bp, a CG content of approximately 55%, and the observed/expected ratio of 0.6 for CG dinuleotides. Based on these and other parameters, several data bases are available to identify, plot, and report CpG islands in selected DNA fragments, such as CpGReport, NewCpGReport, CpGplot, and isochore, which are part of the European Molecular Biology Open Software Suite (EMBOSS).
10.2. Databases for Determination of Transcription and Translation Initiation Site
Methylation silencing of DNA occurs most frequently in CpG islands within the promoter region of genes. Additionally, the CG dinuleotides of the first exon and intron also have important biological significance in terms of alternate splicing, multiple transcripts, and protein isoforms and functions. Therefore, determination of the transcription start site and translation start site are required for the DNA-methylation analysis of genes. The functional identity of the predicted transcription or translational start site (TSS) could be experimentally validated. Several databases provide this information including the DataBase of Human Transcription Start Sites (DBTSS), Eukaryotic Promoter Database (EPD), and Ensembl. DBTSS provides the location of TSS, retrieval of promoter sequence, extraction of cDNA sequences upstream or downstream to TSS [221]. DBTSS is based on 400,225 and 580,209 human and mouse full-length cDNA sequences and contains exact information on the genomic positions of the TSSs and the adjacent promoters for 8,793 and 6,875 human and mouse genes, respectively. The EPD is an annotated non-redundant collection of eukaryotic POL II promoters for which the TSS has been experimentally determined [222]. Available information on this site is either extracted from scientific literature or obtained by in silico primer-extension method. Access to promoter sequences is provided by pointers to positions in nucleotide sequence entries. The annotation part of an entry includes description of the initiation site mapping data, cross-references to other databases, and bibliographic references. EPD is structured in a way that facilitates dynamic extraction of biologically meaningful promoter subsets for comparative sequence analysis. Ensembl is established as a joint initiative between EMBL-EBI and the Wellcome Trust Sanger Institute to develop a software system for automatic annotation on promoters, TSSs, exonic and intronic positions of the eukaryotic genomes in more than 50 species (e.g., human, rat, mouse, zebrafish) [223].
10.3. Analytical Tools for Experimental Design of DNA Methylation
Epigenetics research has benefited vastly from molecular biology methodologies including PCR, DNA sequencing, and restriction enzyme digestion and analysis. Most commonly used modified versions of these techniques are bisulfite conversion and sequencing, methylation-specific PCR, and combined bisulfite restriction analysis (COBRA). DNA treatment with bisulfite leads to deamination of unmethylated cytosine residues into uracil. Therefore, bisulfite treatment is the introductory to determine the overall CpG methylation rate or status of any suspected DNA fragment. In silico conversion of DNA fragments is made possible by Web-based software such as BiQ Analyzer that provides bisulfite conversion in addition to reversed sequence and retrieval of the complementary strand [224]. Following bisulfite conversion, discrimination between methylated and unmethylated cytosine residues could be accomplished by designing the methylation-specific or bisulfite sequencing primers using software such as MethPrimer, MethMarker, or MethBLAST. MethPrimer detects CpG-specific parameters and islands and is essentially based on the primer3 algorithms for designing primers [225, 226]. In addition, MethMarker used as epigenetic primer design software can be used for epigenetic biomarker optimization and the design of assays such as bisulfite single-nucleotide primer extension (SNuPE), bisulfite pyrosequencing, MSP, methylation analysis by DNA immunoprecipitation (MeDIP)-qPCR. MethBLAST software uses a BLAST search on a bisulfite converted product of methylated and unmethylated cytosine residues of the database sequences. It can identify amplicons in the converted genome, and determine the specificity of generated MSP or bisulfite sequencing primers. COBRA is a DNA-methylation analysis strategy that uses methylation-sensitive restriction enzymes. Consequently, the length of the resulting fragments will be used to determine the methylation state and site-specificity [227]. The restriction enzyme catalogue of New England Biolab (NEB), NEB database REBASE [228] and MethMarker can provide a comprehensive list of methylation-sensitive restriction enzymes to be used in setting up a COBRA experiment.
10.4. DNA Methylation Database
Several databases provide general or specific information for epigenetic research. Methdb is a general purpose methylation database listing the results of methylation experiments ranging from overall genome methylation status to site-specific measurements; it covers several species and a wide range of methodologies and sample types for broad and general methylation studies [229]. MethprimerDB is a database linked with MethDB and contains the published primers that could be used as positive control in experimental settings [230]. This database contains probe and primers to test methylation in techniques such as COBRA, bisulfite-PCR-SSCP, bisulfite sequencing, MSP, and Ms-SNuPE.
11. FUTURE CHALLENGES AND CONCLUDING REMARKS
PCa epigenome is a prototypic catastrophic model of epigenetic alterations in tumorigenesis and disease progression. Alterations in DNA methylation status in tumor-suppressor genes, oncogenes, and other regulatory or structural genes are the most frequent epigenetic processes identified in PCa. The importance of promoter hypermethylation in silencing critical tumor-suppressors (e.g., APC, RASSF1) in PCa is well documented. Global hypomethylation has been identified in primary and metastatic PCa. Genome-wide hypomethylation has been associated with high grade and advanced stage tumors. Overall, the combination of promoter hypermethylation and genome-wide hypomethylation might act favorably in prostate carcinogenesis and progression. The epigenome offers a number of promising prognostic, diagnostic, and therapeutic targets for PCa. However, several obstacles need to be addressed. The plasticity of epigenetic processes might open therapeutic opportunities, but at the same time the reversibility of these processes after treatment is of great concern. In addition, non-specific targeting of epigenetic process by available drugs might lead to short-term or long-term undesirable side effects. This could be resolved significantly by developing gene-specific sustainable demethylation approaches. It is not surprising that the field of PCa research has focused on identifying “epigenetic signature” that could be used for prevention, early detection, progression, or distinction between organ-confined indolent and aggressive disease. At least for early detection, aberrant promoter methylation appears to be the most promising early indicator of PCa. Overall, epigenetics is a fascinating and expanding field in modern biology that has turned scientific, clinical, and public attention to its practical applications for cancer prevention, detection, and therapy.
CONFLICT OF INTEREST
None.
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health/National Center for Research Resources (NIH/NCRR; 2P20 RR021970; subproject #1 to S.K), R21CA149137 (to S.K.), R21CA143589 (to S.K.), P20MD004817 (subproject #1 to S.K.), R01MD005824 (to S.K.) and by Roswell Park Cancer Institute and National Cancer Institute (NCI) grant #P30 CA016056. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or National Cancer Institute. We specially thank Ms. Jonna Ellis and Paula Jones for their editorial assistance.
REFERENCES
- 1.Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol. Biomarkers Prev. 2010;19(8 ):1893–1907. doi: 10.1158/1055-9965.EPI-10-0437. [DOI] [PubMed] [Google Scholar]
- 2.Feinberg AP. The epigenetics of cancer etiology. Sem. Cancer Biol. 2004;14:427–432. doi: 10.1016/j.semcancer.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Schulz WA, Hatina J. Epigenetics of prostate cancer: beyond DNA methylation. J. Cell Mol. Med. 2006;10:100–125. doi: 10.1111/j.1582-4934.2006.tb00293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rennie PS, Nelson CC. Epigenetic mechanisms for progression of prostate cancer. Cancer Metastasis Rev. 1998. 1999;17:401–409. doi: 10.1023/a:1006121219097. [DOI] [PubMed] [Google Scholar]
- 5.Li LC, Carroll PR, Dahiya R. Epigenetic changes in prostate cancer: implication for diagnosis and treatment. J. Natl. Cancer Inst. 2005;97:103–115. doi: 10.1093/jnci/dji010. [DOI] [PubMed] [Google Scholar]
- 6.Ballestar E. An introduction to epigenetics. Adv. Exp. Med. Biol. 2011;711:1–11. doi: 10.1007/978-1-4419-8216-2_1. [DOI] [PubMed] [Google Scholar]
- 7.Jones PA. An epigenetic approach for finding tumor suppressors. Cell Cycle. 2003;2:25–26. doi: 10.4161/cc.2.1.251. [DOI] [PubMed] [Google Scholar]
- 8.Prendergast G C, Ziff E B. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991;251:186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- 9.Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 10.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 11.Baylin S B. DNA methylation and gene silencing in cancer. Nat. Clin. Pract. Oncol. 2005;2:S4–S11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 12.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 13.Bird A P. DNA methylation—how important in gene control? Nature. 1984;307:503–504. doi: 10.1038/307503a0. [DOI] [PubMed] [Google Scholar]
- 14.Cross SH, Bird AP. CpG islands and genes. Curr. Opin. Genet. Dev. 1995;5:309–14. doi: 10.1016/0959-437x(95)80044-1. [DOI] [PubMed] [Google Scholar]
- 15.Okano M, Bell DW, Haber D A, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 16.Clarke M F. Epigenetic regulation of normal and cancer stem cells. Ann. N Y Acad. Sci. 2005;1044:90–93. doi: 10.1196/annals.1349.012. [DOI] [PubMed] [Google Scholar]
- 17.Laird PW. The power and the promise of DNA methylation markers. Nat. Rev. Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 18.Weaver J R, Susiarjo M, Bartolomei M S. Imprinting and epigenetic changes in the early embryo. Mamm. Genome. 2009;20:532–543. doi: 10.1007/s00335-009-9225-2. [DOI] [PubMed] [Google Scholar]
- 19.Ogino S, Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Kirkner GJ, Fuchs CS. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int. J. Cancer. 2008;122:2767– 2773. doi: 10.1002/ijc.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Röhrs S, Dirks WG, Meyer C, Marschalek R, Scherr M, Slany R, Wallace A, Drexler HG, Quentmeier H. Hypomethylation and expression of BEX2, IGSF4 and TIMP3 indicative of MLL translocations in Acute Myeloid Leukemia. Mol. Cancer. 2009;8:86. doi: 10.1186/1476-4598-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feinberg A P. Cancer epigenetics takes center stage. PNAS. 2001;98:392–394. doi: 10.1073/pnas.98.2.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ehrlich M. DNA methylation and cancer-associated genetic instability. Adv. Exp. Med. Biol. 2005;570:363–392. doi: 10.1007/1-4020-3764-3_13. [DOI] [PubMed] [Google Scholar]
- 23.Niculescu MD, Zeisel SH. Diet, methyl donors and DNA methylation: interactions between dietary folate, methionine and choline. J. Nutr. Suppl. 2002;132:23333S–23335S. doi: 10.1093/jn/132.8.2333S. [DOI] [PubMed] [Google Scholar]
- 24.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 25.Doerfler W. Patterns of DNA methylation--evolutionary vestiges of foreign DNA inactivation as a host defense mechanism. A proposal. Biol. Chem. Hoppe. Seyler. 1991;372:557–564. [PubMed] [Google Scholar]
- 26.Schulz WA. DNA methylation in urological malignancies. Int. J. Oncol. 1998;13:151–167. [PubMed] [Google Scholar]
- 27.Gama-Sosa MA, Slagel VA, Trewyn RW, Oxenhandler R, Kuo KC, Gehrke CW, Ehrlich M. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 1983;11:6883–6894. doi: 10.1093/nar/11.19.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bedford MT, van Helden PD. Hypomethylation of DNA in pathological conditions of the human prostate. Cancer Res. 1987;47:5274–5276. [PubMed] [Google Scholar]
- 29.Sakai T, Toguchida J, Ohtani N, Yandell DW, Rapaport JM, Dryja TP. Allele-specific hypermethylation of the retinoblastoma tumor-suppressor gene. Am. J. Hum. Genet. 1991;48:880–888. [PMC free article] [PubMed] [Google Scholar]
- 30.Herman JG, Latif F, Weng Y, Lerman MI, Zbar B, Liu S, Samid D, Duan DS, Gnarra JR, Linehan WM, Baylin SB. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. PNAS. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graff JR, Herman JG, Lapidus RG, Chopra H, Xu R, Jarrard DF, Isaacs WB, Pitha PM, Davidson NE, Baylin SB. E-Cadherin expression is silenced by DNA hypermethylation in human breast and prostate carcinomas. Cancer Res. 1995;55:5195–5199. [PubMed] [Google Scholar]
- 32.Reik W, Surani MA. Cancer genetics.Genomic imprinting and embryonal tumours. . Nature. 1989;338:112–113. doi: 10.1038/338112a0. [DOI] [PubMed] [Google Scholar]
- 33.Rainier S, Johnson LA, Dobry CJ, Ping AJ, Grundy PE, Feinberg AP. Relaxation of imprinted genes in human cancer. Nature. 1993;362:747–749. doi: 10.1038/362747a0. [DOI] [PubMed] [Google Scholar]
- 34.Jarrard DF, Bova GS, Isaacs WB. DNA methylation, molecular genetics, and linkage studies in prostate cancer. Prostate. 1996;6:36–44. [PubMed] [Google Scholar]
- 35.Paul R, Ewing CM, Jarrard DF, Isaacs WB. The Cadherin cell-cell adhesion pathway in prostate cancer progression. Br. J. Urol. 1997;79:37–43. doi: 10.1111/j.1464-410x.1997.tb00799.x. [DOI] [PubMed] [Google Scholar]
- 36.Jarrard DF, Paul R, van Bokhoven A, Nguyen SH, Bova GS, Wheelock MJ, Johnson KR, Schalken J, Bussemakers M, Isaacs WB. P-Cadherin is a basal cell-specific epithelial marker that is not expressed in prostate cancer. Clin. Cancer Res. 1997;3:2121–2128. [PubMed] [Google Scholar]
- 37.Li LC, Zhao H, Shiina H, Kane CJ, Dahiya R. PGDB: a curated and integrated database of genes related to the prostate. Nucleic Acids Res. 2003;31:291– 293. doi: 10.1093/nar/gkg008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489 –92. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 39.Strange RC, Spiteri MA, Ramachandran S, Fryer AA. Glutathione-S-transferase family of enzymes. Mutat. Res. 2001;482:21–6. doi: 10.1016/s0027-5107(01)00206-8. [DOI] [PubMed] [Google Scholar]
- 40.Henderson CJ, McLaren AW, Moffat GJ, Bacon EJ, Wolf CR. Pi-class glutathione S-transferase: regulation and function. Chem. Biol. Interact. 1998;111-112:69–82. doi: 10.1016/s0009-2797(97)00176-2. [DOI] [PubMed] [Google Scholar]
- 41.Brooks JD, Weinstein M, Lin X, Sun Y, Pin SS, Bova GS, Epstein JI, Isaacs WB, Nelson WG. CG island methylation changes near the GSTP1 gene in prostatic intraepithelial neoplasia. Cancer Epidemiol. Biomarkers Prev. 1998;7:531–536. [PubMed] [Google Scholar]
- 42.Nakayama M, Bennett CJ, Hicks JL, Epstein JI, Platz EA, Nelson WG, De Marzo AM. Hypermethylation of the human glutathione S-transferase-pi gene (GSTP1) CpG island is present in a subset of proliferative inflammatory atrophy lesions but not in normal or hyperplastic epithelium of the prostate: a detailed study using laser-capture microdissection. Am. J. Pathol. 2003;163:923– 933. doi: 10.1016/s0002-9440(10)63452-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Esteller M, Herman JG. Generating mutations but providing chemosensitivity: the role of O6-methylguanine DNA methyltransferase in human cancer. Oncogene. 2004;23:1– 8. doi: 10.1038/sj.onc.1207316. [DOI] [PubMed] [Google Scholar]
- 44.Kang GH, Lee S, Lee HJ, Hwang KS. Aberrant CpG island hypermethylation of multiple genes in prostate cancer and prostatic intraepithelial neoplasia. J. Pathol. 2004;202:233– 240. doi: 10.1002/path.1503. [DOI] [PubMed] [Google Scholar]
- 45.Konishi N, Nakamura M, Kishi M, Nishimine M, Ishida E, Shimada K. DNA hypermethylation status of multiple genes in prostate adenocarcinomas. Jpn. J. Cancer Res. 2002;93:767–773. doi: 10.1111/j.1349-7006.2002.tb01318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamanaka M, Watanabe M, Yamada Y, Takagi A, Murata T, Takahashi H, Suzuki H, Ito H, Tsukino H, Katoh T, Sugimura Y, Shiraishi T. Altered methylation of multiple genes in carcinogenesis of the prostate. Int. J. Cancer. 2003;106:382–387. doi: 10.1002/ijc.11227. [DOI] [PubMed] [Google Scholar]
- 47.Maruyama R, Toyooka S, Toyooka KO, Virmani AK, Zochbauer-Muller S, Farinas AJ, Minna JD, McConnell J, Frenkel EP, Gazdar AF. Aberrant promoter methylation profile of prostate cancers and its relationship to clinicopathological features. Clin. Cancer Res. 2002;8:514 –519. [PubMed] [Google Scholar]
- 48.Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr. Rev. 2004;25:276–308. doi: 10.1210/er.2002-0032. [DOI] [PubMed] [Google Scholar]
- 49.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261– 282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 50.Jarrard DF, Kinoshita H, Shi Y, Sandefur C, Hoff D, Meisner LF, Chang C, Herman JG, Isaacs WB, Nassif N. Methylation of the androgen receptor promoter CpG island is associated with loss of androgen receptor expression in prostate cancer cells. Cancer Res. 1998;58:5310– 5314. [PubMed] [Google Scholar]
- 51.Newmark JR, Hardy DO, Tonb DC, Carter BS, Epstein JI, Isaacs WB, Brown TR, Barrack ER. Androgen receptor gene mutations in human prostate cancer. Proc. Natl. Acad. Sci. 1992;89:6319 –6323. doi: 10.1073/pnas.89.14.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Visakorpi T, Hyytinen E, Koivisto P, Tanner M, Keinanen R, Palmberg C, Palotie A Tammela T, Isola J Kallioniemi OP. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat. Genet. 1995;9:401– 406. doi: 10.1038/ng0495-401. [DOI] [PubMed] [Google Scholar]
- 53.Gonzalgo ML, Pavlovich CP, Lee SM, Nelson WG. Prostate cancer detection by GSTP1 methylation analysis of postbiopsy urine specimens. Clin. Cancer Res. 2003;9:2673–2677. [PubMed] [Google Scholar]
- 54.Kinoshita H, Shi Y, Sandefur C, Meisner LF, Chang C, Choon A, Reznikoff CR, Bova GS, Friedl A, Jarrard DF. Methylation of the androgen receptor minimal promoter silences transcription in human prostate cancer. Cancer Res. 2000;60:3623–3630. [PubMed] [Google Scholar]
- 55.Nakayama T, Watanabe M, Suzuki H, Toyota M, Sekita N, Hirokawa Y, Mizokami A, Ito H, Yatani R, Shiraishi T. Epigenetic regulation of androgen receptor gene expression in human prostate cancers. Lab. Invest. 2000;80:1789–1796. doi: 10.1038/labinvest.3780190. [DOI] [PubMed] [Google Scholar]
- 56.Sasaki M, Tanaka Y, Perinchery G, Dharia A, Kotcherguina I, Fujimoto S, Dahiya R. Methylation and inactivation of estrogen, progesterone, and androgen receptors in prostate cancer. J. Natl. Cancer Inst. 2002;94:384–90. doi: 10.1093/jnci/94.5.384. [DOI] [PubMed] [Google Scholar]
- 57.Huggins C. Two principles in endocrine therapy of cancers: hormone deprival and hormone interference. Cancer Res. 1965;25:1163–1167. [PubMed] [Google Scholar]
- 58.Castagnetta LA, Carruba G. Human prostate cancer: a direct role for oestrogens. Ciba Found Symp. 1995;191:269– 286. doi: 10.1002/9780470514757.ch16. [DOI] [PubMed] [Google Scholar]
- 59.Carruba G, Pfeffer U, Fecarotta E, Coviello DA, D’Amato E, Lo Castro M, Vidali G, Castagnetta L. Estradiol inhibits growth of hormone-nonresponsive PC3 human prostate cancer cells. Cancer Res. 1994;54:1190– 1193. [PubMed] [Google Scholar]
- 60.Jarred RA, McPherson SJ, Bianco JJ, Couse JF, Korach KS, Risbridger GP. Prostate phenotypes in estrogen-modulated transgenic mice. Trends Endocrinol. Metab. 2002;13:163–168. doi: 10.1016/s1043-2760(02)00575-1. [DOI] [PubMed] [Google Scholar]
- 61.Leav I, Lau KM, Adams JY, McNeal JE, Taplin ME, Wang J, Singh H, Ho SM. Comparative studies of the estrogen receptors ß and a and the androgen receptor in normal human prostate glands, dysplasia, and in primary and metastatic carcinoma. Am. J. Pathol. 2001;159:79 –92. doi: 10.1016/s0002-9440(10)61676-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brolin J, Skoog L, Ekman P. Immunohistochemistry and biochemistry in detection of androgen, progesterone, and estrogen receptors in benign and malignant human prostatic tissue. Prostate. 1992;20:281–295. doi: 10.1002/pros.2990200404. [DOI] [PubMed] [Google Scholar]
- 63.Hobisch A, Hittmair A, Daxenbichler G, Wille S, Radmayr C, Hobisch-Hagen P, Bartsch G, Klocker H, Culig Z. Metastatic lesions from prostate cancer do not express oestrogen and progesterone receptors. J. Pathol. 1997;182:356–361. doi: 10.1002/(SICI)1096-9896(199707)182:3<356::AID-PATH863>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 64.Horvath LG, Henshall SM, Lee CS, Head DR, Quinn DI, Makela S, Delprado W, Golovsky D, Brenner PC, O'Neill G, Kooner R, Stricker PD, Grygiel JJ, Gustafsson JA, Sutherland RL. Frequent loss of estrogen receptor-beta expression in prostate cancer. Cancer Res. 2001;61:5331–5335. [PubMed] [Google Scholar]
- 65.Pasquali D, Rossi V, Esposito D, Abbondanza C, Puca GA, Bellastella A, Sinisi A. Loss of estrogen receptor beta expression in malignant human prostate cells in primary cultures and in prostate cancer tissues. J. Clin. Endocrinol. Metab. 2001;86:2051–2055. doi: 10.1210/jcem.86.5.7441. [DOI] [PubMed] [Google Scholar]
- 66.Li LC, Chui R, Nakajima K, Oh BR, Au HC, Dahiya R. Frequent methylation of estrogen receptor in prostate cancer: correlation with tumor progression. Cancer Res. 2000;60:702–706. [PubMed] [Google Scholar]
- 67.Li LC, Yeh CC, Nojima D, Dahiya R. Cloning and characterization of human estrogen receptor beta promoter. Biochem. Biophys. Res. Commun. 2000;275:682–689. doi: 10.1006/bbrc.2000.3363. [DOI] [PubMed] [Google Scholar]
- 68.Zhang J, Liu L, Pfeifer GP. Methylation of the retinoid response gene TIG1 in prostate cancer correlates with methylation of the retinoic acid receptor beta gene. Oncogene. 2004;23:2241–2249. doi: 10.1038/sj.onc.1207328. [DOI] [PubMed] [Google Scholar]
- 69.Nojima D, Li LC, Dharia A, Perinchery G, Ribeiro-Filho L, Yen TS, Dahiya R. CpG hypermethylation of the promoter region inactivates the estrogen receptor-beta gene in patients with prostate carcinoma. Cancer. 2001;92:2076–2083. doi: 10.1002/1097-0142(20011015)92:8<2076::aid-cncr1548>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 70.Okuno M, Kojima S, Matsushima-Nishiwaki R, Tsurumi H, Muto Y, Friedman SL, Moriwaki H. Retinoids in cancer chemoprevention. Curr. Cancer Drug Targets. 2004;3:285–298. doi: 10.2174/1568009043333023. [DOI] [PubMed] [Google Scholar]
- 71.Van der Leede BJ, Folkers GE, Kruyt FA, Van der Saag PT. Genomic organization of the human retinoic acid receptor beta 2. Biochem. Biophys. Res. Commun. 1992;188:695–702. doi: 10.1016/0006-291x(92)91112-4. [DOI] [PubMed] [Google Scholar]
- 72.Nakayama T, Watanabe M, Yamanaka M, Hirokawa Y, Suzuki H, Ito H, Yatani R, Shiraishi T. The role of epigenetic modifications in retinoic acid receptor beta2 gene expression in human prostate cancers. Lab. Invest. 2001;81:1049 –1057. doi: 10.1038/labinvest.3780316. [DOI] [PubMed] [Google Scholar]
- 73.Woodson K, Gillespie J, Hanson J, Emmert-Buck M, Phillips JM, Linehan WM, Tangrea JA. Heterogeneous gene methylation patterns among pre-invasive and cancerous lesions of the prostate: a histopathologic study of whole mount prostate specimens. Prostate. 2004;60:25–31. doi: 10.1002/pros.20013. [DOI] [PubMed] [Google Scholar]
- 74.Woodson K, Hanson J, Tangrea J. A survey of gene-specific methylation in human prostate cancer among black and white men. Cancer Lett. 2004;205:181–188. doi: 10.1016/j.canlet.2003.11.027. [DOI] [PubMed] [Google Scholar]
- 75.Tokumaru Y, Harden SV, Sun DI, Yamashita K, Epstein JI, Sidransky D. Optimal use of a panel of methylation markers with GSTP1 hypermethylation in the diagnosis of prostate adenocarcinoma. Clin. Cancer Res. 2004;10:5518–5522. doi: 10.1158/1078-0432.CCR-04-0108. [DOI] [PubMed] [Google Scholar]
- 76.Singal R, Ferdinand L, Reis IM, Schlesselman JJ. Methylation of multiple genes in prostate cancer and the relationship with clinicopathological features of disease. Oncol. Rep. 2004;12:631–637. [PubMed] [Google Scholar]
- 77.Jerónimo C, Henrique R, Hoque MO, Ribeiro FR, Oliveira J, Fonseca D, Teixeira MR, Lopes C, Sidransky D. Quantitative RARbeta2 hypermethylation: a promising prostate cancer marker. Clin Cancer Res. 2004;10:4010–4014. doi: 10.1158/1078-0432.CCR-03-0643. [DOI] [PubMed] [Google Scholar]
- 78.Li LC, Okino ST, Dahiya R. DNA methylation in prostate cancer. Biochim. Biophys. Acta. 2004;1704:87–102. doi: 10.1016/j.bbcan.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 79.Jerónimo C, Henrique R, Hoque MO, Mambo E, Ribeiro FR, Varzim G, Oliveira J, Teixeira MR, Lopes C, Sidransky D. A quantitative promoter methylation profile of prostate cancer. Clin. Cancer Res. 2004;10:8472– 8478. doi: 10.1158/1078-0432.CCR-04-0894. [DOI] [PubMed] [Google Scholar]
- 80.Nguyen TT, Nguyen CT, Gonzales FA, Nichols PW, Yu MC, Jones PA. Analysis of cyclindependent kinase inhibitor expression and methylation patterns in human prostate cancers. Prostate. 2000;43:233–242. doi: 10.1002/(sici)1097-0045(20000515)43:3<233::aid-pros10>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 81.Yegnasubramanian S, Kowalski J, Gonzalgo ML, Zahurak M, Piantadosi S, Walsh PC, Bova GS, De Marzo AM, Isaacs WB, Nelson WG. Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res. 2004;64:1975–1986. doi: 10.1158/0008-5472.can-03-3972. [DOI] [PubMed] [Google Scholar]
- 82.Jarrard DF, Bova GS, Ewing CM, Pin SS, Nguyen SH, Baylin SB, Cairns P, Sidransky D, Herman JG, Isaacs WB. Deletional, mutational, and methylation analyses of CDKN2 (p16/MTS1) in primary and metastatic prostate cancer. Genes Chromo. Cancer. 1997;19:90–96. [PubMed] [Google Scholar]
- 83.Gu K, Mes-Masson AM, Gauthier J, Saad F. Analysis of the p16 tumor suppressor gene in early-stage prostate cancer. Mol. Carcinog. 1998;21:164–170. [PubMed] [Google Scholar]
- 84.Hoque MO, Topaloglu O, Begum S, Henrique R, Rosenbaum E, Van Criekinge W, Westra WH, Sidransky D. Quantitative methylation specific polymerase chain reaction gene patterns in urine sediment distinguish prostate cancer patients from control subjects. J. Clin. Oncol. 2005;23:6569–6575. doi: 10.1200/JCO.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 85.Florl AR, Steinhoff C, Müller M, Seifert HH, Hader C, Engers R, Ackermann R, Schulz WA. Coordinate hypermethylation at specific genes in prostate carcinoma precedes LINE-1 hypomethylation. Br. J. Cancer. 2004;91:985–994. doi: 10.1038/sj.bjc.6602030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Herman JG, Merlo A, Mao L, Lapidus RG, Issa JP, Davidson NE, Sidransky D, Baylin SB. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 87.Vos MD, Ellis CA, Bell A, Birrer MJ, Clark GJ. Ras uses the novel tumor suppressor RASSF1 as an effector to mediate apoptosis. J. Biol. Chem. 2000;275:35669–35672. doi: 10.1074/jbc.C000463200. [DOI] [PubMed] [Google Scholar]
- 88.Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21 . Nat. Genet. 2000;25:315– 319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 89.Liu L, Yoon JH, Dammann R, Pfeifer GP. Frequent hypermethylation of the RASSF1A gene in prostate cancer. Oncogene. 2002;21:6835–6840. doi: 10.1038/sj.onc.1205814. [DOI] [PubMed] [Google Scholar]
- 90.Greger V, Debus N, Lohmann D, Hopping W, Passarge E, Horsthemke B. Frequency and parental origin of hypermethylated RB1 alleles in retinoblastoma. Hum. Genet. 1994;94:491– 496. doi: 10.1007/BF00211013. [DOI] [PubMed] [Google Scholar]
- 91.Veigl ML, Kasturi L, Olechnowicz J, Ma AH, Lutterbaugh JD, Periyasamy S, Li GM, Drummond J, Modrich PL, Sedwick WD, Markowitz SD. Biallelic inactivation of hMLH1 by epigenetic gene silencing, a novel mechanism causing human MSI cancers. Proc. Natl. Acad. Sci. 1998;95:8698– 8702. doi: 10.1073/pnas.95.15.8698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li LC, Zhao H, Nakajima K, Oh BR, Ribeiro Filho LA, Carroll P, Dahiya R. Methylation of the E-Cadherin gene promoter correlates with progression of prostate cancer. Urology. 2001;166:705–709. [PubMed] [Google Scholar]
- 93.Watanabe M, Kakiuchi H, Kato H, Shiraishi T, Yatani R, Sugimura T, Nagao M. APC gene mutations in human prostate cancer. Jpn. J. Clin. Oncol. 1996;26:77–81. doi: 10.1093/oxfordjournals.jjco.a023188. [DOI] [PubMed] [Google Scholar]
- 94.Gerstein AV, Almeida TA, Zhao G, Chess E, Shih IeM, Buhler K, Pienta K, Rubin MA, Vessella R, Papadopoulos N. APC/CTNNB1 (beta-catenin) pathway alterations in human prostate cancers. Genes Chromosomes Cancer. 2002;34:9–16. doi: 10.1002/gcc.10037. [DOI] [PubMed] [Google Scholar]
- 95.Bastian PJ, Ellinger J, Wellmann A, Wernert N, Heukamp LC, Müller SC, Von Ruecker A. Diagnostic and prognostic information in prostate cancer with the help of a small set of hypermethylated gene loci. Clin. Cancer Res. 2005;11:4097–4106. doi: 10.1158/1078-0432.CCR-04-1832. [DOI] [PubMed] [Google Scholar]
- 96.Enokida H, Shiina H, Urakami S, Igawa M, Ogishima T, Li LC, Kawahara M, Nakagawa M, Kane CJ, Carroll PR, Dahiya R. Multigene methylation analysis for detection and staging of prostate cancer. Clin. Cancer Res. 2005;11:6582–6588. doi: 10.1158/1078-0432.CCR-05-0658. [DOI] [PubMed] [Google Scholar]
- 97.Shiina H, Breault JE, Basset WW, Enokida H, Urakami S, Li LC, Okino ST, Deguchi M, Kaneuchi M, Terashima M, Yoneda T, Shigeno K, Carroll PR, Igawa M, Dahiya R. Functional Loss of the gamma-catenin gene through epigenetic and genetic pathways in human prostate cancer. Cancer Res. 2005;65:2130–2138. doi: 10.1158/0008-5472.CAN-04-3398. [DOI] [PubMed] [Google Scholar]
- 98.Sekita N, Suzuki H, Ichikawa T, Kito H, Akakura K, Igarashi T, Nakayama T, Watanabe M, Shiraishi T, Toyota M, Yoshie O, Ito H. Epigenetic regulation of the KAI1 metastasis suppressor gene in human prostate cancer cell lines. Jpn. J. Cancer Res. 2001;92:947–951. doi: 10.1111/j.1349-7006.2001.tb01185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schmitt JF, Millar DS, Pedersen JS, Clark SL, Venter DJ, Frydenberg M, Molloy PL, Risbridger GP. Hypermethylation of the inhibin alpha-subunit gene in prostate carcinoma. Mol. Endocrinol. 2002;16:213–220. doi: 10.1210/mend.16.2.0771. [DOI] [PubMed] [Google Scholar]
- 100.Chen H, Toyooka S, Gazdar AF, Hsieh JT. Epigenetic regulation of a novel tumor suppressor gene (hDAB2IP) in prostate cancer cell lines. J. Biol. Chem. 2003;278:3121–3130. doi: 10.1074/jbc.M208230200. [DOI] [PubMed] [Google Scholar]
- 101.Murphy TM, Perry AS, Lawler M. The emergence of DNA methylation as a key modulator of aberrant cell death in prostate cancer. Endocr. Relat. Cancer. 2008;15:11–25. doi: 10.1677/ERC-07-0208. [DOI] [PubMed] [Google Scholar]
- 102.Rouprêt M, Hupertan V, Yates DR, Catto JW, Rehman I, Meuth M, Ricci S, Lacave R, Cancel-Tassin G, de la Taille A, Rozet F, Cathelineau X, Vallancien G, Hamdy FC, Cussenot O. Molecular detection of localized prostate cancer using quantitative methylation-specific PCR on urinary cells obtained following prostate massage. Clin. Cancer Res. 2007;13:1720–1725. doi: 10.1158/1078-0432.CCR-06-2467. [DOI] [PubMed] [Google Scholar]
- 103.Park J, Brena RM, Gruidl M, Zhou J, Huang T, Plass C, Tockman MS. CpG island hypermethylation profiling of lung cancer using restriction landmark genomic scanning (RLGS) analysis. Cancer Biomarkers. 2005;1:193–200. doi: 10.3233/cbm-2005-12-307. [DOI] [PubMed] [Google Scholar]
- 104.Sørensen KD, Wild PJ, Mortezavi A, Adolf K, Tørring N, Heebøll S, Ulhøi BP, Ottosen P, Sulser T, Hermanns T, Moch H, Borre M, Ørntoft TF, Dyrskjøt L. Genetic and epigenetic SLC18A2 silencing in prostate cancer is an independent adverse predictor of biochemical recurrence after radical prostatectomy. Clin. Cancer Res. 2009;15:1400–1410. doi: 10.1158/1078-0432.CCR-08-2268. [DOI] [PubMed] [Google Scholar]
- 105.Shivapurkar N, Toyooka S, Toyooka KO, Reddy J, Miyajima K, Suzuki M, Shigematsu H, Takahashi T, Parikh G, Pass HI, Chaudhary PM, Gazdar AF. Aberrant methylation of trail decoy receptor genes is frequent in multiple tumor types. Int. J. Cancer. 2004;109:786–792. doi: 10.1002/ijc.20041. [DOI] [PubMed] [Google Scholar]
- 106.Van Noesel MM, Van Bezouw S, Salomons GS, Voûte PA, Pieters R, Baylin SB, Herman JG, Versteeg R. Tumor-specific down-regulation of the tumor necrosis factor-related apoptosis-inducing ligand decoy receptors DcR1 and DcR2 is associated with dense promoter hypermethylation. Cancer Res. 2002;62:2157–2161. [PubMed] [Google Scholar]
- 107.Park JY, Zheng W, Kim D, Cheng JQ, Kumar N, Ahmad N, Pow-Sang J. Candidate tumor suppressor gene SLC5A8 is frequently down-regulated by promoter hypermethylation in prostate tumor. Cancer Detect. Prev. 2007;31:359–365. doi: 10.1016/j.cdp.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 108.Cho NY, Kim BH, Choi M, Yoo EJ, Moon KC, Cho YM, Kim D, Kang GH. Hypermethylation of CpG island loci and hypomethylation of LINE-1 and Alu repeats in prostate adenocarcinoma and their relationship to clinicopathological features. J. Pathol. 2007;211:269–277. doi: 10.1002/path.2106. [DOI] [PubMed] [Google Scholar]
- 109.Cheng Y, Kim JW, Liu W, Dunn TA, Luo J, Loza MJ, Kim ST, Zheng SL, Xu J, Isaacs WB, Chang BL. Genetic and epigenetic inactivation of TNFRSF10C in human prostate cancer. Prostate. 2009;69:327–335. doi: 10.1002/pros.20882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hornstein M, Hoffmann MJ, Alexa A, Yamanaka M, Müller M, Jung V, Rahnenführer J, Schulz WA. Protein phosphatase and TRAIL receptor genes as new candidate tumor genes on chromosome 8p in prostate cancer. Cancer Genomics Proteomics. 2008;5:123–136. [PubMed] [Google Scholar]
- 111.Stetler-Stevenson WG, Aznavoorian S, Liotta LA. Tumor cell interactions with the extracellular matrix during invasion and metastasis. Annu. Rev. Cell. Biol. 1993;9:541– 573. doi: 10.1146/annurev.cb.09.110193.002545. [DOI] [PubMed] [Google Scholar]
- 112.Takeichi M. Cadherin cell adhesion receptors as a morphogenetic regulator. Science. 1991;251:1451– 1455. doi: 10.1126/science.2006419. [DOI] [PubMed] [Google Scholar]
- 113.Hirohashi S, Kanai Y. Cell adhesion system and human cancer morphogenesis. Cancer Sci. 2003;94:575–581. doi: 10.1111/j.1349-7006.2003.tb01485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Quinn DI, Henshall SM, Sutherland RL. Molecular markers of prostate cancer outcome. Eur. J. Cancer. 2005;41:858–887. doi: 10.1016/j.ejca.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 115.Kallakury BV, Sheehan CE, Ross JS. Co-downregulation of cell adhesion proteins alpha- and beta-catenins, p120CTN, E-Cadherin, and CD44 in prostatic adenocarcinomas. Hum. Pathol. 2001;32:849–855. doi: 10.1053/hupa.2001.26463. [DOI] [PubMed] [Google Scholar]
- 116.Kallakury BV, Sheehan CE, Winn-Deen E, Oliver J, Fisher HA, Kaufman R P , Jr, Ross JA. Decreased expression of catenins (a and ß), p120 CTN, and E-Cadherin cell adhesion proteins and E-Cadherin gene promoter methylation in prostatic adenocarcinomas. Cancer. 2001;92:2786 –2795. doi: 10.1002/1097-0142(20011201)92:11<2786::aid-cncr10128>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 117.Gokaslan ZL, Chintala SK, York JE, Boyapati V, Jasti S, Sawaya R, Fuller G, Wildrick DM, Nicolson GL, Rao JS. Expression and role of matrix metalloproteinases MMP-2 and MMP-9 in human spinal column tumors. Clin. Exp. Metastasis. 1998;16:721–728. doi: 10.1023/a:1006580728338. [DOI] [PubMed] [Google Scholar]
- 118.Gomez DE, Alonso DF, Yoshiji H, Thorgeirsson UP. Tissue inhibitors of metalloproteinases: structure, regulation and biological functions. Eur. J. Cell Biol. 1997;74:111–122. [PubMed] [Google Scholar]
- 119.Ross JS, Kaur P, Sheehan CE, Fisher HA, Kaufman R.A, Jr, Kallakury BV. Prognostic significance of matrix metalloproteinase 2 and tissue inhibitor of metalloproteinase 2 expression in prostate cancer. Mod. Pathol. 2003;16:198–205. doi: 10.1097/01.MP.0000056984.62360.6C. [DOI] [PubMed] [Google Scholar]
- 120.Gao AC, Lou W, Dong JT, Isaacs JT. CD44 is a metastasis suppressor gene for prostatic cancer located on human chromosome 11p13. Cancer Res. 1997;57:846–849. [PubMed] [Google Scholar]
- 121.Noordzij MA, van Steenbrugge GJ, Verkaik NS, Schröder FH, van der Kwast TH. The prognostic value of CD44 isoforms in prostate cancer patients treated by radical prostatectomy. Clin. Cancer Res. 1997;3:805–815. [PubMed] [Google Scholar]
- 122.Aaltomaa S, Lipponen P, Ala-Opas M, Kosma VM. Expression and prognostic value of CD44 standard and variant v3 and v6 isoforms in prostate cancer. Eur. Urol. 2001;39:138–144. doi: 10.1159/000052428. [DOI] [PubMed] [Google Scholar]
- 123.Harden SV, Guo Z, Epstein JI, Sidransky D. Quantitative GSTP1 methylation clearly distinguishes benign prostatic tissue and limited prostate adenocarcinoma. J. Urol. 2003;169:1138–1142. doi: 10.1097/01.ju.0000049627.90307.4d. [DOI] [PubMed] [Google Scholar]
- 124.Lou W, Krill D, Dhir R, Becich MJ, Dong JT, Frierson HF, Jr, Isaacs WB, Isaacs JT, Gao AC. Methylation of the CD44 metastasis suppressor gene in human prostate cancer. Cancer Res. 1999;59:2329–2331. [PubMed] [Google Scholar]
- 125.Verkaik NS, van Steenbrugge GJ, van Weerden WM, Bussemakers MJ, van der Kwast TH. Silencing of CD44 expression in prostate cancer by hypermethylation of the CD44 promoter region. Lab. Invest. 2000;80:1291–1298. doi: 10.1038/labinvest.3780137. [DOI] [PubMed] [Google Scholar]
- 126.Kito H, Suzuki H, Ichikawa T, Sekita N, Kamiya N, Akakura K, Igarashi T, Nakayama T, Watanabe M, Harigaya K, Ito H. Hypermethylation of the CD44 gene is associated with progression and metastasis of human prostate cancer. Prostate. 2001;49:110–115. doi: 10.1002/pros.1124. [DOI] [PubMed] [Google Scholar]
- 127.Woodson K, Hayes R, Wideroff L, Villaruz L, Tangrea J. Hypermethylation of GSTP1, CD44, and E-Cadherin genes in prostate cancer among US Blacks and Whites. Prostate. 2003;55:199–205. doi: 10.1002/pros.10236. [DOI] [PubMed] [Google Scholar]
- 128.Padar A, Sathyanarayana UG, Suzuki M, Maruyama R, Hsieh JT, Frenkel EP, Minna JD, Gazdar AF. Inactivation of cyclin D2 gene in prostate cancers by aberrant promoter methylation. Clin. Cancer Res. 2003;9:4730–4734. [PubMed] [Google Scholar]
- 129.Bachmann N, Haeusler J, Luedeke M, Kuefer R, Perner S, Assum G, Paiss T, Hoegel J, Vogel W, Maier C. Expression changes of CAV1 and EZH2, located on 7q31 approximately q36, are rarely related to genomic alterations in primary prostate carcinoma. Cancer Genet. Cytogenet. 2008;182:103–110. doi: 10.1016/j.cancergencyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 130.Cui J, Rohr LR, Swanson G, Speights VO, Maxwell T, Brothman AR. Hypermethylation of the caveolin-1 gene promoter in prostate cancer. Prostate. 2001;46:249–256. doi: 10.1002/1097-0045(20010215)46:3<249::aid-pros1030>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 131.Alumkal JJ, Zhang Z, Humphreys EB, Bennett C, Mangold LA, Carducci MA, Partin AW, Garrett-Mayer E, DeMarzo AM, Herman JG. Effect of DNA methylation on identification of aggressive prostate cancer. Urology. 2008;72:1234–1239. doi: 10.1016/j.urology.2007.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Oudes AJ, Roach JC, Walashek LS, Eichner LJ, True LD, Vessella RL, Liu AY. Application of Affymetrix array and Massively Parallel Signature Sequencing for identification of genes involved in prostate cancer progression. BMC Cancer. 2005;5:86. doi: 10.1186/1471-2407-5-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rehman I, Cross SS, Catto JW, Leiblich A, Mukherjee A, Azzouzi AR, Leung HY, Hamdy FC. Promoter hyper-methylation of calcium binding proteins S100A6 and S100A2 in human prostate cancer. Prostate. 2005;65:322–330. doi: 10.1002/pros.20302. [DOI] [PubMed] [Google Scholar]
- 134.Guan M, Xu C, Zhang F, Ye C. Aberrant methylation of EphA7 in human prostate cancer and its relation to clinicopathologic features. Int. J. Cancer. 2009;124:88–94. doi: 10.1002/ijc.23890. [DOI] [PubMed] [Google Scholar]
- 135.Karam JA, Lotan Y, Roehrborn CG, Ashfaq R, Karakiewicz PI, Shariat SF. Caveolin-1 overexpression is associated with aggressive prostate cancer recurrence. Prostate. 2007;67:614–622. doi: 10.1002/pros.20557. [DOI] [PubMed] [Google Scholar]
- 136.Lee SW. H-Cadherin, a novel Cadherin with growth inhibitory functions and diminished expression in human breast cancer. Nat. Med. 1996;2:776–782. doi: 10.1038/nm0796-776. [DOI] [PubMed] [Google Scholar]
- 137.Urnov FD. Methylation and the genome: the power of a small amendment. J. Nutr. 2002;132:2450S–2456S. doi: 10.1093/jn/132.8.2450S. [DOI] [PubMed] [Google Scholar]
- 138.Dunn BK. Hypomethylation: one side of a larger picture. Ann. N.Y. Acad. Sci. 2003;983:28–42. doi: 10.1111/j.1749-6632.2003.tb05960.x. [DOI] [PubMed] [Google Scholar]
- 139.Santourlidis S, Florl A, Ackermann R, Wirtz HC, Schulz WA. High frequency of alterations in DNA methylation in adenocarcinoma of the prostate. Prostate. 1999;39:166–174. doi: 10.1002/(sici)1097-0045(19990515)39:3<166::aid-pros4>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 140.Schulz WA, Elo JP, Florl AR, Pennanen S, Santourlidis S, Engers R, Buchardt M, Seifert HH, Visakorpi T. Genomewide DNA hypomethylation is associated with alterations on chromosome 8 in prostate carcinoma. Genes Chromosomes Cancer. 2002;35:58–65. doi: 10.1002/gcc.10092. [DOI] [PubMed] [Google Scholar]
- 141.Brothman AR, Swanson G, Maxwell TM, Cui J, Murphy KJ, Herrick J, Speights VO, Isaac J, Rohr LR. Global hypomethylation is common in prostate cancer cells: a quantitative predictor for clinical outcome? Cancer Genet. Cytogenet. 2005;156:31–36. doi: 10.1016/j.cancergencyto.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 142.Ogishima T, Shiina H, Breault JE, Tabatabai L, Bassett WW, Enokida H, Li LC, Kawakami T, Urakami S, Ribeiro-Filho LA, Terashima M, Fujime M, Igawa M, Dahiya R. Increased heparanase expression is caused by promoter hypomethylation and up-regulation of transcriptional factor early growth response-1 in human prostate cancer. Clin. Cancer Res. 2005;11:1028–1036. [PubMed] [Google Scholar]
- 143.Tokizane T, Shiina H, Igawa M, Enokida H, Urakami S, Kawakami T, Ogishima T, Okino ST, Li LC, Tanaka Y, Nonomura N, Okuyama A, Dahiya R. Cytochrome P450 1B1 is overexpressed and regulated by hypomethylation in prostate cancer. Clin. Cancer Res. 2005;11:5793–5801. doi: 10.1158/1078-0432.CCR-04-2545. [DOI] [PubMed] [Google Scholar]
- 144.Sassaman DM, Dombroski BA, Moran JV, Kimberland ML, Naas TP, DeBerardinis RJ, Gabriel A, Swergold GD, Kazazian HH., Jr Many human L1 elements are capable of retrotransposition. Nat. Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- 145.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89 –92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 146.Feinberg AP, Vogelstein B. Hypomethylation of ras oncogenes in primary human cancers. Biochem. Biophys. Res. Commun. 1983;111:47–54. doi: 10.1016/s0006-291x(83)80115-6. [DOI] [PubMed] [Google Scholar]
- 147.Van Veldhuizen PJ, Sadasivan R, Cherian R, Wyatt A. Urokinase-type plasminogen activator expression in human prostate carcinomas. Am. J. Med. Sci. 1996;312:8–11. doi: 10.1097/00000441-199607000-00002. [DOI] [PubMed] [Google Scholar]
- 148.Helenius MA, Saramaki OR, Linja MJ, Tammela TL, Visakorpi T. Amplification of urokinase gene in prostate cancer. Cancer Res. 2001;61:5340–5344. [PubMed] [Google Scholar]
- 149.Pakneshan P, Xing RH, Rabbani SA. Methylation status of uPA promoter as a molecular mechanism regulating prostate cancer invasion and growth in vitro and in vivo . FASEB.J. 2003;17:1081–1088. doi: 10.1096/fj.02-0973com. [DOI] [PubMed] [Google Scholar]
- 150.Cho B, Lim Y, Lee DY, Park SY, Lee H, Kim WH, Yang H, Bang YJ, Jeoung DI. Identification and characterization of a novel cancer/testis antigen gene CAGE Biochem. Biophys. Res. Commun. 2002;292:715–726. doi: 10.1006/bbrc.2002.6701. [DOI] [PubMed] [Google Scholar]
- 151.Cho B, Lee H, Jeong S, Bang YJ, Lee HJ, Hwang KS, Kim HY, Lee YS, Kang GH, Jeoung DI. Promoter hypomethylation of a novel cancer/testis antigen gene CAGE is correlated with its aberrant expression and is seen in premalignant stage of gastric carcinoma. Biochem. Biophys. Res. Commun. 2003;307:52– 63. doi: 10.1016/s0006-291x(03)01121-5. [DOI] [PubMed] [Google Scholar]
- 152.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273(5281):1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 153.Reams R R, Agrawal D, Davis M B, Yoder S, Odedina F T, Kumar N, Higginbotham J M, Akinremi T, Suther S, Soliman K F. Microarray comparison of prostate tumor gene expression in African-American and Caucasian American males: a pilot project study. Infect. Agent Cancer. 2009;10(4 Suppl 1: S3) doi: 10.1186/1750-9378-4-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kwabi-Addo B, Wang S, Chung W, Jelinek J, Patierno S R, Wang B D, Andrawis R, Lee N H, Apprey V, Issa J P, Ittmann M. Identification of differentially methylated genes in normal prostate tissues from African American and Caucasian men. Clin. Cancer Res. 2010;15(14 ):3539–3547. doi: 10.1158/1078-0432.CCR-09-3342. [DOI] [PubMed] [Google Scholar]
- 155.Woodson K, Hayes R, Wideroff L, Villaruz L, Tangrea J. Hypermethylation of GSTP1, CD44, and E-Cadherin genes in prostate cancer among US Blacks and Whites. Prostate. 2003;55(3 ):199–205. doi: 10.1002/pros.10236. [DOI] [PubMed] [Google Scholar]
- 156.Kito H, Suzuki H, Ichikawa T, Sekita N, Kamiya N, Akakura K. Igarashi T, Nakayama T, Watanabe M, Harigaya K, Ito H. Hypermethylation of the CD44 gene is associated with progression and metastasis of human prostate cancer. Prostate. 2001;49(2 ):110–115. doi: 10.1002/pros.1124. [DOI] [PubMed] [Google Scholar]
- 157.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 158.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 159.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 160.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 161.Rando OJ, Chang HY. Genome-wide views of chromatin structure. Annu. Rev. Biochem. 2009;78:245–271. doi: 10.1146/annurev.biochem.78.071107.134639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kustatscher G, Ladurner AG. Modular paths to ‘decoding’ and ‘wiping’ histone lysine methylation. Curr. Opin. Chem. Biol. 2007;11:628–635. doi: 10.1016/j.cbpa.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 163.McKinsey TA, Zhang CL, Olson EN. Control of muscle development by dueling HATs and HDACs. Curr. Opin. Genet. Dev. 2001;11:497–504. doi: 10.1016/s0959-437x(00)00224-0. [DOI] [PubMed] [Google Scholar]
- 164.Gray SG, Ekstrom TJ. The human histone deacetylase family. Exp. Cell Res. 2001;262:75–83. doi: 10.1006/excr.2000.5080. [DOI] [PubMed] [Google Scholar]
- 165.Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 166.Goll MG, Bestor TH. Histone modifi cation and replacement in chromatin activation. Genes Dev. 2002;16:1739–1742. doi: 10.1101/gad.1013902. [DOI] [PubMed] [Google Scholar]
- 167.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 168.Fahrner JA, Eguchi S, Herman JG, Baylin SB. Dependence of histone modifi cations and gene expression on DNA hypermethylation in cancer. Cancer Res. 2002;62:7213–7218. [PubMed] [Google Scholar]
- 169.Nguyen CT, Weisenberger DJ, Velicescu M, Gonzales FA, Lin JC, Liang G, Jones PA. Histone H3- lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2'-deoxycytidine. Cancer Res. 2002;62:6456–6461. [PubMed] [Google Scholar]
- 170.Dobosy JR, Selker EU. Emerging connections between DNA methylation and histone acetylation. Cell Mol. Life Sci. 2001;58:721–727. doi: 10.1007/PL00000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Halkidou K, Gnanapragasam VJ, Mehta PB, Logan IR, Brady ME, Cook S, Leung HY, Neal DE, Robson CN. Expression of Tip60, an androgen receptor coactivator, and its role in prostate cancer development. Oncogene. 2003;22:2466–2477. doi: 10.1038/sj.onc.1206342. [DOI] [PubMed] [Google Scholar]
- 172.Debes JD, Schmidt LJ, Huang H, Tindall DJ. p300 mediates androgen-independent transactivation of the androgen receptor by interleukin 6. Cancer Res. 2002;62:5632–5636. [PubMed] [Google Scholar]
- 173.Yegnasubramanian S, Lin X, Haffner MC, DeMarzo AM, Nelson WG. Combination of methylated-DNA precipitation and methylation-sensitive restriction enzymes (COMPARE-MS) for the rapid, sensitive and quantitative detection of DNA methylation. Nucleic Acids Res. 2006;34:e19. doi: 10.1093/nar/gnj022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Lee WH, Isaacs WB, Bova GS, Nelson WG. CG island methylation changes near the GSTP1 gene in prostatic carcinoma cells detected using the polymerase chain reaction: a new prostate cancer biomarker. Cancer Epidemiol. Biomarkers Prev. 1997;6:443–450. [PubMed] [Google Scholar]
- 175.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad.Sci. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Miyamoto K, Ushijima T. Diagnostic and therapeutic applications of epigenetics. Jpn. J. Clin. Oncol. 2005;35:293–301. doi: 10.1093/jjco/hyi088. [DOI] [PubMed] [Google Scholar]
- 178.Lee WH, Morton RA, Epstein JI, Brooks JD, Campbell PA, Bova GS, Hsieh WS, Isaacs WB, Nelson WG. Cytidine methylation of regulatory sequences near the pi-class glutathione S-transferase gene accompanies human prostatic carcinogenesis. Proc. Natl. Acad. Sci. 1994;91:11733–11737. doi: 10.1073/pnas.91.24.11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Harden SV, Guo Z, Epstein JI, Sidransky D. Quantitative GSTP1 methylation clearly distinguishes benign prostatic tissue and limited prostate adenocarcinoma. J. Urol. 2003;169:1138–1142. doi: 10.1097/01.ju.0000049627.90307.4d. [DOI] [PubMed] [Google Scholar]
- 180.Jerónimo C, Usadel H, Henrique R, Silva C, Oliveira J, Lopes C, Sidransky D. Quantitative GSTP1 hypermethylation in bodily fluids of patients with prostate cancer. Urology. 2002;60:1131–1135. doi: 10.1016/s0090-4295(02)01949-0. [DOI] [PubMed] [Google Scholar]
- 181.Gonzalgo ML, Nakayama M, Lee SM, De Marzo AM, Nelson WG. Detection of GSTP1 methylation in prostaticm secretions using combinatorial MSP analysis. Urology. 2004;63:414–418. doi: 10.1016/j.urology.2003.08.039. [DOI] [PubMed] [Google Scholar]
- 182.Goessl C, Muller M, Heicappell R, Krause H, Miller K. DNA-based detection of prostate cancer in blood, urine, and ejaculates. Ann. NY. Acad. Sci. 2001;945:51–58. doi: 10.1111/j.1749-6632.2001.tb03863.x. [DOI] [PubMed] [Google Scholar]
- 183.Bastian PJ, Ellinger J, Schmidt D, Wernert N, Wellmann A, Muller SC, von Rücker A. GSTP1 hypermethylation as a molecular marker in the diagnosis of prostatic cancer: is there a correlation with clinical stage, Gleason grade, PSA value or age? Eur. J. Med. Res. 2004;9:523–527. [PubMed] [Google Scholar]
- 184.Köllermann J, Müller M, Goessl C, Krause H, Helpap B, Pantel K, Miller K. Methylation-specific PCR for DNAbased detection of occult tumor cells in lymph nodes of prostate cancer patients. Eur. Urol. 2003;44:533–538. doi: 10.1016/s0302-2838(03)00361-0. [DOI] [PubMed] [Google Scholar]
- 185.Das PM, Singal R. DNA methylation and cancer. J. Clin. Oncol. 2004;22:4632–4642. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- 186.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 187.Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A, MacLeod AR. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat. Genet. 2003;33:61–65. doi: 10.1038/ng1068. [DOI] [PubMed] [Google Scholar]
- 188.Lin X, Tascilar M, Lee WH, Vles WJ, Lee BH, Veeraswamy R, Asgari K, Freije D, van Rees B, Gage WR, Bova GS, Isaacs WB, Brooks JD, DeWeese TL, De Marzo AM, Nelson WG. GSTP1 CpG island hypermethylation is responsible for the absence of GSTP1 expression in human prostate cancer cells. Am. J. Pathol. 2001;159:1815–1826. doi: 10.1016/S0002-9440(10)63028-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Taylor SM, Jones PA. Changes in phenotypic expression in embryonic and adult cells treated with 5-azacytidine. J. Cell Phys. 1982;111:187–194. doi: 10.1002/jcp.1041110210. [DOI] [PubMed] [Google Scholar]
- 190.Friedman S. The irreversible binding of azacytosine containing DNA fragments to bacterial DNA (cytosine-5) methyltransferases. J. Biol. Chem. 1985;260:5698–5705. [PubMed] [Google Scholar]
- 191.Cheng JC, Weisenberger DJ, Gonzales FA, Liang G, Xu GL, Hu YG, Marquez VE, Jones PA. Continuous zebularine treatment effectively sustains demethylation in human bladder cancer cells. Mol. Cell Biol. 2004;24:1270–1278. doi: 10.1128/MCB.24.3.1270-1278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lin X, Asgari K, Putzi MJ, Gage WR, Yu X, Cornblatt BS, Kumar A, Piantadosi S, DeWeese TL, De Marzo AM, Nelson WG. Reversal of GSTP1 CpG island hypermethylation and reactivation of pi-class glutathione S-transferase (GSTP1) expression in human prostate cancer cells by treatment with procainamide. Cancer Res. 2001;61:8611–8616. [PubMed] [Google Scholar]
- 193.Villar-Garea A, Fraga MF, Espada J, Esteller M. Procaine is a DNAdemethylating agent with growth-inhibitory effects in human cancer cells. Cancer Res. 2003;63:4984–4989. [PubMed] [Google Scholar]
- 194.Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H, Welsh W, Yang CS. Tea polyphenol (-) epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003;63:7563–7570. [PubMed] [Google Scholar]
- 195.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 196.Chinnaiyan P, Vallabhaneni G, Armstrong E, Huang SM, Harari PM. Modulation of radiation response by histone deacetylase inhibition. Int. J. Radiat. Oncol. Biol. Phys. 2005;62:223–229. doi: 10.1016/j.ijrobp.2004.12.088. [DOI] [PubMed] [Google Scholar]
- 197.Deroanne CF, Bonjean K, Servotte S, Devy L, Colige A, Clausse N, Blacher S, Verdin E, Foidart JM, Nusgens BV, Castronovo V. Histone deacetylases inhibitors as antiangiogenic agents altering vascular endothelial growth factor signaling. Oncogene. 2002;21:427–436. doi: 10.1038/sj.onc.1205108. [DOI] [PubMed] [Google Scholar]
- 198.Butler LM, Agus DB, Scher HI, Higgins B, Rose A, Cordon-Cardo C, Thaler HT, Rifkind RA, Marks PA, Richon VM. Suberoylanilide hydroxamic acid, an inhibitor of histone deacetylase, suppresses the growth of prostate cancer cells in vitro and in vivo. Cancer Res. 2000;60:5165–5170. [PubMed] [Google Scholar]
- 199.Marks PA, Richon VM, Miller T, Kelly WK. Histone deacetylase inhibitors. Adv. Cancer Res. 2004;91:137–168. doi: 10.1016/S0065-230X(04)91004-4. [DOI] [PubMed] [Google Scholar]
- 200.Qian DZ, Kato Y, Shabbeer S, Wei Y, Verheul HM, Salumbides B, Sanni T, Atadja P, Pili R. Targeting tumor angiogenesis with histone deacetylase inhibitors: the hydroxamic acid derivative LBH589. Clin. Cancer Res. 2006;12:634–642. doi: 10.1158/1078-0432.CCR-05-1132. [DOI] [PubMed] [Google Scholar]
- 201.Burgess A, Ruefli A, Beamish H, Warrener R, Saunders N, Johnstone R, Gabrielli B. Histone deacetylase inhibitors specifically kill non proliferating tumour cells. Oncogene. 2004;23:6693 – 6701. doi: 10.1038/sj.onc.1207893. [DOI] [PubMed] [Google Scholar]
- 202.Frommer M, McDonald L E, Millar D S, Collis C M, Watt F, Grigg G W, Molloy P L, Paul C L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Huang TH, Laux DE, Hamlin BC, Tran P, Tran H, Lubahn D B. Identification of DNA methylation markers for human breast carcinomas using the methylation-sensitive restriction fingerprinting technique. Cancer Res. 1997;57:1030–1034. [PubMed] [Google Scholar]
- 204.Kuras L. Characterization of protein-DNA association in vivo by chromatin immunoprecipitation. Methods Mol. Biol. 2004;284:147–162. doi: 10.1385/1-59259-816-1:147. [DOI] [PubMed] [Google Scholar]
- 205.Karimi M, Johansson S, Ekström T J. LUMA (LUminometric Methylation Assay)-A high throughput method to the analysis of genomic DNA methylation. Exp. Cell. Res. 2006;312:1989–1995. doi: 10.1016/j.yexcr.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 206.Cedar H, Solage A, Glaser G, Razin A. Direct detection of methylated cytosine in DNA by use of the restriction enzyme MspI. Nucleic Acids Res. 1979;6:2125–32. doi: 10.1093/nar/6.6.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ronaghi M. Pyrosequencing sheds light on DNA sequencing. Genome Res. 2001;11:3–11. doi: 10.1101/gr.11.1.3. [DOI] [PubMed] [Google Scholar]
- 208.Tost J, El Abdalaoui H, Gut I G. Serial pyrosequencing for quantitative DNA methylation analysis. Biotechniques. 2006;40:721–722. doi: 10.2144/000112190. [DOI] [PubMed] [Google Scholar]
- 209.Shiao YH, Crawford E B, Anderson LM, Patel P, Ko K. Allele-specific germ cell epimutation in the spacer promoter of the 45S ribosomal RNA gene after Cr(III) exposure. Toxicol. Appl. Pharmacol. 2005;205:290–296. doi: 10.1016/j.taap.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 210.Smith LM. The future of DNA sequencing. Science. 1993;262:530–532. doi: 10.1126/science.8211178. [DOI] [PubMed] [Google Scholar]
- 211.Hillenkamp F, Karas M, Beavis R C, Chait B T. Matrix-assisted laser desorption/ionization mass spectrometry of biopolymers. Anal. Chem. 1991;63:1193A–1203A. doi: 10.1021/ac00024a002. [DOI] [PubMed] [Google Scholar]
- 212.Fan JB, Oliphant A, Shen R, Kermani BG, Garcia F, Gunderson KL, Hansen M, Steemers F, Butler SL, Deloukas P, Galver L, Hunt S, McBride C, Bibikova M, Rubano T, Chen J, Wickham E, Doucet D, Chang W, Campbell D, Zhang B, Kruglyak S, Bentley D, Haas J, Rigault P, Zhou L, Stuelpnagel J, Chee MS. Highly parallel SNP genotyping. Cold Spring Harb. Symp. Quant. Biol. 2003;68:69–78. doi: 10.1101/sqb.2003.68.69. [DOI] [PubMed] [Google Scholar]
- 213.Fan JB, Gunderson KL, Bibikova M, Yeakley JM, Chen J, Wickham Garcia E, Lebruska LL, Laurent M, Shen R, Barker D. Illumina universal bead arrays. Methods Enzymol. 2006;410:57–73. doi: 10.1016/S0076-6879(06)10003-8. [DOI] [PubMed] [Google Scholar]
- 214.Aggerholm A, Holm MS, Guldberg P, Olesen LH, Hokland P. Promoter hypermethylation of p15INK4B, HIC1, CDH1, and ER is frequent in myelodysplastic syndrome and predicts poor prognosis in early-stage patients. Eur. J. Haematol. 2006;76:23–32. doi: 10.1111/j.1600-0609.2005.00559.x. [DOI] [PubMed] [Google Scholar]
- 215.Cremonesi L, Firpo S, Ferrari M, Righetti PG, Gelfi C. Double-gradient DGGE for optimized detection of DNA point mutations. Biotechniques. 1997;22:326–330. doi: 10.2144/97222rr01. [DOI] [PubMed] [Google Scholar]
- 216.Reibenwein J, Pils D, Horak P, Tomicek B, Goldner G, Worel N, Elandt K, Krainer M. Promoter hypermethylation of GSTP1, AR, and 14-3-3sigma in serum of prostate cancer patients and its clinical relevance. Prostate. 2007;67:427–432. doi: 10.1002/pros.20533. [DOI] [PubMed] [Google Scholar]
- 217.Jarrard D F, Modder J, Fadden P, Fu V, Sebree L, Heisey D, Schwarze S R, Friedl A. Alterations in the p16/pRb cell cycle checkpoint occur commonly in primary and metastatic human prostate cancer. Cancer Lett. 2002;185:191–199. doi: 10.1016/s0304-3835(02)00282-3. [DOI] [PubMed] [Google Scholar]
- 218.Lodygin D, Epanchintsev A, Menssen A, Diebold J, Hermeking H. Functional epigenomics identifies genes frequently silenced in prostate cancer. Cancer Res. 2005;65:4218– 4227. doi: 10.1158/0008-5472.CAN-04-4407. [DOI] [PubMed] [Google Scholar]
- 219.Pulukuri SM, Patibandla S, Patel J, Estes N, Rao J S. Epigenetic inactivation of the tissue inhibitor of metalloproteinase-2 (TIMP-2) gene in human prostate tumors. Oncogene. 2007;26:5229–5237. doi: 10.1038/sj.onc.1210329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Pakneshan P, Szyf M, and Rabbani SA. Hypomethylation of urokinase (uPA) promoter in breast and prostate cancer: prognostic and therapeutic implications. Curr. Cancer Drug Targets. 2005;5:471–488. doi: 10.2174/156800905774574011. [DOI] [PubMed] [Google Scholar]
- 221.Wakaguri H, Yamashita R, Suzuki Y, Sugano S, Nakai K. DBTSS: database of transcription start sites, progress report 2008. Nucleic Acids Res. 2008;36(Database issue):D97–101. doi: 10.1093/nar/gkm901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Schmid C D, Perier R, Praz V, Bucher P. EPD in its twentieth year: towards complete promoter coverage of selected model organisms. Nucleic Acids Res. 2006;34 (Database issue):D82–85. doi: 10.1093/nar/gkj146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Flicek P, Aken B L, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, Fernandez-Banet J, Gordon L, Gräf S, Haider S, Hammond M, Howe K, Jenkinson A, Johnson N, Kähäri A, Keefe D, Keenan S, Kinsella R, Kokocinski F, Koscielny G, Kulesha E, Lawson D, Longden I, Massingham T, McLaren W, Megy K, Overduin B, Pritchard B, Rios D, Ruffier M, Schuster M, Slater G, Smedley D, Spudich G, Tang Y A, Trevanion S, Vilella A, Vogel J, White S, Wilder S P, Zadissa A, Birney E, Cunningham F, Dunham I, Durbin R, Fernández-Suarez X M, Herrero J, Hubbard T J, Parker A, Proctor G, Smith J, Searle S M. Ensembl's 10th year. Nucleic Acids Res. 2010;38(Database issue):557–D562. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Bock C, Reither S, Mikeska T, Paulsen M, Walter J, Lengauer T. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformtics. 2005;21(21 ):4067–4068. doi: 10.1093/bioinformatics/bti652. [DOI] [PubMed] [Google Scholar]
- 225.Li L C, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18(11 ):1427–31. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 226.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 227.Eads C A, Laird P W. Combined bisulfite restriction analysis (COBRA) Methods Mol. Biol. 2002;200:71–85. doi: 10.1385/1-59259-182-5:071. [DOI] [PubMed] [Google Scholar]
- 228.Roberts R J, Vincze T, Posfai J, Macelis D. REBASE--a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2010;38(Database issue):D234–D236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Grunau C, Renault E, Rosenthal A, Roizes G. MethDB--a public database for DNA methylation data. Nucleic Acids Res. 2001;29(1 ):270–274. doi: 10.1093/nar/29.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Pattyn F, Hoebeeck J, Robbrecht P, Michels E, De Paepe A, Bottu G, Coornaert , Herzog R, Speleman F, Vandesompele J. methBLAST and methPrimerDB: web-tools for PCR based methylation analysis. BMC Bioinformatics. 2006;7:496. doi: 10.1186/1471-2105-7-496. [DOI] [PMC free article] [PubMed] [Google Scholar]