Abstract
Lead (Pb) is known to alter the functions of numerous organ systems, including the hematopoietic and immune systems. Pb can induce anemia and can lower host resistance to bacterial and viral infections. The anemia is due to Pb’s inhibition of hemoglobin synthesis and Pb’s induction of membrane changes, leading to early erythrocyte senescence. Pb also increases B-cell activation/proliferation and skews T-cell help (Th) toward Th2 subset generation. The specific mechanisms for many of the Pb effects are, as yet, not completely understood. Therefore, we performed gene expression analysis, via microarray, on RNA from the spleens of developmentally Pb-exposed mice, in order to gain further insight into these Pb effects. Splenic RNA microarray analysis indicated strong up-regulation of genes coding for proteolytic enzymes, lipases, amylase, and RNaseA. The data also showed that Pb affected the expression of many genes associated with innate immunity. Analysis of the microarray results via GeneSifter software indicated that Pb increased apoptosis, B-cell differentiation, and Th2 development. Direct up-regulation by Pb of expression of the gene encoding the heme-regulated inhibitor (HRI) suggested that Pb can decrease erythropoiesis by blocking globin mRNA translation. Pb’s high elevation of digestive/catabolizing enzymes could generate immunogenic self peptides. With Pb’s potential to induce new self-peptides and to enhance the expression of caspases, cytokines, and other immunomodulators, further evaluation of Pb’s involvement in autoimmune phenomena, especially Th2-mediated autoantibody production, and alteration of organ system activities is warranted.
Keywords: B-cells, cytokines, erythrocytes, gene expression, inflammation, lead, macrophages, microarray, spleen, T-helper cells
Introduction
The acute toxic effects of the heavy metal Pb, including activation/inhibition of certain enzymes, induction of hemolytic anemia, and inhibition of Na+/K+ ATPase leading to membrane fragility, were reviewed almost 4 decades ago (Valle and Ulmer, 1972). However, the actual mechanisms responsible for the toxic effects of low-level chronic Pb exposure (blood levels 10–30 μg/dl) are just beginning to be assessed, especially for developmental exposures. Most orally ingested Pb is excreted, but a portion is absorbed and is transferred to the blood (where Pb binds to hemoglobin in the erythrocytes). Because Pb is carried through the circulatory system by erythrocytes, virtually all tissues in the body can become exposed to the toxic metal (Goering, 1993; Gidlow 2004). Although Pb can damage most of the organ systems, those particularly susceptible to the toxic effects of Pb exposure are the hematopoietic, immune, renal, and nervous systems (Lawrence and McCabe, 1995).
As noted above, acute Pb exposure causes hemolytic anemia (Vallee and Ulmer, 1972), and previous reports have indicated that the Pb-induced anemia stems from a number of factors, among which are decreased hemoglobin synthesis, increased hemolysis arising from membrane fragility, and increased erythrocyte destruction in the spleen (Woods, 1995). In hemoglobin synthesis, Pb interferes with the production of heme, both by inhibiting δ-aminolevulinic acid dehydrase (ALA-D) activity (Rubino, 1962; Ahamed et al., 2006; Ahamed and Siddiqui, 2007) and by blocking incorporation of iron into the protoporphyrin ring during the final step of heme synthesis (Woods, 1995). In addition, Pb has a direct effect on alpha and beta globin synthesis; this effect can be overcome by addition of heme (Waxman and Rabinovitz, 1966). With respect to decreased red cell survival, Rubino (1962) noted that in cases of Pb poisoning, the erythrocyte survival time was one-sixth of the cell’s normal lifetime. It was suggested that this decrease in red cell survival time was due to increased fragility of the red cell membrane upon exposure to Pb (Hernberg and Nikkanen, 1970; Simons, 1993). Erythrocytes that are recognized as fragile or senescent undergo destruction by the macrophages present in the red pulp of the spleen (Kuypers and DeJong, 2004; Mebius and Kraal, 2005).
One of the first observations about the effect of Pb on the immune system was that Pb lowered host defenses against a number of infectious agents (Hemphil et al., 1971; Gainer, 1974; Cook et al., 1975; Lawrence, 1985). In the spleen, phagocytes (macrophages and polymorphonuclear cells) are responsible for slowing the propagation of an invading pathogen, while an antigen-specific adaptive immune response (antibody- or cell-mediated) is being established. Pb was reported to inhibit macrophage function (Kowolenko et al., l988; Mauel et al., 1989) possibly by overloading macrophages with cellular debris and inhibiting macrophage production of nitric oxide (Tian and Lawrence, 1995). In the context of adaptive humoral and cellular immune responses, Pb increased both B-cell and T-cell in vitro proliferation (Lawrence, 1981a–c; Warner and Lawrence, 1986; Razani-Bordoujerdi et al., 1999). Pb also enhanced major histocompatibility complex class II (MHC class II) expression by B-cells (McCabe and Lawrence, 1990; Guo et al., 1996). Furthermore, T-cell experiments performed on cultured spleen cells and on Th1 (IFN-γ expressing) and Th2 (IL-4 expressing) cell clones indicated that Pb skews T-cell help toward that of Th2 responses (Heo et al., 1996). In vivo, Pb exposure of mice increased plasma IL-4 and IgE levels, indicating that Pb skews naïve T-cells toward Th2 (Heo et al., 1996; Heo et al., 1997). Heo et al. (1998) suggested that Pb inhibits Th1 development by increasing adenylate cyclase activity, and thereby increasing cyclic AMP levels. These effects of Pb on the immune system suggested that Pb could promote antibody production, including autoantibodies. The influence of Pb on autoimmunity has been suggested (Wedeen et al., 1978 and 1979); Pb workers with dysfunctional renal function had antibodies deposited in their kidneys. A mouse model also has shown that Pb exacerbates systemic lupus erythematosis (SLE) symptoms in lupus-prone NZM mice (Hudson et al., 2003).
To explore the effects of developmental Pb exposure on the hematopoietic and immune systems in a more global manner, we performed gene-expression microarray analysis on RNA isolated from the spleens of female mouse pups at post-natal day 21 (pnd21). The toxic effects of Pb are most pronounced on the female immune system during development (Bunn et al., 2001). For these experiments, we chose a Pb dose (0.1 mM) and length of exposure (gestation-day 8 (gd8) to postnatal-day 21 (pnd21)) of BALB/c mice that had been previously shown to enhance Th2 immunity of neonates (Snyder et al., 2000); this developmental Pb exposure results in environmentally relevant blood Pb levels (Snyder et al., 2000; Needleman, 2004). Our microarray results are indicating direct effects of Pb exposure on the expression of genes pertinent to hematopoiesis and regulation of immune system processes.
Further studies addressing the mechanism of Th2 skewing were performed with splenocytes from OVA-tg mice deficient in the expression of STAT4 or STAT6. IL-4 and IL-12 are the most influential factors in determining development of Th precursor cells (Thp) toward IL-4 producing Th2 and IFN-γ producing Th1 cells, respectively (Sedar et al., 1992; Hsieh et al., 1992; Sedar et al., 1993; Macatonia et al., 1995). Deficiencies of STAT6 and STAT4 had been reported to severely curtail development of IL-4-mediated Th2 cells (Shimoda et al., 1996; Kaplan et al., 1996a) and IL-12-mediated Th1 cells (Thierfelder et al., 1996; Kaplan et al., 1996b), respectively. These effects were demonstrated through in vitro differentiation assays using polyclonal activators, such as anti-CD3 mAb. Preliminary data showed that disruption of STAT4 or STAT6 expression in the OVA-tg mice did not cause any significant alteration in the distribution of lymphoid cell populations (CD4+ and CD8+ T-cells, B-cells, and NK-cells), but STAT6 deficiency did result in lower serum levels of IgG1 and IgE.
The present study, by examining Pb’s impact on gene expression in the spleen, has focused on developmental Pb exposure and its effects on the hematopoietic and immune systems. The spleen is a major secondary lymphoid organ, and it influences the development of both the hematopoietic and immune systems (Cyster, 2003; Mebius and Kraal, 2005). We paired gene expression data with results from in vitro experiments with splenocytes from STAT4- and STAT6-deficient mice, in order to develop a better understanding of the mechanisms underlying T-helper 2 cell skewing. Our microarray results, in addition to supporting findings from earlier reports concerning the effects of Pb on the immune system, such as, increased B cell activation/proliferation and skewing of T-helper cell subsets toward Th2, revealed unexpectedly high up-regulation of amylase, RNase, lipases, and proteolytic enzymes. Several possible explanations for this enzymatic up-regulation are proposed in the discussion.
Materials and Methods
Animals and Treatments
Experiments were performed with BALB/c mouse pups exposed to 0.1 mM Pb acetate administered via the dam’s drinking water from gd8 to pnd21. BALB/c mice were obtained from Taconic Farms (Germantown, NY). Mice were housed two females with one male. Once females with vaginal plugs were identified, they were housed separately and were considered pregnant (gestational day 0, gd0). At gd8, these dams were placed on drinking water with or without 0.1 mM Pb acetate. Pregnant females were housed one per standard cage and allowed free access to food and water in our specialized pathogen-free, AAALAC-approved animal facility. All mice were maintained on a 12-h light/dark cycle and were seronegative for known background viral agents. Once born, each litter was designated as N=1. All of the mouse pups were euthanized by CO2 asphyxiation at 21 days of age (pnd21), and spleens were aseptically collected from each of three litters and immediately frozen at −80°C. The Pb acetate dose was based on the determination by Snyder et al. (2000) that this concentration produces blood Pb levels comparable to levels of children exposed to low-level environmental Pb, i.e., about 10–30 μg/dl (Needleman, 2004). The timespan of gd8 to pd21 was chosen to allow for embryo implantation in utero, and to maximize the effect of developmental Pb exposure, given that Bunn et al. (2001) found higher blood Pb levels at the later stages of gestational exposure. Animals were treated humanely under protocol procedures approved by our Wadsworth Center IACUC.
Pb Concentration Analysis of Spleen Tissue
An aliquot of 25 μl of spleen homogenate (prepared as for the amylase assay described later) was diluted to 4 ml with a H2O containing 2 % (v/v) of concentrated HNO3, 0.005% (v/v) Triton X-100, and 10 μg/l Ir (internal standard). Samples were analyzed in duplicate, with three replicates each, using ICP-MS instrumentation (Elan DRCll). The limit of detection, LOD, was calculated using the spleen homogenization buffer alone and was determined to be 1.7 μg/l.
RNA Isolation
Total RNA was isolated from spleens of three control and three experimental mice, with the aid of a RNeasy Midi kit obtained from Qiagen, Inc. (Valencia, CA). Each mouse was considered to be representative of one litter. Spleens were homogenized, using a rotor-stator homogenizer, with RLT reagent from Qiagen containing beta-mercaptoethanol (2-ME) (one spleen per 4.0 ml). The homogenate was cleared of debris by centrifugation at 4,000 × g for 20 min at room temperature. Each supernatant was applied to a Midi column for extraction of total RNA as per manufacturer protocols. The RNA was further purified on a Qiagen Mini column, which included an “On-Column DNase digestion” for removal of contaminating genomic DNA. To measure the RNA concentration, we took absorbance readings at 260 and 280 nm on a Beckman DU640 spectrophotometer; RNA was considered pure if the 260/280 ratio was >1.8. Two procedures were used to assess RNA sample integrity. First, RNA degradation was determined by gel electrophoresis using 1% agarose, and second, RNA integrity was checked with an Agilent bioanalzyer.
Microarray Analysis
Total RNA (2 μg) was reverse-transcribed to double-stranded cDNA and then subjected to an in vitro transcription process in the presence of biotin-11-UTP from PerkinElmer Life and Analytical Sciences (Boston, MA). The biotinylated amplified RNA was then fluorescently labeled by attachment of a Cy-5 streptavidin fluor conjugate from Amersham Biosciences (Piscataway, NJ). The Cy-5 labeled RNA was then hybridized at 37°C for 18 hr to Mouse Whole Genome Codelink slides, according to the manufacturer-recommended protocol. The slides were scanned on an Axon 4000B scanner at 600V and 635 nm wavelength, using the Codelink image and data analysis software. Raw data from scanned results were median-normalized, and duplicate positive controls and negative controls were analyzed to check the quality of the data. Negative controls were required to have a signal to noise ratio of ≤3, while positive controls were checked for reproducibility of signal and low standard deviation between replicates. The microarray data were characterized for overall quality through generation of scatter plots of the data. The signal intensities all fell along a 45° straight line in a symmetrical pattern, indicating that the data were of sufficient quality for further analysis.
Software Analysis of Microarray Data
Statistical analysis of the microarray data was done using GeneSifter software from VizX Labs (Seattle, WA). Relevant settings included a lower fold-change cut-off of 1.2, a quality cut-off of 0.6, log transformation of the raw data, and a t-test P value of ≤0.05. Control genes on the slide were excluded from the analysis. Statistical analysis was performed using the Hochberg and Benjamini (1990) restrictions.
Absolute-Quantification by Real-Time RT-PCR
A two-step process was employed for quantification of the cytokine mRNA. First, cDNA was prepared from 1μg of total RNA, using a first-strand cDNA synthesis kit from Roche Applied Science (Indianapolis, IN). Following the synthesis reaction, 200 μl of PCR-grade water was added to each tube. The second step involved separate amplification for each cytokine gene sequence, using primer kits from Search-LC (Heidelberg, Germany). Amplifications were performed in duplicate with an Applied Biosystems 7500 real-time PCR instrument under the following conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 30 sec, 68°C for 40 sec, and 72°C for 50 sec. Readings were taken at the third step of each cycle. Following amplification, melting curves were generated with 1 cycle of 95°C for 15 sec, 60°C for 1 min, and 95°C for 15 min. Standard curves were generated for each run, using standards of known copy number supplied by Search-LC. Quantification was performed for the mRNAs of the cytokines IL-4 and IFN-γ. RT-PCR results were recorded as copy number per microliter (CN), and all cytokine results were normalized to the copy number (CN) obtained for glyceraldehyde 3-phosphate dehydrogenase (GAPDH) from the same cDNA synthesis sample, according to the formula (cytokine CN/GAPDH CN) X 100.
Relative-Quantification RT-PCR
A two-step process was again employed to determine relative quantity of selected genes. First, cDNA was synthesized from 2 μg of total RNA by use of the cDNA archive kit from Applied Biosystems, Inc. (Foster City, CA), according to the manufacturer’s directions. After the 2-hr incubation at 37°C, the 100 μl reaction mixtures were diluted with an equal volume of PCR-grade water. Relative quantification was then carried out by use of a TaqMan procedure, with primers, probes, and master mixes obtained from Applied Biosystems. Gene expression TaqMan kits from Applied Biosystems were for mouse chymotrypsin C (Mm013488833_m1), jumonji (Mm00524465_m1), macrophage stimulating 1 receptor (Mm00436365_m1), and GAPDH (Mm99999915_g1). The house-keeping gene, gapdh (GenBank: NM_008084) was again used as the endogenous control for normalization of the cytokine mRNA quantities. We performed amplification in an Applied Biosystems 7500 real-time PCR instrument under conditions specified by the manufacturer. Relative gene expression was calculated according to the following equation: 2−(Exp Ct−GAPDH Ct) X 1000.
Amylase Assay
Amylase activity was detected via a starch plate radial diffusion assay. The starch plate was prepared with 13 ml of a mixture of 1 % agarose and 0.1 % soluble starch dissolved in 0.1 M phosphate buffer (0.045M NaH2PO4, 0.055M Na2HPO4, 0.007M NaCl, pH 6.9). Wells, 1.5 mm in diameter, were punched in the gel, spaced ~ 1.5 cm apart. Samples were prepared by homogenization of each mouse spleen in 1 ml of M-PER mammalian protein extraction reagent (Pierce Biotechnology, Rockford, IL) containing protease inhibitors from Sigma (St. Louis, MO), or HALT from Pierce Biotechnology. Following the homogenization procedure, the homogenates were spun at 8,000 × g for 20 min at 4°C. The clarified homogenates were diluted 1:10, 1:100, and 1:1000 with the 0.1 M phosphate buffer. Diluted and undiluted homogenates, plus positive and negative controls, were loaded in duplicate, at 2 μl/well. Amylase activity in the homogenates was quantitated by comparison with purified human saliva amylase Type IX-A (Sigma, St. Louis, MO). A standard curve was generated for each plate. The plates were incubated in a moisture chamber at 37°C overnight. To stain, the gel was flooded with an iodine stain solution (1.65g KI, 2.5g I2 dissolved in 30 ml of warm water, and then filtered) diluted 1:100 with phosphate buffer. The gel was scanned with a Fujifilm LAS 3000 imager, and relative amylase activity was determined with the aid of the Multi Gauge V3.0 software.
Trypsin Detection
Trypsin inhibitor was coupled to flow-cell 2 of a CM5 sensor chip (GE Healthcare, Biacore Division, Uppsala, Sweden) by means of an amine coupling procedure according to the manufacturer’s directions, using a BIA3000 instrument. The coupling procedure was performed at pH 4.5, and the final ligand density was 1,400 response units (RU). Flow-cell 1 was activated and blocked with ethanolamine for use as a blank surface binding control. The running buffer used for the experiment was HBS-EP, pH 7.4 (10mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005 % (v/v) Surfactant P20). Spleen homogenate supernatants (from two control mice and two Pb-treated mice, representing two control and two Pb litters), prepared with M-PER reagent plus a Sigma protease inhibitor cocktail, were diluted 1:10 with running buffer, and 20 μl of this diluted sample was run over flow-cells 1 and 2 at a flow rate of 10 μl/min. The response units were read 15 sec after the injection of sample ceased. Purified trypsin was run as a positive control. The chip surface was regenerated with 10 mM glycine, pH 2.0.
Measurement of Trypsin Enzymatic Activity
Mouse spleens from control and 0.1 mM Pb-exposed mice were homogenized individually on ice in 1 ml of M-PER lysis reagent. For measurement of enzyme activity, the homogenates were diluted 1:10 or 1:100 with 1 mM HCl. Following dilution, the samples were spun for 20 min at 4°C in a microcentrifuge at ~10,000 × g, to pellet the precipitated material. The trypsin standard was prepared in 1 mM HCl at a concentration of 0.5 mg/ml. The substrate BAPNA (N-α-benzoyl-DL-arginine-p-nitroanilide hydrochloride), obtained from Roche Applied Science (Indianapolis, IN), was prepared by dissolving 10 mg in 125 μl of dimethysulfoxide (DMSO); the solution was brought up to 25 ml with Tris buffer, 0.05 M, pH 8.5; CaCl2, 0.02 M. This solution was prepared just before use. For the assay standards, the trypsin stock solution (0.5 mg/ml) was diluted 1:10; then, 1:2 serial dilutions were performed to obtain concentrations of 0.025, 0.0125, 0.00625, 0.00313, 0.00156, and 0.00078 mg/ml, in 1 mM HCl. The assay was set up in duplicate according to Page et al. (2000). Trypsin activity was detected by measurement of the absorbance at 405 nm in a Bio-Tek Instruments, Inc. EL808 Ultra microplate reader (Winooski, VT). A trypsin unit is defined, according to Gaborit et al., (1993), as an absorbance increase of 0.01 at 410 nm. The method described here is an adaptation of that reported by Kakade et al. (1974) and Page et al. (2000).
In vitro differentiation of naïve CD4+ T cells from OVA-tg mice
In vitro antigen-dependent, short-term differentiation was performed as described previously (Heo et al. 1998). Briefly, splenocytes (2 × 106/ml) from unimmunized stat4−/− DO11.10+/− mice, stat6−/− DO11.10+/− mice, wild-type DO11.10+/− mice, or DO11.10 OVA-tg mice, were stimulated with OVA (0.5 mg/ml) in the presence of various additives, such as purified TGF-β2 (10 ng/ml; Genzyme), anti-IL-4 (10 μg/ml; NIH Biorepository), anti-IL-12 (10 μg/ml; PharMingen) mAbs, and PbCl2 (25 μM). T-cells were expanded under the same culture conditions at 72 hr as the primary stimulation, and were harvested on day 7. The cells were counted following washing with PBS, and 2 X 105 cells were re-stimulated with 0.5 mg OVA and 20 Gy-irradiated BALB/c mouse splenocytes (2.5 X 106/ml) in the absence of the experimental additives. Culture supernatants were collected at 48 hr after re-stimulation, and were then used for IL-4 and IFN-γ quantification by ELISA assay as described previously (Heo et al., 1996, 1997, 1998).
Results
Splenic Pb concentration at pnd21
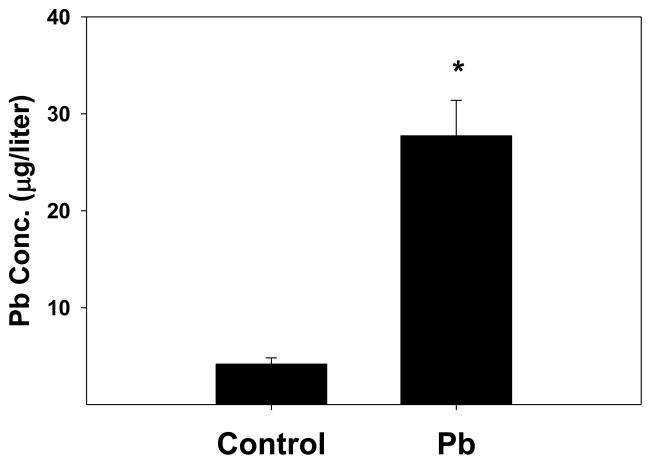
In order to assess the uptake of Pb into the spleen, we performed elemental Pb analysis on spleen homogenates made from tissues of control and Pb-exposed mouse pups. There was a significant elevation of Pb in the spleen homogenates prepared from the Pb-exposed group (Fig. 1). A second group of spleen homogenates prepared and Pb levels were calculated based on tissue weights; again the Pb-exposed offspring had higher Pb levels (13.77 ± 2.29 μg/g) than than those of offspring from the dams that drank distilled water (4.46 ± 0.2μg/g).
Figure 1.
Spleen tissue from Pb-exposed pnd21 mice had significantly higher levels of Pb than did tissue from non-exposed mice. Experimental mice were exposed to 0.1 mM Pb through the dam’s drinking water from gd8 to pnd21. Spleens were homogenized in 1 ml of M-PER reagent including HALT (a cocktail of protease inhibitors), and were analyzed for elemental Pb as described in the methods section. The data represent an n of 4 litters for control and 4 litters for the Pb-exposed mice; significance of the difference between the groups, indicated by the asterisk was determined by the Student’s t-test at p ≤ 0.05.
Genes whose expression was most significantly changed by the developmental Pb exposure
Pair-wise analysis of the microarray data was performed using the GeneSifter software with a fold-change cut-off set at ≥1.2, a p-value of ≤ 0.05 by Student’s t-test, and a quality restriction parameter value of 0.6. Genes with a ≥3-fold expression change induced by Pb were ranked in order of increasing P-value for presentation (Table S1, in Supplemental section). The list indicates that the most strongly up-regulated genes were those coding for enzymes necessary for the breakdown of proteins (e.g., carboxypeptidases and serine proteases), lipids (e.g., pancreatic lipase and phospholipase, and carboxyl ester lipase), and starches (amylase 2). Table 1 lists the genes whose expression was most significantly changed by the Pb exposure. The most significantly up-regulated gene induced by Pb was amylase, but the most highly represented among the genes up-regulated by Pb exposure were the proteolytic enzymes, such as carboxypeptidases, chymotrypsin, trypsin, and elastase. Several lipases and RNase A were also significantly increased by Pb. Listed are additional genes associated with signal transduction, including C-src tyrosine kinase, N-myc downstream regulated gene 1, and Src family associated phosphoprotein 1. Pb also had substantial effects on the expression of genes coding for several calcium modulation proteins, such as calcineurin binding protein, protocadherin 9, and Pellino 2, a modulator of IL-1 signaling (Yu et al., 2002; Jensen and Whitehead, 2003).
Table 1.
Genes in the spleen most significantly affected by developmental Pb exposurea
| Name | Gene ID | p-valueb | Ratioc | Direction | Gene Identifier |
|---|---|---|---|---|---|
| Amylase 2, pancreatic | Amy2 | 0.0000220 | 149.14 | Up | BG142417 |
| Pancreatic lipase | Pnlip | 0.0000658 | 8.2 | Up | NM_026925 |
| Carboxypeptidase A2, pancreatic | Cpa2 | 0.0000715 | 41.54 | Up | BE850208 |
| Chymotrypsin C (caldecrin) | Ctrc | 0.0000829 | 31.72 | Up | NM_007919 |
| Carboxypeptidase A1 | Cpa1 | 0.0001760 | 167.2 | Up | BC052661 |
| Trypsin 4 | Try4 | 0.0006028 | 47.58 | Up | AI386046 |
| Chymotrypsin C (caldecrin) | Ctrc | 0.0006873 | 48.22 | Up | CF580524 |
| Pellino 2 | Peli2 | 0.0007767 | 3.08 | Up | NM_033602 |
| Vomeronasal 1 receptor, G3 | V1rg3 | 0.0010779 | 7.07 | Up | NM_134204 |
| Indoleamine-pyrrole 2,3 dioxygenase | Indo | 0.0020459 | 3.07 | Up | NM_008324 |
| Amylase 2, pancreatic | Amy2 | 0.0022797 | 21.05 | Up | BM503680 |
| Olfactory receptor 1223 | Olfr1223 | 0.0022916 | 3.28 | Up | NM_146892 |
| Carboxypeptidase B1 (tissue) | Cpb1 | 0.0023351 | 28.91 | Up | AK003061 |
| C-src tyrosine kinase | Csk | 0.0024317 | 3.32 | Up | AK047069 |
| Protease, serine, 3 | Prss3 | 0.0024751 | 5.1 | Up | CF580442 |
| Calcineurin binding protein 1 | Cabin1 | 0.0036986 | 3.93 | Up | BY571589 |
| Ring finger protein 138 | Rnf138 | 0.0040949 | 3.88 | Up | NM_019706 |
| Olfactory receptor 1225 | Olfr1225 | 0.0047093 | 3.72 | Up | NM_146891 |
| CUB and zona pellucida-like domains 1 | Cuzd1 | 0.0052438 | 3.85 | Up | NM_008411 |
| Cytochrome P450, family 3, subfamily a, polypeptide 44 | Cyp3a44 | 0.0059048 | 3.79 | Up | NM_177380 |
| Protocadherin 9 | Pcdh9 | 0.0065029 | 3.36 | Up | BB188861 |
| Histone cluster 1, H2aa | Hist1h2aa | 0.0067366 | 3.59 | Down | NM_175658 |
| Ribonuclease, RNase A family, 1 (pancreatic) | Rnase1 | 0.0080881 | 4.05 | Up | NM_011271 |
| Carboxyl ester lipase | Cel | 0.0080991 | 6.68 | Up | NM_009885 |
| Olfactory receptor 643 | Olfr643 | 0.0092722 | 4.34 | Up | NM_147077 |
| N-myc downstream regulated gene 1 | Ndrg1 | 0.0092926 | 5.14 | Up | NM_008681 |
| Kyphoscoliosis peptidase | Ky | 0.0095591 | 3.27 | Up | NM_024291 |
| Tight junction protein 3 | Tjp3 | 0.0100910 | 3.83 | Up | NM_013769 |
Pair-wise comparison of mouse spleen codelink data from 3 control litters and 3 Pb-exposed litters.
Student’s t-test, significant for P ≤ 0.05.
Ratio = Pb signal/control signal when direction is Up, and control signal/Pb signal when direction is Down.
Real-Time RT-PCR results show increased chymotrypsin gene expression
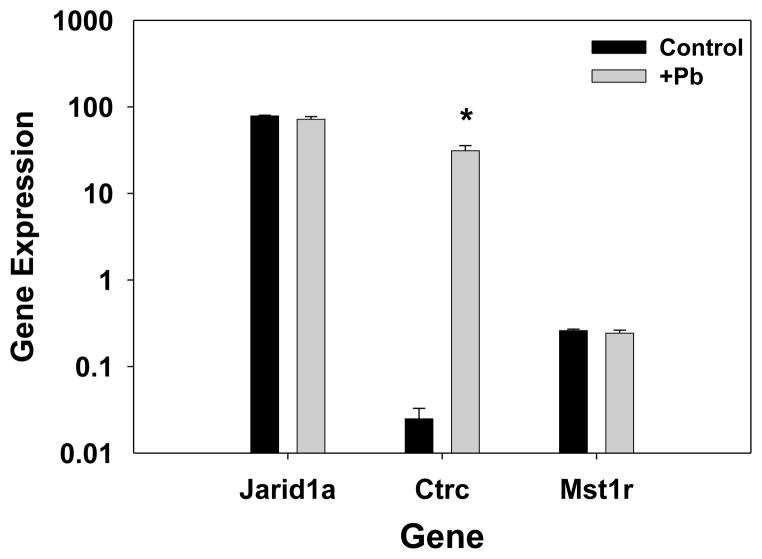
Expression of the ctrc gene that codes for chymotrypsin was measured by real-time RT-PCR, along with expression of two other genes whose expression via microarray analysis was either slightly changed or unchanged by Pb. These genes with low-fold changes were analyzed as further controls for the validation of the microarray results. As shown (Fig. 2), Pb increased the quantity of mRNA for chymotrypsin C to a much greater extent than it did for either jumonji (Jarid1a), a protein involved in DNA structural modification and methylation, or a macrophage stimulatory receptor (MSt1r). These results are in agreement with the microarray data, which indicated that chymotrypsin gene expression was strongly increased by Pb (>2000 fold, by RT-PCR), whereas jumonji gene expression and macrophage stimulatory receptor gene expression were not highly changed by Pb.
Figure 2.
Real-time RT-PCR analysis confirms the microarray analysis of increased expression of chymotrypsin upon developmental Pb exposure. Relative gene expression in the spleen was determined for control (n=3) and 0.1mM Pb-exposed mouse pups (n=3) representative of separate litters. The TaqMan method was performed as described in the methods. Relative gene expression was calculated with respect to the Ct for the gene normalized to gapdh, as described in the methods. Gene IDs indicate the following: Jarid1a, jumonji; Ctrc, chymotrypsin C; and Mst1r, macrophage stimulating 1 receptor. Significance of the difference between the groups, indicated by the asterisk was determined by the Student’s t-test at p ≤ 0.05.
Increased amylase and trypsin enzyme activity after Pb exposure
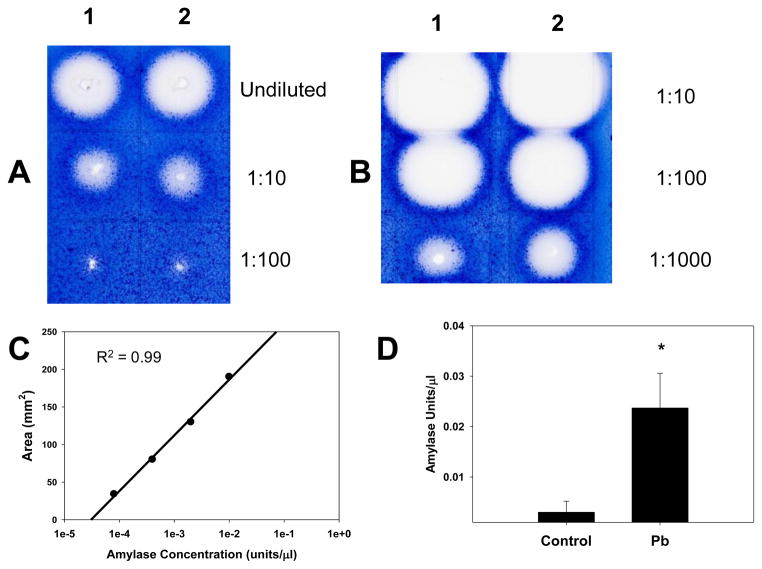
As indicated in Table 1, Pb exposure greatly increased expression of the amylase gene. In order to determine whether there was a corresponding increase in amylase activity in the presence of Pb, we performed a simple enzyme assay for amylase, using a starch gel. Amylase digests starch down to its components, glucose and fructose. Therefore, if amylase activity is enhanced, more starch will be digested. Staining of the gel with iodine, which stains starch blue, indicates the extent of amylase activity, based on plaque area (Fig. 3). Spleen homogenates from Pb-exposed offspring had more amylase activity than did homogenates from the H2O (control) offspring. Imager software analysis of the starch gels indicated that the Pb-exposed pups (N = 5) had about 10 times more amylase activity than did the control pups (N = 5) (Fig. 3D).
Figure 3.
Splenic amylase activity was increased by developmental Pb-exposure. Mouse pups were exposed to 0.1mM Pb from gd8 to pnd21. Spleen homogenates shown (2 control and 2 Pb-exposed) were prepared and assayed for amylase activity as described in the methods. Each spleen homogenate result shown here was from one spleen of a mouse representative of one litter. The gel photographs display the raw data, A) Control and B) Pb-exposed. Homogenates were diluted 1:10, 1:100, and 1:1000 with 0.1M phosphate buffer (pH 6.9) before applying to the gel. C shows a sample standard curve generated with purified amylase, and D shows the relative splenic amylase activity of control and 0.1 mM Pb-exposed mouse pups. Significance of the difference between the groups, indicated by the asterisk was determined by the Student’s t-test at p ≤ 0.05.
Pb also increased trypsin expression in the mouse spleens. Surface plasmon resonance measurements were employed to determine the relative trypsin protein content in the control and Pb-exposed spleens of the offspring. Trypsin inhibitor was bound to flow-cell 2, in order to capture trypsin present in the spleen homogenates; flow-cell 1 was a blank surface control to measure non-specific binding to the chip surface. As shown (Table 2), homogenates from the Pb-exposed offspring had several-fold higher amounts of trypsin protein than did homogenates from control offspring, as indicated by the increase in RU. Trypsin enzyme activity present in spleen homogenates from control and Pb-exposed pups was quantified: the spleen homogenates from control offspring had 55.00 ± 1.41 units/ml of trypsin, and the spleen homogenates of the Pb-exposed offspring had 86.13 ± 3.36 units/ml of trypsin activity, P < 0.01.
Table 2.
Effect of Pb exposure on trypsin protein content in mouse pup spleena
| Sample | Blank Surface RUb | Trypsin Inhib. RU | Trypsin Inhib. minus Blank RU | Average RU |
|---|---|---|---|---|
| Control #1 | 119.3 | 204.9 | 85.6 | 92.2 |
| 85.5 | 184.3 | 98.8 | ||
| Control #2 | 78.3 | 149.7 | 71.4 | 73.2 |
| 74.5 | 149.4 | 74.9 | ||
| +Pb #1 | 123.3 | 321.4 | 198.1 | 198.1 |
| 97.9 | 296 | 198.1 | ||
| +Pb #2 | 88.9 | 299.1 | 210.2 | 206.2 |
| 86.4 | 288.5 | 202.1 |
Spleen homogenates were prepared, as described in the methods (protease inhibitors present). Spleen homogenates were prepared separately from two control and two Pb exposed mice, each representative of a litter.
RU, response unit.
Pb modulated expression of genes associated with the innate immune response
Although it has been well established that Pb has certain strong effects on cell-mediated immunity, its effects on the innate immune response have not been delineated. Gene expression changes by Pb exposure of genes included in the immune-system biological process ontology are listed in Table S2 according to ascending P-value. The list includes many genes that encoding for proteins that are part of the innate immune response, such as, members of the complement system (C4bp and Cfp), Toll-like receptors (Tlr8 and Tlr9), and other immediate pathogen response molecules, such as Nod2, Clec4n, Ulbp1, Rya3, Cbl, and Ly96. Lymphoid cell recruitment by Pb exposure is indicated by chemokines and chemokine receptors, including Ccl1, Ccr4, Ccl12, and Ccr7. Additionally, Pb up-regulated expression of the gene for indoleamine-pyrrole 2,3 dioxygenase (IDO), an enzyme important during innate immunity, for its function to deplete L-Trp from local cell microenvironments and to promote formation of kynurenine pathway metabolites (King and Thomas, 2007). Depletion of L-Trp slows the growth of the pathogens, because it depletes local stores of L-Trp needed for protein synthesis. Interestingly, IDO also inhibits Th1 activity and enhances Th2 activity (Xu et al., 2008).
Impact of Pb on apoptosis in the spleen
Table 3 lists the genes, whose expression was changed by Pb exposure, which are associated with apoptosis according to the GeneSifter software analysis. Pb caused decreased expression of genes for Bcl2 and Akt1, known inhibitors of apoptosis. However, Pb increased the expression of genes that code for proteins associated with increased apoptosis; these include caspase 7, traf2, Fadd, and Trail. Although the gene for caspase 12 is included here, the function of its product is more closely linked to the innate immune response, given that it blunts the signaling of the NOD2 pathway, leading to NF-κB activation (LeBlanc et al., 2008). Bcl10 is also up-regulated by Pb; this protein at low levels promotes the binding of Traf2 to c-IAPs. Overproduction of Bcl2 will block formation of the Traf2/c-IAP complexes and decrease apoptosis (Yoneda et al., 2000), since Pb decreases Bcl2 expression, Traf2/c-IAP complex formation should remain unblocked. Interestingly, caspase 6 expression is down-regulated by Pb. Caspase 6 is a member of the intrinsic pathway of apoptosis, and it and caspase 2 can be activated by caspase 7. Activated caspase 6 will then go on to cleave caspase 8 and other caspase 6 substrates, such as lamin A/C (Inoue et al., 2009).
Table 3.
Effect of developmental Pb exposure on genes in the spleen associated with apoptosisa
| Gene Name | Gene ID | p-valueb | Ratioc | Direction | Gene Identifier |
|---|---|---|---|---|---|
| B-cell leukemia/lymphoma 2 | Bcl2 | 0.00237 | 1.39 | Down | NM_009741 |
| Phosphatidylinositol 3 kinase, regulatory subunit, polypeptide 3 (p55) | Pik3r3 | 0.00533 | 1.56 | Up | NM_181585 |
| Caspase 12 | Casp12 | 0.01226 | 2.12 | Up | NM_009808 |
| B-cell leukemia/lymphoma 10 | Bcl10 | 0.01252 | 1.69 | Up | NM_009740 |
| Tnf receptor-associated factor 2 | Traf2 | 0.01697 | 5.89 | Up | NM_009422 |
| Thymoma viral proto-oncogene 1 | Akt1 | 0.01752 | 1.28 | Down | NM_009652 |
| Baculoviral IAP repeat-containing 2 | Birc2 | 0.02239 | 2.07 | Up | NM_007465 |
| RIKEN cDNA 1500003O03 gene | 1500003O03Rik | 0.02432 | 2.24 | Up | NM_019769 |
| Colony stimulating factor 2 receptor, beta 1, | Csf2rb1 | 0.02465 | 2.86 | Up | BY529130 |
| CASP8 and FADD-like apoptosis regulator | Cflar | 0.02502 | 2.11 | Up | BC023121 |
| Baculoviral IAP repeat-containing 3 | Birc3 | 0.02803 | 2.4 | Up | NM_007464 |
| Phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 1 (p85 alpha) | Pik3r1 | 0.02956 | 1.76 | Up | AW824975 |
| Fas (TNFRSF6)-associated via death domain | Fadd | 0.03329 | 2.25 | Up | NM_010175 |
| Protein kinase, cAMP dependent, catalytic, alpha | Prkaca | 0.03852 | 1.46 | Up | BI991369 |
| Protein kinase, cAMP dependent regulatory, type II beta | Prkar2b | 0.03992 | 2.75 | Up | NM_011158 |
| tumor necrosis factor (ligand) superfamily, member 10 | Trail | 0.04009 | 2.39 | Up | NM_009425 |
| Caspase 7 | Casp7 | 0.04100 | 1.5 | Up | NM_007611 |
| Apoptotic peptidase activating factor 1 | Apaf1 | 0.04378 | 3.08 | Up | NM_009684 |
| Caspase 6 | Casp6 | 0.04481 | 2.84 | Down | NM_009811 |
| V-rel reticuloendotheliosis viral oncogene homolog A (avian) | Rela | 0.04625 | 1.78 | Down | BY184365 |
Mouse spleen data from 3 control and 3 Pb-exposed whole mouse genome microarrays.
Results were analyzed by pair-wise comparison using GeneSifter software.
Significance was determined by Student’s t-test, p≤0.05.
Ratio=Pb signal/control signal when the direction is Up, and control signal/Pb signal when the direction is Down
Increased B-cell activation and proliferation by Pb
The microarray results presented here are suggestive of increased B-cell function, activation, and proliferation (Tables S2 and 4). Table 4 lists the genes affected by Pb exposure that are B-cell associated. Prominent among the genes up-regulated by Pb are those that code for IL-7 (pre-B-cell growth factor; 3.42-fold change), MHC class II (2.65-fold change), the heavy chain of IgM (3.63-fold change), Notch2, needed for production of marginal zone B-cells, CD27 and IL-7 receptor (CD127), needed for generation of memory B-cells, and Bcl6, which is essential for the differentiation of germinal center B cells (Fukuda et al., 1997; Arguni et al., 2006). Other genes up-regulated by Pb (not specific to B-cell activity, but necessary for B-cell function) are those genes involved in DNA recombination (e.g. Rag2) and transcription regulation (e.g. histone deacetylase 7A).
Table 4.
Effect of developmental Pb exposure on B-cell associated genes in the spleena
| Gene Name | Gene ID | p-valueb | Ratioc | Direction | Gene Identifier |
|---|---|---|---|---|---|
| Recombination activating gene 2 | Rag2 | 0.00207 | 1.83 | Up | NM_009020 |
| Interleukin 7 | Il7 | 0.01355 | 3.42 | Up | NM_008371 |
| Leukocyte immunoglobulin-like receptor, subfamily B | Lilrb3 | 0.01695 | 1.54 | Up | NM_008848 |
| B-cell leukemia/lymphoma 6 | Bcl6 | 0.01800 | 2.58 | Up | BE985970 |
| B and T lymphocyte associated | Btla | 0.01864 | 2.58 | Up | NM_177584 |
| RNA binding motif protein 24 | Rbm24 | 0.02275 | 2.35 | Up | BM943034 |
| Immunoglobulin heavy chain 6 (heavy chain of IgM) | Igh-6 | 0.02972 | 3.63 | Up | X56392 |
| CD antigen 27 | Cd27 | 0.03047 | 1.96 | Up | L24495 |
| Nonhomologous end-joining factor 1 | Nhej1 | 0.03146 | 2.45 | Down | NM_010544 |
| Histone deacetylase 9 | Hdac9 | 0.03271 | 1.81 | Up | AF324492 |
| Histocompatibility 2, class II antigen A, beta 1 | H2-Ab1 | 0.03447 | 2.65 | Up | NM_207105 |
| Notch gene homolog 2 (Drosophila) | Notch2 | 0.03941 | 2.25 | Up | NM_010928 |
| Interleukin 4 | Il4 | 0.04138 | 2.24 | Up | NM_021283 |
| Protein kinase, DNA activated, catalytic polypeptide | Prkdc | 0.04352 | 1.44 | Up | NM_011159 |
| Tumor necrosis factor (ligand) superfamily, member 13b | Tnfsf13b | 0.04670 | 1.67 | Up | NM_033622 |
| Histone deacetylase 7A | Hdac7a | 0.04715 | 2.71 | Up | NM_019572 |
| Interleukin 7 receptor | Il7r | 0.04741 | 1.87 | Up | NM_008372 |
| Interleukin 21 | Il21 | 0.04863 | 2.09 | Up | NM_021782 |
Mouse spleen data from 3 control and 3 Pb-exposed whole mouse genome microarrays.
Results were analyzed by pairwise comparison using GeneSifter software.
Significance was determined by Student’s t-test, p≤0.05.
Ratio=Pb signal/control signal when the direction is Up, and control signal/Pb signal when the direction is Down
Microarrary data reaffirm that Pb skews T-helper cell subsets toward a Th2 immune response
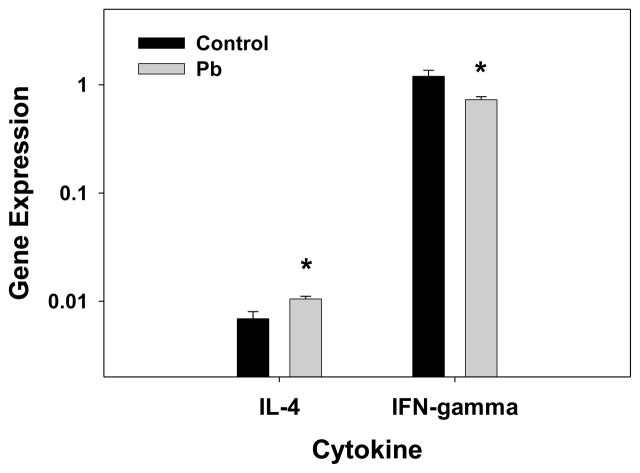
Developmental Pb exposure increased expression of IL-4 and decreased expression of IFN-γ in the spleen (Fig. 4), as determined by quantitative real-time RT-PCR. Th2 skewing was observed as an increase in production of IL-4 by splenocytes in culture, in the presence of Pb (Heo et al., 1996). An inspection of the microarray results (Table S1 and S2) indicated that Pb up-regulated expression of adenylate cyclase 8 (3.66-fold); this up-regulation correlates with our earlier finding that Pb increased cAMP levels (Heo et al., 1998). With respect to targeting of il-4 gene expression, we found that phosphatidylinositol 3-kinase (PI3K), the co-regulator of cAMP activated protein kinase A, was up-regulated (1.76-fold) (Table S2). Therefore, the increase in adenylate cyclase and PI3K expression by Pb could promote an increase in il-4 gene expression and Th2 skewing. Table 5 lists cytokine genes affected by Pb exposure. As pointed out in Table 5, interferon-α expression was highly down-regulated by Pb. In accordance with these results (i.e., the microarray data demonstrating Th2 skewing), two interferon-induced proteins, interferon-activated gene 203 and interferon-induced protein 35 (Table S2), were down-regulated by Pb. Overall, the cytokine expression pattern shown in Table 5 reflects the Th2 skewing due to Pb exposure. However, the data indicate that Pb increased the expression of CD247, the T-cell receptor zeta chain, but Pb decreased expression of Zap70, the zeta chain-associated protein kinase. This out-come suggests that T-cell activity (the cell-mediated-immune response) is actually down-regulated by Pb.
Figure 4.
Pb promotes Th2 skewing in the spleen as indicated by relative Th1 and Th2 cytokine mRNA levels. Cytokine transcripts for IL-4 and IFN-gamma were measured by an absolute quantification RT-PCR method and were normalized to gapdh transcript level. Results are presented as average (cytokine CN/GAPDH CN) X 100 ± SEM. The results represent data from 3 control and 3 0.1 mM Pb treated mouse pup spleens, representative of 3 litters per condition. Significance of the difference between the groups, indicated by the asterisk was determined by the Student’s t-test at p ≤ 0.05.
Table 5.
List of cytokines in the spleen affected by Pb exposurea
| Gene Name | Gene ID | p-valueb | Ratioc | Direction | Gene Identifier |
|---|---|---|---|---|---|
| Interferon alpha 1 | Ifna1 | 0.0094344 | 1.33 | Down | NM_010502 |
| Inhibin beta-A | Inhba | 0.0129858 | 2.01 | Up | NM_008380 |
| Interleukin 7 | Il7 | 0.0135505 | 3.42 | Up | NM_008371 |
| Interleukin 20 | Il20 | 0.0193603 | 1.66 | Up | NM_021380 |
| Bone morphogenetic protein 8a | Bmp8a | 0.0222596 | 3.35 | Down | NM_022033 |
| Interleukin 5 | Il5 | 0.0231758 | 1.99 | Up | NM_010558 |
| Oncostatin M | Osm | 0.0293897 | 1.52 | Up | AK087945 |
| Interleukin 4 | Il4 | 0.0413793 | 2.24 | Up | NM_021283 |
| Interleukin 31 | Il31 | 0.0451831 | 2.15 | Up | AK005939 |
| Interleukin 21 | Il21 | 0.0486276 | 2.09 | Up | NM_021782 |
Mouse spleen data from 3 control and 3 Pb-exposed whole mouse genome microarrays.
Results were analyzed by pair-wise comparison using GeneSifter software.
Significance was determined by Student’s t-test, p≤0.05.
Ratio=Pb signal/control signal when the direction is Up, and control signal/Pb signal when the direction is Down
Effects of Pb on T cell development in the absence of STAT4 or STAT6
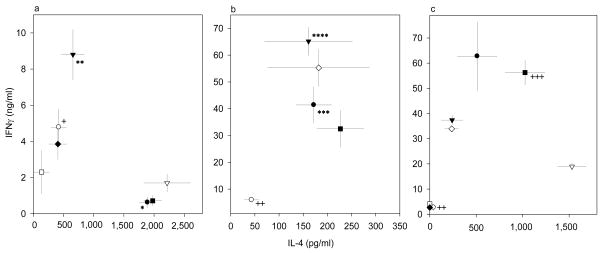
The microarray data did not point to a direct effect of Pb exposure on expression of any of the STAT proteins. Since T-helper subsets Th1 and Th2 convey their activities through modulation of STAT4 and STAT6, respectively, we performed experiments to obtain more information about Pb’s ability to skew T-cell help toward a Th2 (IL-4) response. The effects of Pb were compared with those of antibodies to IL-12 or IL-4, which signal through STAT4 or STAT6, respectively. Additionally, TGF-β was evaluated, since it can skew CD4 cells toward Th17 or Treg cells. The consequences of STAT4 or STAT6 deficiency with antigen (OVA)-driven differentiation of Thp were examined, as described in the Methods. Our assay design is perhaps closer to the in vivo environmental conditions, in which differentiation occurs, than was true for assays that used purified CD4+ T cells and different types of antigen presenting cells (APCs) (Sedar et al. 1992; Hsieh et al. 1992; Sedar et al. 1993; Macatonia et al. 1995). Predominant development of one Th effector subset was determined through comparison of the levels of IL-4 and IFN-γ produced in response to OVA re-stimulation. Spleen cells from OVA-tg mice lacking STAT4 produced substantially larger amounts of IL-4 and significantly reduced amounts of IFN-γ (Fig 5a) relative to the amount produced by WT cells (Fig. 5c); in contrast, IL-4 production in the absence of STAT6 was inhibited, but IFN-γ production was not significantly altered (Fig. 5b), relative to the response of WT cells (Fig. 5c). TGF-β inhibited the production of IFN-γ and IL-4 by the WT and STAT6-deficient cells, but surprisingly, TGF-β enhanced production of IFN-γ by the STAT4-deficient cells. In the absence of STAT6, anti-IL4 still enhanced the Th1 (IFN-γ) response, but Pb, either with or without anti-IL-4, continued to slightly enhance the IL-4 response and inhibit the IFNγ response. In the absence of STAT4, Pb had no effect on IL-4 or IFNγ production, but anti-IL-4 was able to enhance IFN-γ production and inhibit IL-4 production.
Figure 5.
Modification of OVA-tg T-cell differentiation with spleen cells from stat4−/− DO11.10+/− (a), stat6−/− DO11.10+/− (b), or wild type DO11.10+/− (c) mice. Cells were differentiated with OVA in the absence of various additives (black circle) or presence of TGF-β (open circle), anti-IL-4 (black triangle), anti-IL-12 (open triangle), Pb (black square), TGF-β+anti-IL-4 (open square), TGF-β+anti-IL-12 (black diamond), or Pb+anti-IL-4 (open diamond). Data were obtained from 6 mice assayed individually in six separate experiments, and the results are expressed as the mean amounts (± SEM) of IL-4 and IFN-γ produced.
Discussion
The persistence of the immunotoxic effects of Pb after developmental exposures have been documented for studies performed in rats, mice, and chickens (Miller et al., 1998; Snyder et al., 2000; Bunn et al., 2001; Lee et al., 2002). Interestingly, most of these studies indicated that developmental Pb exposure decreased, to some extent, spleen cell numbers and blood leukocyte numbers. In rats, gestationally exposed female offspring measured at 13–14 weeks of age showed decreased leukocyte counts in the blood, decreased interferon-gamma production, increased IgE secretion, and decreased delayed-type hypersensitivity (DTH) (Miller et al., 1998). After gestational and lactational exposure to Pb, mice had decreased cell numbers for all lymphoid subsets and elevated plasma IgE levels at 2 weeks of age (Snyder et al., 2000). Pb exposure during early (3–9 days) and late (15–21 days) gestation differentially affected the immune system in male and female rats (Bunn et al., 2001). Increases in plasma IgE levels and decreases in DTH indicate skewing toward a Th2 mediated immune response. Using exposure periods and Pb levels similar to Snyder et al. (2000), we now provide some new insight into how developmental Pb exposure can affect immunity.
Decreased splenocyte cell counts could be the result of increased apoptosis in the spleen due to the Pb exposure. Microarray results suggest that Pb increases apoptosis, since Bcl2 expression was decreased, and a number of genes associated with apoptosis were up-regulated. Recently, Xu, J. et al. (2008) reported that Pb increases DNA damage via induction of oxidative stress, as measured by production of reactive oxygen species (ROS) and malondialdehyde (MDA). Such DNA damage leads to apoptosis or to mutations due to inaccurate DNA repair. They observed that Pb raised the Bax/Bcl2 ratio and that the Pb-induced apoptosis was mainly mitochondrion mediated. Apoptotic cells are targeted for phagocytosis by macrophages in the spleen, through the presence of phosphatidylserine on the cells’ outer membranes (Kuypers and DeJong, 2004). The combination of elevated levels of catabolizing enzymes and increased rate of cell death could lead to release of numerous self peptides, as well as DNA fragments, thus potentially enhancing induction of self immunity. It has been recently reported (Zhang et al., 2009) that antibodies to dsDNA cross-react with self proteins, causing physiological changes. Such systemic lupus erythematosus (SLE) “polyreactive” antibodies were demonstrated to bind glomerular structures in vitro, suggestive of their potential for pathological effects on the kidney in vivo. Pb does enhance the level of anti-dsDNA in lupus-prone mice (Hudson et al., 2003), which extends the possibility of its potential promotion of autoimmunity.
Our results indicated that developmental Pb exposure markedly increased expression of amylase, peptidases (exo- and endopeptidases), and lipases in the spleen. A possible explanation for the up-regulation of amylase activity is Pb-induced stress. Secretion of α-amylase has been associated with a number of forms of stress. For example, salivary amylase activity was significantly elevated for several days after surgical procedures in children (Groza et al., 1971). In other studies, serum levels of pancreatic amylase have been observed to increase due to various forms of stress (Kubisch and Logsdon, 2007; Cosen-Binker et al., 2004; Takano et al., 1992). Up-regulation of chymotrypsin, elastase, carboxypeptidases and trypsin 4 by Pb could be indications that Pb is causing inflammation or injury to the spleen, since proteases have been found to be released from lysosomes during inflammatory reactions (Vischer and Bertrand, 1976; Wang et al., 1998). In addition to activation of the complement system, these proteases can function in the activation and proliferation of lymphocytes, as was demonstrated by Vischer and Bertrand (1976) and Waldo et al. (2003). Increases in these enzymes could be in response to increased apoptosis in the spleen.
Microarray results from our present study point to a marked involvement of Pb in the innate immune response. For example, C4bp was highly up-regulated (5.15-fold) by Pb exposure. One of the functions of this protein is to regulate the classical complement pathway. C4bp binds to 4b and blocks the formation of C3/C5 convertase, which is (4b, C2a). In this manner, C4bp can down-regulate the activity of the complement pathway (Rawal and Pangburn, 2007; Rawal et al., 2009). Another function of C4bp is to compensate for the loss of membrane-bound complement inhibitors on apoptotic cells. C4bp can bind to apoptotic cells via exposed phosphatidylserine, and, thus, can block the lysis of these cells by complement. This protective effect helps to limit the release of pro-inflammatory factors caused by the lysis of apoptotic cells by complement. Increases in C4bp expression during developmental Pb exposure could be an indication that the spleen is trying to decrease the amount of cells undergoing lysis by complement. Accumulation of apoptotic cells is believed to be the underlying cause of certain autoimmune diseases, including SLE (Trouw et al., 2007). Further evidence of developmental Pb involvement in the innate immune system response is the increase in the expression of caspase-12. Recent reports have indicated that caspase-12 is detrimental to clearance of bacterial pathogens, both systemic and abdominal. Mice deficient in caspase-12 were able to clear Listeria monocytogenes from the spleen more efficiently than were wild type mice (casp12+/+) (Saleh et al., 2006). Caspase-12 also regulates the formation of mature IL-1β and IL-18, through direct association and inhibition of caspase-1 (Saleh et al., 2006). Since IL-18 has a powerful effect on induction of IFN-γ, a decrease in the active form of IL-18 could decrease IFN-γ production. Down regulation of IFN-γ production has been shown to occur due to developmental Pb exposure (Miller et al., 1998).
The microarray data provide further insight into Pb’s impact on hemoglobin synthesis and red cell life span in the spleen. The developmental Pb exposure described here produced a 5.74-fold increase (p=0.03) in expression of the gene coding for eukaryotic translation initiation factor 2-alpha kinase 2 [eif2ak2 or heme-regulated inhibitor (HRI)], an enzyme important for the regulation of globin synthesis (Chen et al., 1990). Pb has been shown to impair globin synthesis by inhibiting cellular reductases, such as thioredoxin and/or thioredoxin reductase; these enzymes are responsible for maintaining HRI in an inactive state (Matts et al., 1991). However, the appearance of increased levels of HRI in the blood of rabbits during Pb-induced anemia (Anand and Pal, 2002) suggested that Pb also inhibits globin synthesis by increasing HRI expression. Our microarray results give support to the findings of Anand and Pal (2002), because they show that Pb can increase HRI gene expression. With respect to the impact of Pb on erythrocyte life-span, our data indicate that Pb decreased expression of CD47 by 1.86-fold (p=0.02). Interaction between CD47 on the erythrocyte and SHPS-1 on the macrophage inhibits phagocytosis of the erythrocyte, whereas a decrease in this interaction increases phagocytosis (Ishikawa-Sekigami et al., 2006). Senescent erythrocytes possess less CD47 than do young erythrocytes (Sparrow and Healey, 2006). Pb can also promote phosphatidylserine exposure on the outer membrane surface of erythrocytes (Kempe et al., 2005). By decreasing CD47 expression and increasing phosphatidylserine exposure, Pb can decrease the life-span of erythrocytes.
Our gene expression results support earlier findings of Pb-induced increases of IL-4 expression and development of Th2 cells. Experiments performed with STAT4- and STAT6-deficient mice indicated that Pb can alter the development of Th1 and Th2 cells even in the absence of the STAT4 and STAT6 signaling pathways. Heo et al. (1998) made the connection between Th2-subtype skewing and the elevation of cAMP levels in the presence of Pb, suggesting that Pb inhibits Th1 development by enhancing adenylate cyclase activity. The microarray data from our present study indicates that Pb exposure increases the expression of an adenylate cyclase (Adcy8), and also the expression of PI3K. However, Th2 skewing may not be due to a direct effect of Pb on the T-cells; alternatively, the skewing could be mediated through APCs. Our microarray data suggest that Pb has a strong effect on promotion of B-cell differentiation and proliferation (Tables S1, 4, and S2). APC involvement in the mechanism of Th2 skewing by Pb was observed in experiments employing bone-marrow-derived dendritic cells (BM-DCs) (Gao et al., 2007), in that Pb-exposed BM-DCs had the ability to polarize antigen-specific naïve T cells toward Th2 cells. Pb modification of DC function had been posited to be the reason for the skewing toward the Th2 response (Lawrence and McCabe, 1995).
It is noteworthy that the suppression of Th2 development with cells lacking STAT6 did not favor a dominance of Th1 development, in that IFN-γ production was lower than with the WT cells. Additionally, Pb was still able to slightly inhibit the Th1 response and to enhance the Th2 response, when cells had no STAT6. The ability of anti-IL-4 to enhance the IFN-γ response of the STAT4-deficient cells indicates that IL-4 can interfere with Th1 cell development, even when IL-12 cannot signal through STAT4; therefore, IL-4 inhibits at a level other than just compromising IL-12 signaling. The ability of anti-IL-4 to enhance the IFN-γ response of the STAT6-deficient cells indicates that IL-4 can still be made by cells lacking STAT6, and that T- cells that cannot be signaled by IL-4 through STAT6 can still be skewed away from Th1 responses by IL-4. The later capability cannot be due to preferential development of Th2 cells, since anti-IL-4 had no effect on the development of Th2 cells.
In addition to TGF-β’s commonly known immunosuppressive effects on T-cell proliferation and cytokine production, the cytokine also has been investigated for its regulatory role on the balance between Th1 and Th2 responses (Swain et al., 1991; Prochazkova et al., 2009). To further delineate the effects of TGF-β on in vitro antigen-driven development of Th subsets, and to investigate a potential interaction between STAT and TGF-β, we added TGF-β as a modifier of antigen priming, with the same experimental protocol as was used with the cytokine-neutralizing Abs. As expected, TGF-β induced significant inhibition of both IL-4- and IFN-γ-producing ability, with cells from the stat6−/− mice and the WT mice; however, to our surprise, TGF-β significantly enhanced the production of IFN-γ (Th1 cell response) with the cells from stat4−/−/OVA-tg mice. This unique observation implies that the potential ability of TGF-β to favor a Th1 response can be overshadowed in a Th1-favoring cytokine environment. Interestingly, TGF-β suppressed the Th1 response of the WT cells significantly more than did Pb; however, with the stat4−/− cells, TGF-β allowed a greater IFN-γ response than did Pb.
As was noted earlier, Pb has been reported to preferentially suppress the development of Th1 cells, and to enhance the development of Th2 cells from OVA-tg and OVA-tg/recombinase activator genes (RAG) 2−/− mice (Heo et al., 1998). Now, we have shown that the Pb-induced preferential effects on the development of Th2 cells still exist with stat4−/− and stat6−/− spleen cells, indicating that Pb as well as TGF-β can differentially alter the development of Th1 and Th2 cells, even in the absence of STAT4 and STAT6 signaling pathways. Further analysis will be needed to assess effects of the combination of Pb and TGF-β, and the combination of various cytokines and Pb, on the development of additional CD4+ T cell subsets, such as Th17 and Treg cells.
Our present results highlight the potential of Pb to induce immune responses, likely Th2-mediated responses, to endogenous constituents. The high expression of catabolizing enzymes, the higher levels of apoptosis and cell stress, induced by Pb along with Pb’s ability to alter immunity are proposed to increase any genetic penetrance for an autoimmune disease. Additionally, the well described effects of Pb on multiple organ systems, including the central nervous, cardiovascular, and reproductive systems could be related to systemic inflammation resulting from the described catabolic activities induced by Pb.
Supplementary Material
Table S1: A listing of genes whose expression is changed by developmental Pb exposure greater then 3.0 fold in the spleen.
Table S2: A listing of the immune system biological process genes whose expression is changed 1.2 fold or greater by developmental Pb exposure.
Acknowledgments
This work was supported by NIH grant RO1 ES11135 (to DAL). GE Healthcare generously provided the CodeLink microarray slides used for the present study. We thank Sridar V. Chittur and Marcy Kuentzel from the Microarray Core Facility at the Center for Functional Genomics of SUNY Albany, for technical assistance in performing the microarray assay. We thank the Inorganic and Nuclear Chemistry Laboratory, under the supervision of Dr. Patrick Parsons, within the Wadsworth Center for quantification of elemental Pb in spleen homogenates. We acknowledge the help of the Immunology Core, Wadsworth Center, with the molecular interaction analysis experiments. We thank Donghong Gao and Pamela Kruger for their technical assistance, and Adriana Verschoor for her editorial assistance.
Abbreviations
- APC
antigen presenting cell
- ALA-D
δ-aminolevulinic acid dehydrase
- BAPNA
N-α-benzoyl-DL-arginine-p-nitroanilide hydrochloride
- eIF
eukaryotic initiation factor
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HRI
heme-regulated inhibitor
- IL
interleukin
- MHC
major histocompatibility complex
- Pb
lead
- PDK1
phosphoinositide-dependent kinase 1
- PI3K
phosphatidylinositol-3-kinase
- PMA
phorbol myristate acetate
- PMNs
polymorphonuclear cells
- SLE
systemic lupus erythematosis
- Th
T-helper
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jane Kasten-Jolly, Email: kjolly@wadsworth.org.
Yong Heo, Email: yheo@cu.ac.kr.
David A. Lawrence, Email: david.lawrence@wadsworth.org.
References
- Ahamed M, Verma S, Kumar A, Siddiqui MKJ. Delta-aminolevulinic acid dehydratase inhibition and oxidative stress in relation to blood lead among urban adolescents. Human & Exp Toxicol. 2006;25:547–553. doi: 10.1191/0960327106het657oa. [DOI] [PubMed] [Google Scholar]
- Ahamed M, Siddiqui MKJ. Low level lead exposure and oxidative stress: Current opinions. Clinica Chimica Acta. 2007;383:57–64. doi: 10.1016/j.cca.2007.04.024. [DOI] [PubMed] [Google Scholar]
- Anand S, Pal JK. The haem-regulated eukaryotic initiation factor 2alpha kinase: a molecular indicator of lead-toxicity anaemia in rabbits. Biotechnol Appl Biochem. 2002;36:57–62. doi: 10.1042/ba20020009. [DOI] [PubMed] [Google Scholar]
- Arguni E, Arima M, Tsuruoka N, Sakamoto A, Hatano M, Tokuhisa T. JunD/AP-1 and STAT3 are the major enhancer molecules for high Bcl6 expression in germinal center B cells. International Immunology. 2006;18:1079–1089. doi: 10.1093/intimm/dxl041. [DOI] [PubMed] [Google Scholar]
- Bunn TL, Parsons PJ, Kao E, Dietert RR. Exposure to lead during critical windows of embryonic development: Differential immunotoxic outcome based on stage of exposure and gender. Toxicol Sci. 2001;64:57–66. doi: 10.1093/toxsci/64.1.57. [DOI] [PubMed] [Google Scholar]
- Chen JJ, Yang JM, Petryshyn R, Kosower N, London LM. Disulfide bond formation in the regulation of eIF-2α kinase by heme. J Biol Chem. 1990;264:9559–9564. [PubMed] [Google Scholar]
- Cook JA, Hoffman EO, diLuzio NR. Influence of lead and cadmium on the susceptibility of rats to bacterial challenge (39117) Proc Exp Biol Med. 1975;150:741–747. doi: 10.3181/00379727-150-39117. [DOI] [PubMed] [Google Scholar]
- Cosen-Binker LI, Binker MG, Negri G, Tiscornia O. Influence of stress in acute pancreatitis and correlation with stress-induced gastric ulcer. Pancreatology. 2004;4:470–484. doi: 10.1159/000079956. [DOI] [PubMed] [Google Scholar]
- Cyster JG. Lymphoid organ development and cell migration. Immunol Rev. 2003;195:5–14. doi: 10.1034/j.1600-065x.2003.00075.x. [DOI] [PubMed] [Google Scholar]
- Fukuda T, Yoshida T, Okada S, Hatano M, Miki T, Ishibashi K, Okabe S, Koseki H, Hirosawa S, Taniguchi M, Miyasaka N, Tokuhisa T. Disruption of the Bcl6 gene results in an impaired germinal center formation. J Exp Med. 1997;186:439–448. doi: 10.1084/jem.186.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaborit T, Quillien L, Gueguen J. Determination of trypsin inhibitors activity in seeds by a microtitre plate method. In: Poel AVd., editor. International Workshop on Antinutritional Factors (ANFs) in legume seeds. Wageningen, the Netherlands: 1993. [Google Scholar]
- Gainer JH. Lead aggravates viral diseases and represses antiviral activity. Environ Health Perspect. 1974;7:113–119. doi: 10.1289/ehp.747113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Mondal TK, Lawrence DA. Lead effects on development and function of bone marrow-derived dendritic cells promote Th2 immune responses. Toxicol Appl Pharmacol. 2007;222:69–79. doi: 10.1016/j.taap.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidlow DA. Lead toxicity. Occup Med. 2004;54:76–81. doi: 10.1093/occmed/kqh019. [DOI] [PubMed] [Google Scholar]
- Goering PL. Lead-protein interactions as a basis for lead toxicity. Neurotoxicol. 1993;14:45–60. [PubMed] [Google Scholar]
- Groza P, Zamfir V, Lungu D. Postoperative salivary amylase changes in children. Rev Roum Physiol. 1971;8:307–312. [PubMed] [Google Scholar]
- Guo TL, Mudzinski SP, Lawrence DA. Regulation of HLA-DR and invariant chain expression by human peripheral blood mono-nuclear cells with lead, interferon-γ, or interleukin-4. Cell Immunol. 1996;171:1–9. doi: 10.1006/cimm.1996.0166. [DOI] [PubMed] [Google Scholar]
- Hemphil FE, Kaeberle M, Buck WB. Lead suppression of mouse resistance to Salmonella typhimurium. Science. 1971;172:1031–1032. doi: 10.1126/science.172.3987.1031. [DOI] [PubMed] [Google Scholar]
- Heo Y, Parsons PJ, Lawrence DA. Lead differentially modifies cytokine production in vitro and in vivo. Toxicol Appl Pharmacol. 1996;138:149–157. doi: 10.1006/taap.1996.0108. [DOI] [PubMed] [Google Scholar]
- Heo Y, Lee WT, Lawrence DA. In vivo environmental pollutants lead and mercury induce oligoclonal T cell responses skewed toward type-2 reactivities. Cell Immunol. 1997;179:185–195. doi: 10.1006/cimm.1997.1160. [DOI] [PubMed] [Google Scholar]
- Heo Y, Lee WT, Lawrence DA. Differential effects of lead and cAMP on development and activities of Th1- and Th2-lymphocytes. Toxicol Sci. 1998;43:172–185. doi: 10.1006/toxs.1998.2457. [DOI] [PubMed] [Google Scholar]
- Hernberg S, Nikkanen J. Enzyme inhibition by lead under normal urban conditions. Lancet. 1970;1:63–64. doi: 10.1016/s0140-6736(70)91846-5. [DOI] [PubMed] [Google Scholar]
- Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- Hsieh CS, Heimberger AB, Gold JS, O’Garra A, Murphy KM. Differential regulation of T helper phenotype development by interleukins 4 and 10 in an αβ T-cell-receptor transgenic system. Proc Natl Acad Sci. 1992;89:6065–6069. doi: 10.1073/pnas.89.13.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson CA, Cao L, Kasten-Jolly J, Kirkwood JN, Lawrence DA. Susceptibility of lupus-prone NZM mouse strains to lead exacerbation of systemic lupus erythematosus symptoms. J Toxicol Environ Health A. 2003;66:895–918. doi: 10.1080/15287390306456. [DOI] [PubMed] [Google Scholar]
- Inoue S, Browne G, Melino G, Cohen GM. Ordering of caspases in cells undergoing apoptosis by the intrinsic pathway. Cell Death and Differentiation. 2009;16:1053–1061. doi: 10.1038/cdd.2009.29. [DOI] [PubMed] [Google Scholar]
- Ishikawa-Sekigami T, Kaneko Y, Okazawa H, Tomizawa T, Okajo J, Saito Y, Okuzawa C, Sugawara-Yokoo M, Nishiyama U, Ohnishi H, Matozaki T, Nojima Y. SHPS-1 promotes the survival of circulating erythrocytes through inhibition of phagocytosis by splenic macrophages. Blood. 2006;107:341–348. doi: 10.1182/blood-2005-05-1896. [DOI] [PubMed] [Google Scholar]
- Jensen LE, Whitehead AS. Pellino2 activates the mitogen activated protein kinase pathway. FEBS Letters. 2003;545:199–202. doi: 10.1016/s0014-5793(03)00533-7. [DOI] [PubMed] [Google Scholar]
- Kakade ML, Rackis JJ, McGhee JE, Puski G. Determination of trypsin inhibitor activity of soy products: A collaborative analysis of an improved procedure. Cereal Chem. 1974;51:376–382. [Google Scholar]
- Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for the development of Th2 cells. Immunity. 1996a;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- Kaplan MH, Sun YL, Hoey T, Grusby MJ. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature. 1996b;382:174–177. doi: 10.1038/382174a0. [DOI] [PubMed] [Google Scholar]
- Kempe DS, Lang PA, Eisele K, Klarl BA, Wieder T, Huber SM, Duranton C, Lang F. Stimulation of erythrocyte phosphatidylserine exposure by lead ions. Am J Physiol Cell Physiol. 2005;288:C396–C402. doi: 10.1152/ajpcell.00115.2004. [DOI] [PubMed] [Google Scholar]
- King NJC, Thomas SR. Molecules in focus: Indolamine 2,3-dioxygenase. Intl J Biochem Cell Biol. 2007;39:2167–2172. doi: 10.1016/j.biocel.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Kowolenko M, Tracy L, Mudzinski S, Lawrence DA. Effect of lead on macrophage function. J Leukoc Biol. 1988;43:357–364. doi: 10.1002/jlb.43.4.357. [DOI] [PubMed] [Google Scholar]
- Kubisch CH, Logsdon CD. Secretagogues differentially activate endoplasmic reticulum stress responses in pancreatic acinar cells. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1804–G1812. doi: 10.1152/ajpgi.00078.2007. [DOI] [PubMed] [Google Scholar]
- Kuypers FA, DeJong K. The role of phosphatidylserine in recognition and removal of erythrocytes. Cell Molec Biol. 2004;50:147–158. [PubMed] [Google Scholar]
- Lawrence DA. In vivo and in vitro effects of lead on humoral and cell-mediated immunity. Infect Immun. 1981a;31:136–143. doi: 10.1128/iai.31.1.136-143.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence DA. Heavy metal modulation of lymphocyte activities. Toxicol Appl Pharmacol. 1981b;57:439–451. doi: 10.1016/0041-008x(81)90241-6. [DOI] [PubMed] [Google Scholar]
- Lawrence DA. Heavy metal modulation of lymphocyte activities. II Lead, an in vitro mediator of B-cell activation. Int J Immunopharmacol. 1981c;3:153–161. doi: 10.1016/0192-0561(81)90006-0. [DOI] [PubMed] [Google Scholar]
- Lawrence DA. Immunotoxicity of Heavy Metals. In: Dean J, Luster M, Munson AF, Amos H, editors. Immunotoxicology and Immunopharmacology. Raven Press; New York: 1985. pp. 341–353. [Google Scholar]
- Lawrence DA, McCabe MJ. Immune modulation by toxic metals. In: Goyer RA, Klaassen CD, Waalkes MP, editors. Metal Toxicology. Academic Press; San Diego: 1995. pp. 305–337. [Google Scholar]
- LeBlanc PM, Yeretssian G, Rutherford N, Doiron K, Ndiri A, Zhu L, Green DR, Gruenheid S, Saleh M. Caspase-12 modulates NOD signaling and regulates antimicrobial peptide production and mucosal immunity. Cell Host and Microbe. 2008;3:146–157. doi: 10.1016/j.chom.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Lee JE, Naqi SA, Kao E, Dietert RR. Embryonic exposure to lead: comparison of immune and cellular responses in unchallenged and virally stressed chickens. Arch Toxicol. 2002;75:717–724. doi: 10.1007/s00204-001-0283-9. [DOI] [PubMed] [Google Scholar]
- Macatonia SE, Hosken NA, Litton M, Viera P, Hsieh CS, Culpepper JA, Wysocka M, Trinchieri G, Murphy KM, O’Garra A. Dendritic cells produce IL-12 and direct the development of Th1 cells from naïve CD4+ T cells. J Immunol. 1995;154:5071–5079. [PubMed] [Google Scholar]
- Matts RL, Schatz JR, Hurst R, Kagen R. Toxic heavy metal ions activate the heme-regulated eukaryotic initiation factor-2α kinase by inhibiting the capacity of hemin-supplemented reticulocytes to reduce disulfide bonds. J Biol Chem. 1991;266:12695–12702. [PubMed] [Google Scholar]
- Mauel J, Ransijn A, Buchmuller-Rouiller Y. Lead inhibits intracellular killing of Leishmania parasites and extracellular cytolysis of target cells by macrophages exposed to macrophage activating factor. J Leukoc Biol. 1989;45:401–409. doi: 10.1002/jlb.45.5.401. [DOI] [PubMed] [Google Scholar]
- McCabe MJ, Lawrence DA. The heavy metal lead exhibits B-cell Ia expression and differentiation. J Immunol. 1990;145:671–677. [PubMed] [Google Scholar]
- Mebius RE, Kraal G. Structure and function of the spleen. Nature Rev Immunol. 2005;5:606–616. doi: 10.1038/nri1669. [DOI] [PubMed] [Google Scholar]
- Miller TE, Golemboski KA, Ha RS, Bunn T, Sanders FS, Dietert RR. Developmental exposure to lead causes persistent immunotoxicity in Fischer 344 rats. Toxicol Sci. 1998;42:129–135. doi: 10.1006/toxs.1998.2424. [DOI] [PubMed] [Google Scholar]
- Needleman H. Lead poisoning. Ann Rev Med. 2004;55:209–222. doi: 10.1146/annurev.med.55.091902.103653. [DOI] [PubMed] [Google Scholar]
- Nurieva R, Yang XO, Martinez G, Zhang Y, Panopoulos AD, Ma L, Schluns K, Tian Q, Watowich SS, Jetten AM, Dong C. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 2007;448:480–483. doi: 10.1038/nature05969. [DOI] [PubMed] [Google Scholar]
- Page D, Quillien L, Duc G. Trypsin inhibitory activity measurements: simplifications of the standard procedure used for pea seed. Crop Sci. 2000;40:1482–1485. [Google Scholar]
- Prochazkova J, Fric J, Pokorna K, Neuwirth A, Krulova M, Zajicova A, Holan V. Distinct regulatory roles of transforming growth factor-β and interleukin-4 in the development and maintenance of natural and induced CD4+CD25+Foxp3+ regulatory T cells. Immunology. 2009;128:e670–e678. doi: 10.1111/j.1365-2567.2009.03060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawal N, Pangburn MK. Role of the C3b-binding site on C4b-binding protein in regulating classical pathway C5 convertase. Molecular Immunology. 2007;44:1105–1114. doi: 10.1016/j.molimm.2006.07.282. [DOI] [PubMed] [Google Scholar]
- Rawal N, Rajagopalan R, Salvi VP. Stringent regulation of complement lectin pathway C3/C5 convertase by C4b-binding protein (C4BP) Molecular Immunology. 2009;46:2902–2910. doi: 10.1016/j.molimm.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razani-Boroujerdi S, Edwards B, Sopori ML. Lead stimulates lymphocyte proliferation through enhanced T cell-B-cell interaction. J Pharm Exp Therap. 1999;288:714–719. [PubMed] [Google Scholar]
- Rubino GF. The role of lead in porphyrin metabolism. Panminerva Med. 1962;4:340–344. [PubMed] [Google Scholar]
- Saleh M, Mathison JC, Wolinski MK, Bensinger SJ, Fitzgerald P, Droin N, Ulevitch RJ, Green DR, Nicholson DW. Enhanced bacterial clearance and spsis resistance in caspase-12-deficient mice. Nature. 2006;440(20):1064–1068. doi: 10.1038/nature04656. [DOI] [PubMed] [Google Scholar]
- Seder RA, Gazzinelli R, Sher A, Paul WE. Interleukin 12 acts directly on CD4 + T cells to enhance priming for interferon γ production and diminishes interleukin 4 inhibition of such priming. Proc Natl Acad Sci. 1993;90:10188–10192. doi: 10.1073/pnas.90.21.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seder RA, Paul WE, Davis MM, de St Groth BF. The oresence of interleukin 4 during in vitro priming determines the lymphokine-producing potential of CD4+ T cells from T cell receptor transgenic mice. J Exp Med. 1992;176:1091–1098. doi: 10.1084/jem.176.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda K, van Deursent J, Sangster MY, Sarawar SR, Carson RT, Tripp RA, Chu C, Quelle FW, Nosaka T, Vignali DAA, Doherty PC, Grosveld G, Paul WE, Ihle JN. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted State6 gene. Nature. 1996;380:630–633. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- Simons T. Lead-calcium interactions in cellular lead toxicity. Neurotoxicol. 1993;14:77–86. [PubMed] [Google Scholar]
- Snyder JE, Filipov NM, Parsons PJ, Lawrence DA. The efficiency of maternal transfer of lead and its influence on plasma IgE and splenic cellularity of mice. Toxicol Sci. 2000;57:87–94. doi: 10.1093/toxsci/57.1.87. [DOI] [PubMed] [Google Scholar]
- Sparrow R, Healey G. Reduced espression of CD47 on stored red blood cells. Transfusion. 2006;46:1263. doi: 10.1111/j.1537-2995.2006.00883.x. [DOI] [PubMed] [Google Scholar]
- Swain SL, Huston G, Tonkonogy S, Weinberg A. Transforming growth factor-β and IL-4 cause helper T cell precursors to develop into distinct effector helper cells that differ in lymphokine secretion pattern and cell surface phenotype. J Immunol. 1991;147:2991–3000. [PubMed] [Google Scholar]
- Takano S, Kimura T, Yamaguchi H, Kinjo M, Nawata H. Effects of stress on the development of chronic pancreatitis. Pancreas. 1992;7:548–555. doi: 10.1097/00006676-199209000-00007. [DOI] [PubMed] [Google Scholar]
- Thierfelder WE, van Deursen JM, Yamamoto K, Tripp RA, Sarawar SR, Carson RT, Sangster MY, Vignali DAA, Doherty PC, Grosveld GC, Ihle JN. Requirement for Stat4 in interleukin-12-mediated responses of natural killer and T cells. Nature. 1996;382:171–174. doi: 10.1038/382171a0. [DOI] [PubMed] [Google Scholar]
- Tian L, Lawrence DA. Lead inhibits nitric oxide production in vitro by murine splenic macrophages. Toxicol Appl Pharmacol. 1995;132:156–163. doi: 10.1006/taap.1995.1096. [DOI] [PubMed] [Google Scholar]
- Trouw LA, Bengtsson AA, Gelderman KA, Dahlback B, Sturfelt G, Blom AM. C4b-binding protein and factor H compensate for the loss of membrane-bound complement inhibitors to protect apoptotic cells against excessive complement attack. J Biol Chem. 2007;282:28540–28548. doi: 10.1074/jbc.M704354200. [DOI] [PubMed] [Google Scholar]
- Vallee BL, Ulmer DD. Biochemical effects of mercury, cadmium, and lead. Ann Rev Biochem. 1972;41:91–128. doi: 10.1146/annurev.bi.41.070172.000515. [DOI] [PubMed] [Google Scholar]
- Vischer TL, Bertrand L. Stimulating effect of neutral proteases on cells in vitro. Agents and Actions. 1976;6:180–182. doi: 10.1007/BF01972205. [DOI] [PubMed] [Google Scholar]
- Waldo SW, Rosario HS, Penn AH, Schmid-Schonbein GW. Pancreatic digestive enzymes are potent generators of mediators for leukocyte activation and mortality. Shock. 2003;20:138–143. doi: 10.1097/01.shk.0000073866.47824.ae. [DOI] [PubMed] [Google Scholar]
- Wang XC, Strauss KI, Ha QN, Nagula S, Wolpoe ME, Jacobowitz DM. Chymotrypsin gene expression in rat peripheral organs. Cell Tissue Res. 1998;292:345–354. doi: 10.1007/s004410051065. [DOI] [PubMed] [Google Scholar]
- Warner GL, Lawrence DA. Stimulation of murine lymphocyte responses by cations. Cell Immunol. 1986;101:425–439. doi: 10.1016/0008-8749(86)90155-3. [DOI] [PubMed] [Google Scholar]
- Waxman HS, Rabinovitz M. Control of reticulocyte polysome content and hemoglobin synthesis by heme. Biochem Biophys acta. 1966;129:369–379. [Google Scholar]
- Wedeen RP, Malik DK, Batuman V, Bogden JD. Geographic lead nephropathy: Case report. Environ Res. 1978;17:409–415. doi: 10.1016/0013-9351(78)90044-0. [DOI] [PubMed] [Google Scholar]
- Wedeen RP, Malik DK, Batuman V. Detection and treatment of occupational lead nephropathy. Arch Intern Med. 1979;139:53–57. [PubMed] [Google Scholar]
- Woods JS. Hematopoietic System. In: Goyer RA, Klaassen CD, Waalkes MP, editors. Metal Toxicology. Academic Press; San Diego: 1995. pp. 287–304. [Google Scholar]
- Xu J, Lian LJ, Wu C, Wang XF, Fu WY, Xu LH. Lead induces oxidative stress, DNA damage and alteration of p53, Bax and Bcl-2 expression in mice. Food Chem Toxicol. 2008;46:1488–1494. doi: 10.1016/j.fct.2007.12.016. [DOI] [PubMed] [Google Scholar]
- Xu H, Zhang Gng-X, Ciric B, Rostami A. IDO: a double-edged sword for Th1/Th2 regulation. Immunol Lett. 2008;121:1–6. doi: 10.1016/j.imlet.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda T, Imaizumi K, Maeda M, Yui d, Manabe T, Katayama T, Sato N, Gomi F, Morihara T, Mori Y, Miyoshi K, Hitomi J, Ugawa S, Yamada S, Okabe M, Tohyama M. Regulatory mechanisms of TRAF2-mediated signal transduction by Bcl10, a MALT lymphoma-associated protein. J Biol Chem. 2000;275:11114–11120. doi: 10.1074/jbc.275.15.11114. [DOI] [PubMed] [Google Scholar]
- Yu KY, Kwon HJ, Norman DAM, Vig E, Goebl MG, Harrington MA. Cutting Edge: Mouse pellino-2 modulates IL-1 and lipopolysaccharide signaling. J Immunol. 2002;169:4075–4078. doi: 10.4049/jimmunol.169.8.4075. [DOI] [PubMed] [Google Scholar]
- Zhang J, Jacobi AM, Wang T, Berlin R, Volpe BT, Diamond B. Polyreactive autoantibodies in systemic lupus erythematosus have pathogenic potential. J Autoimmun. 2009;33:270–274. doi: 10.1016/j.jaut.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: A listing of genes whose expression is changed by developmental Pb exposure greater then 3.0 fold in the spleen.
Table S2: A listing of the immune system biological process genes whose expression is changed 1.2 fold or greater by developmental Pb exposure.