Abstract
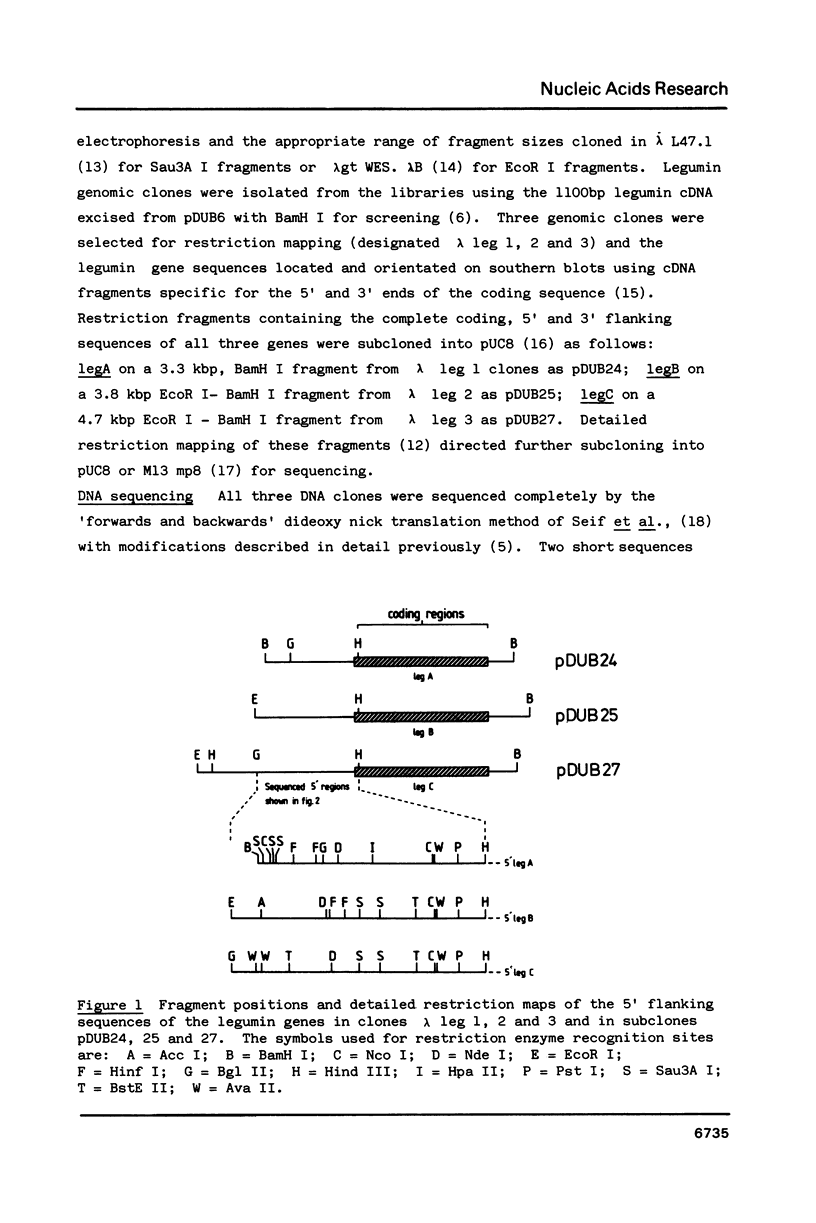
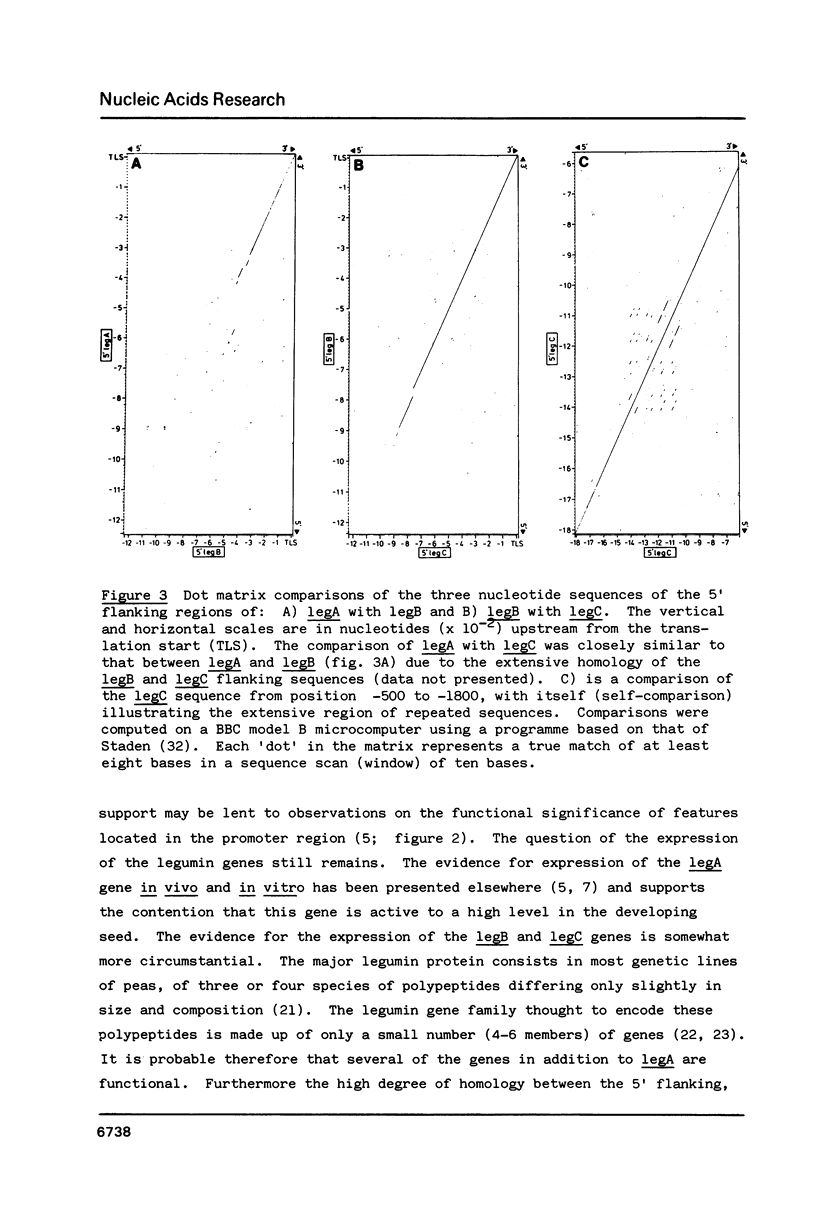
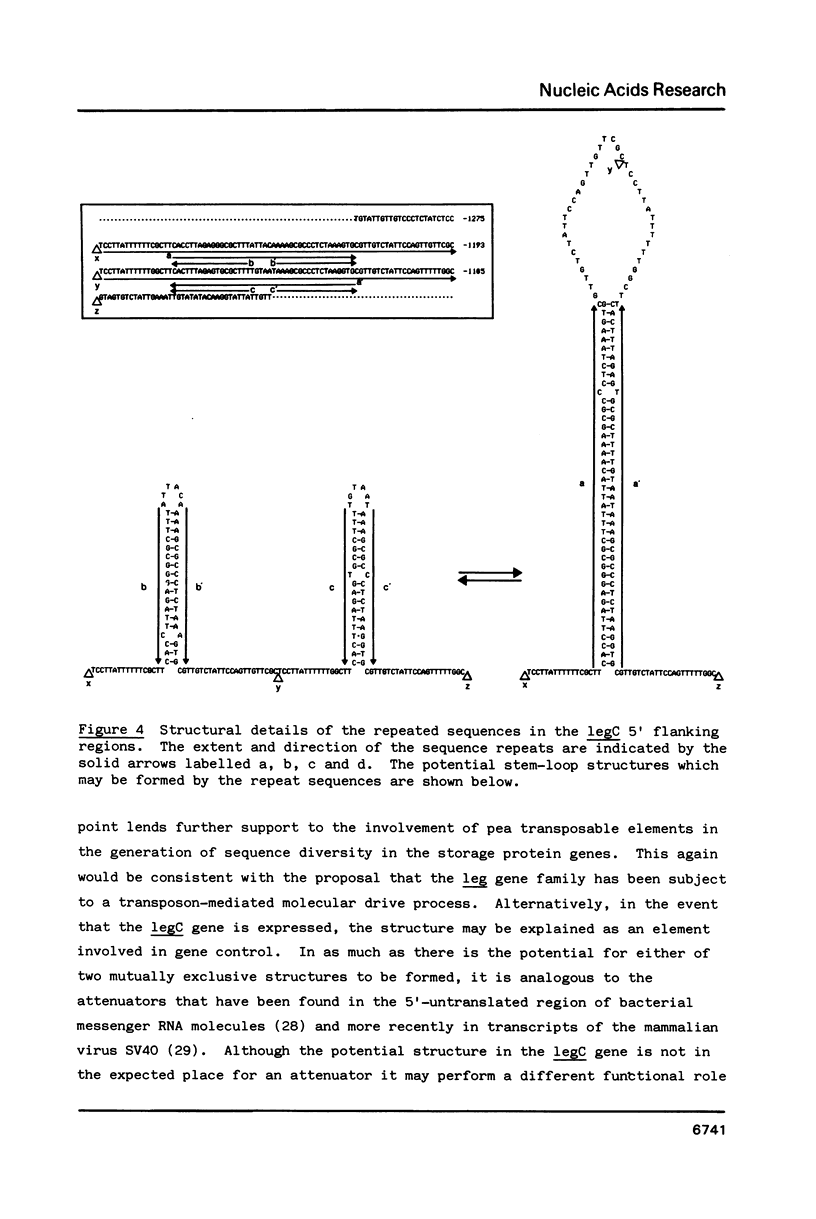
Approximately 1200 nucleotides of sequence data from the promoter and 5'-flanking regions of each of three pea (Pisum sativum L.) legumin genes (legA, legB and legC) are presented. The promoter regions of all three genes were found to be identical including the "TATA box", and "CAAT box', and sequences showing homology to the SV40 enhancers. The legA sequence begins to diverge from the others about 300bp from the start codon, whereas the other two genes remain identical for another 550bp. The regions of partial homology exhibit deletions or insertions and some short, comparatively well conserved sequences. The significance of these features is discussed in terms of evolutionary mechanisms and their possible functional roles. The legC gene contains a region that may potentially form either of two mutually exclusive stem-loop structures, one of which has a stem 42bp long, which suggests that it could be fairly stable. We suggest that a mechanism of switching between such alternative structures may play some role in gene control or may represent the insertion of a transposable element.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Chandrasekaran R., Birdsall D. L., Leslie A. G., Ratliff R. L. Left-handed DNA helices. Nature. 1980 Feb 21;283(5749):743–745. doi: 10.1038/283743a0. [DOI] [PubMed] [Google Scholar]
- Bonas U., Sommer H., Saedler H. The 17-kb Tam1 element of Antirrhinum majus induces a 3-bp duplication upon integration into the chalcone synthase gene. EMBO J. 1984 May;3(5):1015–1019. doi: 10.1002/j.1460-2075.1984.tb01921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler P. M., Higgins T. J., Randall P. J., Spencer D. Regulation of Legumin Levels in Developing Pea Seeds under Conditions of Sulfur Deficiency: Rates of Legumin Synthesis and Levels of Legumin mRNA. Plant Physiol. 1983 Jan;71(1):47–54. doi: 10.1104/pp.71.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler P. M., Spencer D., Randall P. J., Higgins T. J. Influence of Sulfur Nutrition on Developmental Patterns of Some Major Pea Seed Proteins and Their mRNAs. Plant Physiol. 1984 Jul;75(3):651–657. doi: 10.1104/pp.75.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruzzi G., Broglie R., Edwards C., Chua N. H. Tissue-specific and light-regulated expression of a pea nuclear gene encoding the small subunit of ribulose-1,5-bisphosphate carboxylase. EMBO J. 1984 Aug;3(8):1671–1679. doi: 10.1002/j.1460-2075.1984.tb02031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domoney C., Casey R. Measurement of gene number for seed storage proteins in Pisum. Nucleic Acids Res. 1985 Feb 11;13(3):687–699. doi: 10.1093/nar/13.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover G. Molecular drive: a cohesive mode of species evolution. Nature. 1982 Sep 9;299(5879):111–117. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- Gatehouse J. A., Evans I. M., Bown D., Croy R. R., Boulter D. Control of storage-protein synthesis during seed development in pea (Pisum sativum L.). Biochem J. 1982 Oct 15;208(1):119–127. doi: 10.1042/bj2080119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierl A., Schwarz-Sommer Z., Saedler H. Molecular interactions between the components of the En-I transposable element system of Zea mays. EMBO J. 1985 Mar;4(3):579–583. doi: 10.1002/j.1460-2075.1985.tb03669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham D. E. The isolation of high molecular weight DNA from whole organisms or large tissue masses. Anal Biochem. 1978 Apr;85(2):609–613. doi: 10.1016/0003-2697(78)90262-2. [DOI] [PubMed] [Google Scholar]
- Gruss P. Magic enhancers? DNA. 1984;3(1):1–5. doi: 10.1089/dna.1.1984.3.1. [DOI] [PubMed] [Google Scholar]
- Hay N., Skolnik-David H., Aloni Y. Attenuation in the control of SV40 gene expression. Cell. 1982 May;29(1):183–193. doi: 10.1016/0092-8674(82)90102-7. [DOI] [PubMed] [Google Scholar]
- Keller E. B., Calvo J. M. Alternative secondary structures of leader RNAs and the regulation of the trp, phe, his, thr, and leu operons. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6186–6190. doi: 10.1073/pnas.76.12.6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder P., Tiemeier D., Enquist L. EK2 derivatives of bacteriophage lambda useful in the cloning of DNA from higher organisms: the lambdagtWES system. Science. 1977 Apr 8;196(4286):175–177. doi: 10.1126/science.322278. [DOI] [PubMed] [Google Scholar]
- Loenen W. A., Brammar W. J. A bacteriophage lambda vector for cloning large DNA fragments made with several restriction enzymes. Gene. 1980 Aug;10(3):249–259. doi: 10.1016/0378-1119(80)90054-2. [DOI] [PubMed] [Google Scholar]
- Lycett G. W., Croy R. R., Shirsat A. H., Boulter D. The complete nucleotide sequence of a legumin gene from pea (Pisum sativum L.). Nucleic Acids Res. 1984 Jun 11;12(11):4493–4506. doi: 10.1093/nar/12.11.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seif I., Khoury G., Dhar R. A rapid enzymatic DNA sequencing technique: determination of sequence alterations in early simian virus 40 temperature sensitive and deletion mutants. Nucleic Acids Res. 1980 May 24;8(10):2225–2240. doi: 10.1093/nar/8.10.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw C. H., Carter G. H., Watson M. D., Shaw C. H. A functional map of the nopaline synthase promoter. Nucleic Acids Res. 1984 Oct 25;12(20):7831–7846. doi: 10.1093/nar/12.20.7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982 May 11;10(9):2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Vodkin L. O., Rhodes P. R., Goldberg R. B. cA lectin gene insertion has the structural features of a transposable element. Cell. 1983 Oct;34(3):1023–1031. doi: 10.1016/0092-8674(83)90560-3. [DOI] [PubMed] [Google Scholar]
- Weiher H., König M., Gruss P. Multiple point mutations affecting the simian virus 40 enhancer. Science. 1983 Feb 11;219(4585):626–631. doi: 10.1126/science.6297005. [DOI] [PubMed] [Google Scholar]


