Abstract
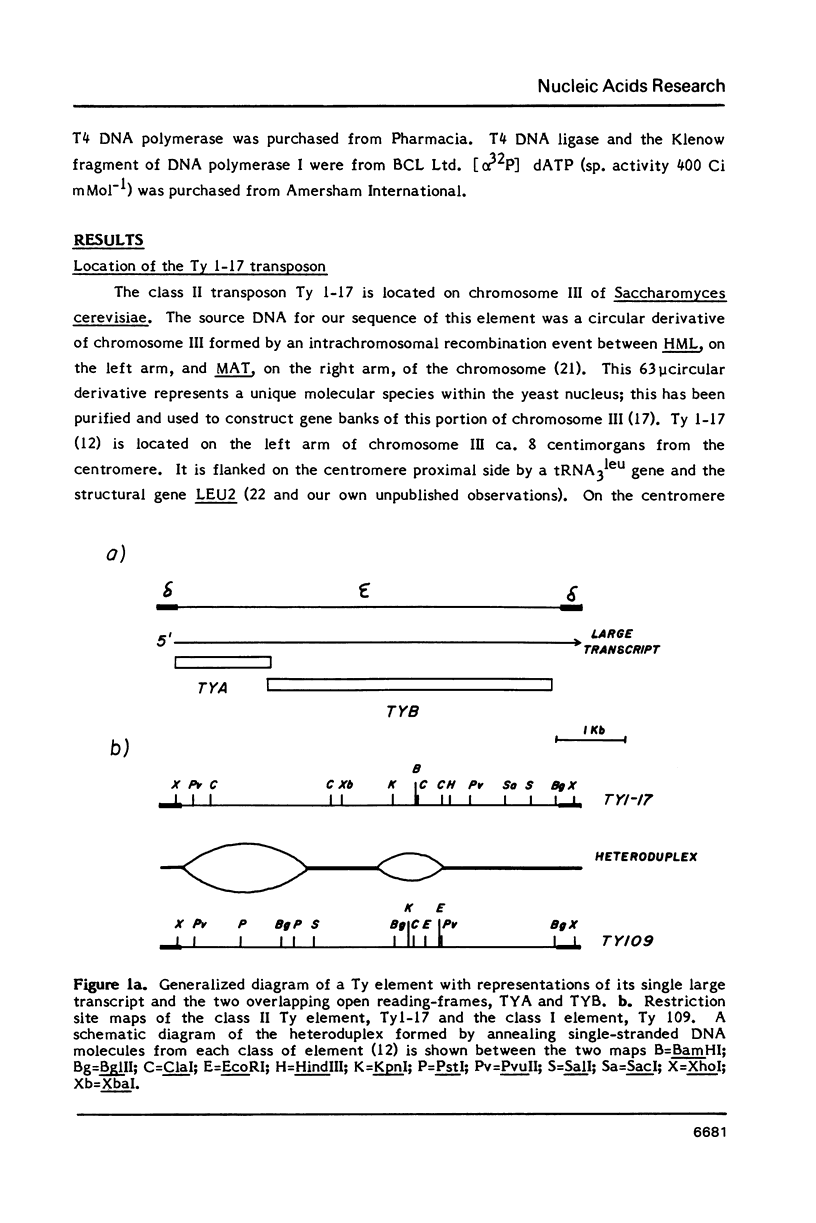
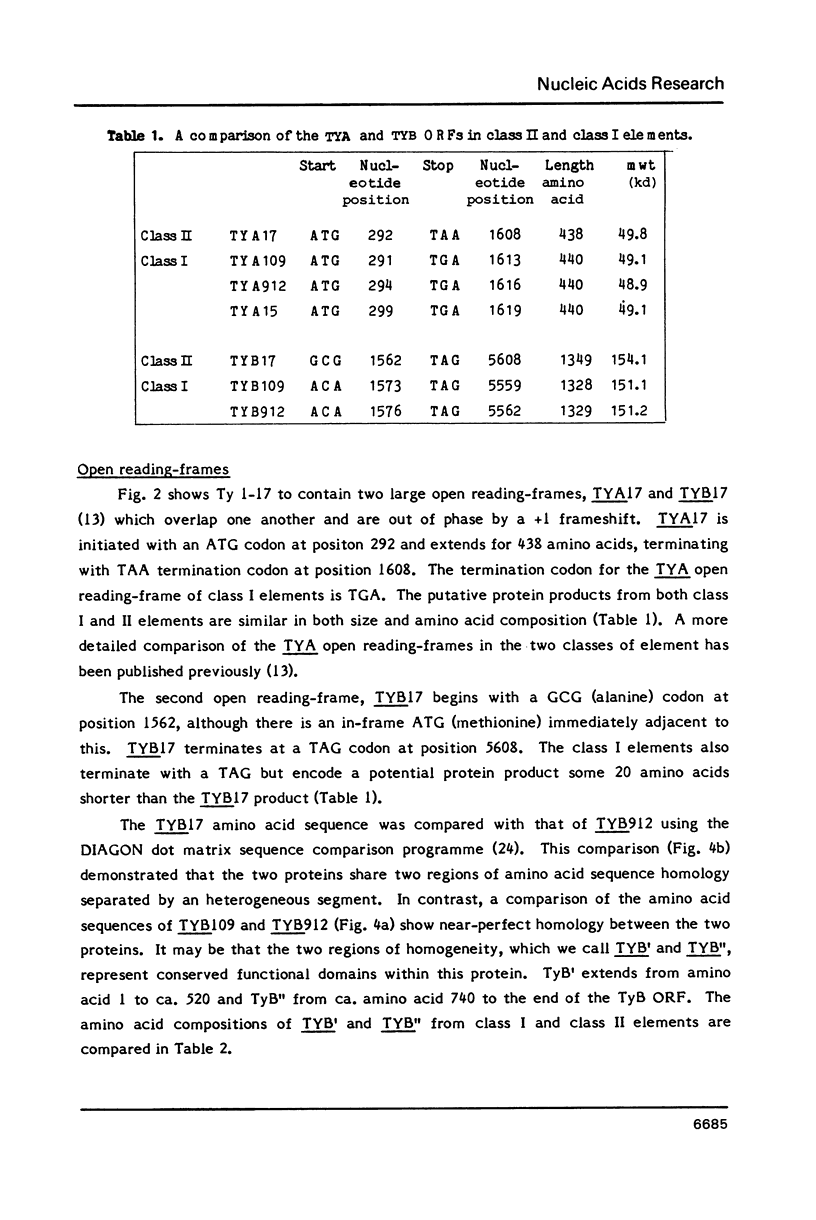
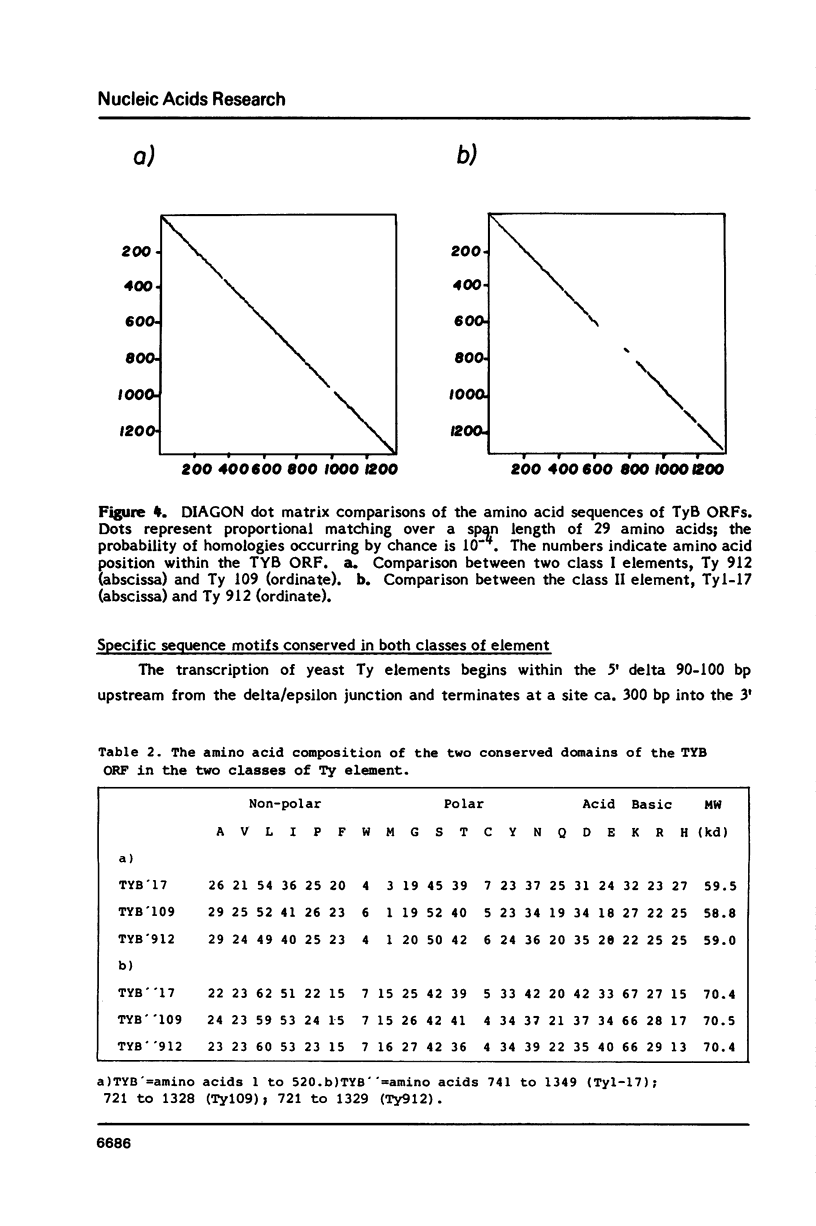
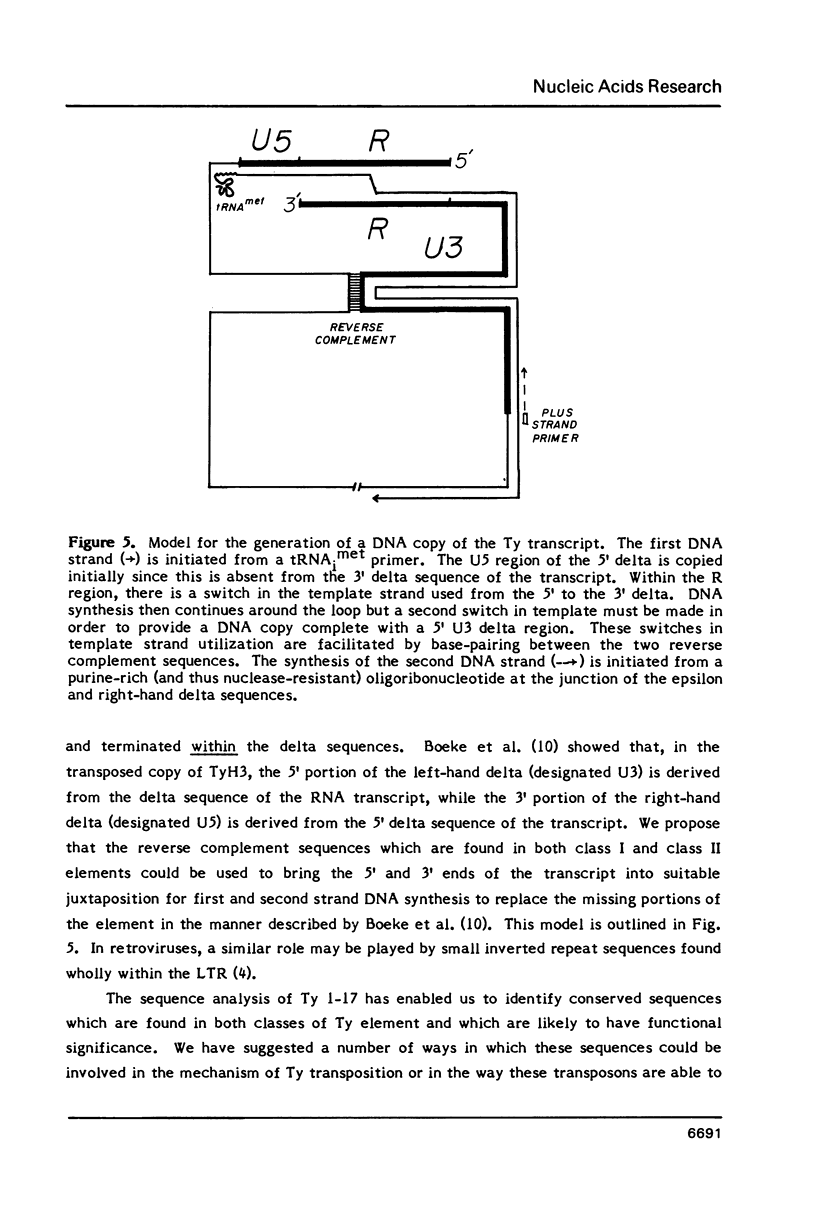
We have determined the nucleotide sequence of a class II yeast transposon (Ty 1-17) which is found just centromere-distal to the LEU2 structural gene on chromosome III of Saccharomyces cerevisiae. The complete element is 5961 bp long and is bounded by two identical, directly repeated, delta sequences of 332 bp each. The sequence organization indicates that Ty 1-17 is a retrotransposon, like the class I elements characterized previously. It contains two long open reading-frames, TyA (439 amino acids) and TyB (1349 amino acids). In this paper, the sequences of the two classes of yeast transposon are compared with one another and with analogous elements, such as retroviral proviruses, cauliflower mosaic virus and copia sequences. Features of the Ty 1-17 sequence which may be important to its mechanism of transposition and its genetic action are discussed.
Full text
PDF










Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andreadis A., Hsu Y. P., Kohlhaw G. B., Schimmel P. Nucleotide sequence of yeast LEU2 shows 5'-noncoding region has sequences cognate to leucine. Cell. 1982 Dec;31(2 Pt 1):319–325. doi: 10.1016/0092-8674(82)90125-8. [DOI] [PubMed] [Google Scholar]
- Baltimore D. Retroviruses and retrotransposons: the role of reverse transcription in shaping the eukaryotic genome. Cell. 1985 Mar;40(3):481–482. doi: 10.1016/0092-8674(85)90190-4. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Garfinkel D. J., Styles C. A., Fink G. R. Ty elements transpose through an RNA intermediate. Cell. 1985 Mar;40(3):491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- Cameron J. R., Loh E. Y., Davis R. W. Evidence for transposition of dispersed repetitive DNA families in yeast. Cell. 1979 Apr;16(4):739–751. doi: 10.1016/0092-8674(79)90090-4. [DOI] [PubMed] [Google Scholar]
- Clare J., Farabaugh P. Nucleotide sequence of a yeast Ty element: evidence for an unusual mechanism of gene expression. Proc Natl Acad Sci U S A. 1985 May;82(9):2829–2833. doi: 10.1073/pnas.82.9.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger P. L. Random subcloning of sonicated DNA: application to shotgun DNA sequence analysis. Anal Biochem. 1983 Feb 15;129(1):216–223. doi: 10.1016/0003-2697(83)90072-6. [DOI] [PubMed] [Google Scholar]
- Dubois E., Jacobs E., Jauniaux J. C. Expression of the ROAM mutations in Saccharomyces cerevisiae: involvement of trans-acting regulatory elements and relation with the Ty1 transcription. EMBO J. 1982;1(9):1133–1139. doi: 10.1002/j.1460-2075.1982.tb01308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigel A., Feldmann H. Ty1 and delta elements occur adjacent to several tRNA genes in yeast. EMBO J. 1982;1(10):1245–1250. doi: 10.1002/j.1460-2075.1982.tb00020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder R. T., Loh E. Y., Davis R. W. RNA from the yeast transposable element Ty1 has both ends in the direct repeats, a structure similar to retrovirus RNA. Proc Natl Acad Sci U S A. 1983 May;80(9):2432–2436. doi: 10.1073/pnas.80.9.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errede B., Cardillo T. S., Wever G., Sherman F., Stiles J. I., Friedman L. R., Sherman F. Studies on transposable elements in yeast. I. ROAM mutations causing increased expression of yeast genes: their activation by signals directed toward conjugation functions and their formation by insertion of Ty1 repetitive elements. II. deletions, duplications, and transpositions of the COR segment that encompasses the structural gene of yeast iso-1-cytochrome c. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):593–607. [PubMed] [Google Scholar]
- Fulton A. M., Mellor J., Dobson M. J., Chester J., Warmington J. R., Indge K. J., Oliver S. G., de la Paz P., Wilson W., Kingsman A. J. Variants within the yeast Ty sequence family encode a class of structurally conserved proteins. Nucleic Acids Res. 1985 Jun 11;13(11):4097–4112. doi: 10.1093/nar/13.11.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauber J., Nelböck-Hochstetter P., Feldmann H. Nucleotide sequence and characteristics of a Ty element from yeast. Nucleic Acids Res. 1985 Apr 25;13(8):2745–2758. doi: 10.1093/nar/13.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury G., Gruss P. Enhancer elements. Cell. 1983 Jun;33(2):313–314. doi: 10.1016/0092-8674(83)90410-5. [DOI] [PubMed] [Google Scholar]
- Kingsman A. J., Gimlich R. L., Clarke L., Chinault A. C., Carbon J. Sequence variation in dispersed repetitive sequences in Saccharomyces cerevisiae. J Mol Biol. 1981 Feb 5;145(4):619–632. doi: 10.1016/0022-2836(81)90306-5. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Mitra S. W., Chow M., Champoux J., Baltimore D. Synthesis of murine leukemia virus plus strong stop DNA initiates at a unique site. J Biol Chem. 1982 Jun 10;257(11):5983–5986. [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Patarca R., Haseltine W. A. Sequence similarity among retroviruses--erratum. Nature. 1984 Jun 21;309(5970):728–728. doi: 10.1038/309728b0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnick T. M., Lerner R. A., Sutcliffe J. G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981 Oct 15;293(5833):543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Siliciano P. G., Tatchell K. Transcription and regulatory signals at the mating type locus in yeast. Cell. 1984 Jul;37(3):969–978. doi: 10.1016/0092-8674(84)90431-8. [DOI] [PubMed] [Google Scholar]
- Smith J. K., Cywinski A., Taylor J. M. Specificity of initiation of plus-strand DNA by Rous sarcoma virus. J Virol. 1984 Nov;52(2):314–319. doi: 10.1128/jvi.52.2.314-319.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R., McLachlan A. D. Codon preference and its use in identifying protein coding regions in long DNA sequences. Nucleic Acids Res. 1982 Jan 11;10(1):141–156. doi: 10.1093/nar/10.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern J. N., Newlon C. S., Herskowitz I., Hicks J. B. Isolation of a circular derivative of yeast chromosome III: implications for the mechanism of mating type interconversion. Cell. 1979 Oct;18(2):309–319. doi: 10.1016/0092-8674(79)90050-3. [DOI] [PubMed] [Google Scholar]
- Taguchi A. K., Ciriacy M., Young E. T. Carbon source dependence of transposable element-associated gene activation in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Jan;4(1):61–68. doi: 10.1128/mcb.4.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. M. An analysis of the role of tRNA species as primers for the transcription into DNA of RNA tumor virus genomes. Biochim Biophys Acta. 1977 Mar 21;473(1):57–71. doi: 10.1016/0304-419x(77)90007-5. [DOI] [PubMed] [Google Scholar]
- Toh H., Kikuno R., Hayashida H., Miyata T., Kugimiya W., Inouye S., Yuki S., Saigo K. Close structural resemblance between putative polymerase of a Drosophila transposable genetic element 17.6 and pol gene product of Moloney murine leukaemia virus. EMBO J. 1985 May;4(5):1267–1272. doi: 10.1002/j.1460-2075.1985.tb03771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson V. M., Cox D., Young E. T., Russell D. W., Smith M. Characterization of transposable element-associated mutations that alter yeast alcohol dehydrogenase II expression. Mol Cell Biol. 1983 Jan;3(1):20–31. doi: 10.1128/mcb.3.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]



