Abstract
The relationships among the extant five gymnosperm groups—gnetophytes, Pinaceae, non-Pinaceae conifers (cupressophytes), Ginkgo, and cycads—remain equivocal. To clarify this issue, we sequenced the chloroplast genomes (cpDNAs) from two cupressophytes, Cephalotaxus wilsoniana and Taiwania cryptomerioides, and 53 common chloroplast protein-coding genes from another three cupressophytes, Agathis dammara, Nageia nagi, and Sciadopitys verticillata, and a non-Cycadaceae cycad, Bowenia serrulata. Comparative analyses of 11 conifer cpDNAs revealed that Pinaceae and cupressophytes each lost a different copy of inverted repeats (IRs), which contrasts with the view that the same IR has been lost in all conifers. Based on our structural finding, the character of an IR loss no longer conflicts with the “gnepines” hypothesis (gnetophytes sister to Pinaceae). Chloroplast phylogenomic analyses of amino acid sequences recovered incongruent topologies using different tree-building methods; however, we demonstrated that high heterotachous genes (genes that have highly different rates in different lineages) contributed to the long-branch attraction (LBA) artifact, resulting in incongruence of phylogenomic estimates. Additionally, amino acid compositions appear more heterogeneous in high than low heterotachous genes among the five gymnosperm groups. Removal of high heterotachous genes alleviated the LBA artifact and yielded congruent and robust tree topologies in which gnetophytes and Pinaceae formed a sister clade to cupressophytes (the gnepines hypothesis) and Ginkgo clustered with cycads. Adding more cupressophyte taxa could not improve the accuracy of chloroplast phylogenomics for the five gymnosperm groups. In contrast, removal of high heterotachous genes from data sets is simple and can increase confidence in evaluating the phylogeny of gymnosperms.
Keywords: phylogenomics, chloroplast genome, gymnosperms, heterotachy, long-branch attraction
Introduction
The growing importance of genome-scale data in addressing deep phylogenies, “phylogenomics,” is well recognized in the plant phylogenetic community. Some studies maintained that phylogenomic analyses not only settled previously debated phylogenies but also improved resolution of trees (e.g., Gee 2003; Rokas et al. 2003; Delsuc et al. 2005; Rodríguez-Ezpeleta et al. 2005; Dunn et al. 2008). However, this view has recently been challenged by several studies that pointed out inconsistencies with use of different tree-building methods (e.g., Philippe et al. 2005; Jeffroy et al. 2006; Cannarozzi et al. 2007). Jeffroy et al. (2006) claimed that the incongruence of phylogenomic estimates can be a result of systematic errors. Importantly, systematic errors cannot be removed by increasing the data because potential systematic errors also grow with increased size of data sets (Rodríguez-Ezpeleta et al. 2007).
Systematic errors may result from sequence composition biases among lineages and sequence heterotachy. Heterotachy portrays different rates of sites / genes in different lineages (Wu and Susko 2009) and has been found in protein-coding genes of chloroplast genomes (cpDNAs) (Lockhart et al. 2006). In model-based phylogenetics, systematic errors can derive from model misspecification, which may result in the artifact of long-branch attraction (LBA) (Kelchner and Thomas 2007), which groups two unrelated long-branched lineages (Bergsten 2005). The LBA artifact was found to adversely influence the accuracy of tree reconstruction in numerous phylogenomic analyses (e.g., Brinkmann et al. 2005; Bleidorn et al. 2009; Hampl et al. 2009; Zhong et al. 2010).
Strategies for mitigating LBA artifacts include removing LBA lineages (Duvall and Bricker Ervin 2004; Hampl et al. 2009) or deleting fast-evolving sites or genes (Hajibabaei et al. 2006; Goremykin et al. 2009; Hampl et al. 2009; Inagaki et al. 2009). Although removal of long-branch lineages is simple, it is impractical when the lineages of interest have long branches. Therefore, adding more taxa to improve tree accuracy has been proposed (e.g., Hendy and Penny 1989; Bergsten 2005; Hedtke et al. 2006; Heath et al. 2008; Pick et al. 2010; Zhong et al. 2010). Reconstructing trees with amino acid rather than nucleotide sequences was also proposed: Analyses of amino acid sequences can avoid the effect—from biases of codon usage (Inagaki et al. 2004) and reduce substitution saturation (Mathews et al. 2010). Whether these alternative methods can improve chloroplast phylogenomics of gymnosperms needs further examinations.
Gymnosperms, a group of seed-bearing plants with seeds developed on the leaf- or scale-like appendages of cones, include more than 1,000 living species in five major groups. They originated in the Carboniferous period (Renner 2009) and now include cycads (ca. 300 spp.), Ginkgo (1 sp.), Pinaceae (ca. 225 spp.), gnetophytes, and cupressophytes. The gnetophytes comprise about 80 spp. in three monotypic families: Ephedraceae, Gnetaceae, and Welwitschiaceae. The cupressophytes are conifers but exclude Pinaceae and include about 405 spp. in six families sensu lato: Araucariaceae, Cephalotaxaceae, Cupressaceae, Podocarpaceae, Sciadopityaceae, and Taxaceae. Morphologies of the five gymnosperm groups are extremely diversified. In the past two decades, molecular phylogenetists had divergent views on the evolutionary relationships among these five gymnosperm groups, especially the phylogenetic position of gnetophytes. Previously, molecular analyses placed gnetophytes as a sister clade to the rest of seed plants (the “gnetales-sister” hypothesis; e.g., Hamby and Zimmer 1992; Albert et al. 1994), to the conifers (the “gnetifers” hypothesis; e.g., Chaw et al. 1997; Soltis et al. 1999), or to the Pinaceae of conifers (the “gnepines” hypothesis; e.g., Bowe et al. 2000; Chaw et al. 2000). Recently, in analyzing 56 chloroplast protein-coding genes, Zhong et al. (2010) recovered that gnetophytes and cupressophytes formed a clade sister to Pinaceae (the “gnecup” hypothesis), but they also reported that the gnecup topology was resulted from the LBA artifact.
Controversies exist in the use of cpDNA structural mutations in inferring phylogenetic relationships of gnetophytes and conifer families. Restriction mapping analyses suggested a common loss of likely the same inverted repeat (IR) copy in conifer families, which provides strong evidence for the monophyly of all conifers (Raubeson and Jansen 1992). In contrast, gnetophytes and Pinaceae were suggested to share some synapomorphic cpDNA features, such as common losses of all ndh (Braukmann et al. 2009) and rps16 (Wu et al. 2007, 2009) genes, and expansion of IRs to the 3′ region of psbA gene (Wu et al. 2007), which implies that Pinaceae might not be monophyletic with the rest of conifer families and that traditional delimitation of the conifer families might have to be revised.
To date, only one complete cpDNA of cupressophytes, Cryptomeria japonica (Hirao et al. 2008), has been used to represent some 400 species in the six families of cupressophytes in a recent phylogenomic study (Zhong et al. 2010). To increase the spectrum of sample diversity, we have sequenced two additional cpDNAs of cupressophytes, Cephalotaxus wilsoniana (Cephalotaxaceae) and Taiwania cryptomerioides (Cupressaceae), and 53 common chloroplast protein-coding genes of a non-Cycadaceae cycad (Bowenia serrulata) and three other cupressophyte representatives (Araucariaceae: Agathis dammara, Podocarpaceae: Nageia nagi, and Sciadopityaceae: Sciadopitys verticillata). Therefore, our data represent five of the six families of cupressophytes (all except Taxaceae). Significantly, our comparative analyses of cpDNAs revealed that Pinaceae and cupressophytes each lost a different IR copy, which suggests that in the two groups, loss of an IR copy is homoplasious rather than synapomorphic. We also demonstrated that high heterotachous genes contributed to incongruence of phylogenetic estimates. Exclusion of high heterotachous genes from data sets mitigated the LBA artifact and congruently generated a gnepines topology, regardless of the tree-building method used. Our analyses robustly support a sisterhood relationship between Pinaceae and gnetophytes and give an example of how extra caution is needed when using cpDNA structural mutations to address phylogeny.
Materials and Methods
Plant Materials and DNA Extraction
Young leaves of 2-year-old A. dammara, B. serrulata, C. wilsoniana, N. nagi, S. verticillata, and T. cryptomerioides growing in the greenhouse of Academia Sinica were harvested for DNA extraction. Two grams of leaves were ground with liquid nitrogen, and then total DNAs were extracted by use of a 2 × cetyltrimethylammonium bromide protocol (Stewart and Via 1993).
Long-Range Polymerase Chain Reaction, Sequencing, and Assembling
A set of specific polymerase chain reaction (PCR) primers, including primers of Wu et al. (2007) and Lin et al. (2010), was used to amplify 3- to 15-kb specific cpDNA fragments following the protocol of long-range PCR (TaKaRa LA Taq; Takara Bio Inc.). The fragments were designed to overlap each other by 300- to 500-bp regions of their boundaries. At least three independent PCR amplicons of each fragment were mixed and purified. All of the purified amplicons were hydrosheared, cloned, sequenced, and assembled as described (Wu et al. 2007). Except for the cpDNAs of C. wilsoniana and T. cryptomerioides, those of A. dammara, B. serrulata, N. nagi, and S. verticillata were incomplete because several gaps had not been filled, and therefore, only 53 protein-coding genes of the cpDNAs were analyzed.
Gene Annotation
Protein-coding, ribosomal RNA (rRNA), and transfer RNA (tRNA) genes were annotated by use of DOGMA (http://dogma.ccbb.utexas.edu/). tRNA genes were verified by tRNAscan (http://lowelab.ucsc.edu/tRNAscan-SE/).
Dot-Plot Analyses
CpDNA dot-plot comparisons between Cycas taitungensis (NC_009618) and the three cupressophytes, C. wilsoniana, T. cryptomerioides, and Cryptomeria japonica (NC_010548), were conducted by the program Mulan (http://mulan.dcode.org/).
Alignments and Phylogenetic Estimates
DNA sequences of the 53 chloroplast protein-coding genes (table 1) in 27 representative taxa were extracted from National Center for Biotechnology Information, and our new data including five angiosperms (Nicotiana tomentosiformis: NC_007602, Typha latifolia: NC_013823, Drimys granadensis: NC_008456, Nymphaea alba: NC_006050, and Amborella trichopoda: NC_005086), seven Pinaceae (Pinus thunbergii: NC_001631, Cathaya argyrophylla: NC_014589, Picea morrisonicola: AB480556, Pseudotsuga wilsoniana: AB601120, Larix decidua: AB501189, Keteleeria davidiana: NC_011930, and Cedrus deodara: AB480043), three gnetophytes (Ephedra equisetina: NC_011954, Gnetum parvifolium: NC_011942, and Welwitschia mirabilis: AP009568), six cupressophytes (A. dammara: AB65053–AB650588, Cryptomeria japonica: NC_010548, C. wilsoniana: AP012265, N. nagi: AB644440–AB644492, S. verticillata: AB645770–AB645822, and T. cryptomerioides: AP012266), Ginkgo (DQ069338–DQ069698), three cycads (B. serrulata: AB645675–AB645727, Cycas taitungensis: (NC_009618), and Cycas micronesis: EU016802−EU016882), and two ferns (Angiopteris evecta: NC_008829 and Psilotum nudum: NC_003386). We translated the nucleotides to amino acids based on the universal codon table. Amino acids of homologous genes were aligned by the MUSCLE program implemented in Mega 5 (Tamura et al. 2011) with the option of removing gaps and ambiguous sites. These aligned genes were concatenated into a 53-gene data set or into 12 data sets of functional categories according to the classification of Race et al. (1999).
Table 1.
Fifty-Three Protein-Coding Genes for Reconstruction of Phylogenetic Trees
| Photosynthetic Electron Transport and Related Processes |
Gene Expression |
||||||
| Photosystem II (psb)a | Cytochrome b6/f Complex (pet)a | Photosystem (psa)a | ATP Synthase (atp)a | CO2 Fixation | RNA Polymerase (rpo)a | Ribosome (rib)a | Other |
| psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbZ | petA, petB, petD, petG, petL, petN | psaA, psaB, psaC, psaI, psaJ, ycf3b, ycf4b | atpA, atpB, atpE, atpF, atpH, atpI | rbcL | rpoA, rpoB, rpoC1, rpoC2 | rpl2, rpl14, rpl16, rpl20, rpl33, rps2, rps3, rps4, rps8, rps11, rps14, rps19 | ccsA, cemA, clpP, matK |
Abbreviations of functional categories in Figure 2.
Function for assemblage of photosystem I complex (Naver et al. 2001; Ozawa et al. 2009).
Maximum likelihood (ML) trees were constructed by use of RA×ML 7.0.4 (Stamatakis 2006) with a general time reversible (GTR) of amino acids and a rate heterogeneity CAT model allowing autoestimates of amino acid substitution matrix. Maximum parsimony (MP) trees were performed in Mega 5 (Tamura et al. 2011) using the method of Close-Neighbor-Interchange on random trees with ten initial trees (random addition). Clade supports (in percentage) were evaluated with 1,000 bootstrap replicates for both ML and MP methods. Bayesian inference (BI) trees were built by use of the PhyloBayes 3.2 (Lartillot et al. 2009) with a mixture of branch length (MBL) and CAT + Γ (four discrete gamma rates) substitution model. Three independent Markov chain Monte Carlo chains were run with at least 35,000 cycles (∼1,420,000 generations). The first 25% cycles were removed as burn-in. Convergence of three chains was checked on the basis of maximum posterior differences (low heterotachous data set [L-data set] = 0.14; high heterotachous data set [H-data set] = 0.27) following the authors’ suggestion.
Pairwise ML distances of the 20 sampled gymnosperms for each functional category were calculated by RA×ML 7.0.4 (Stamatakis 2006) with Amborella used as the outgroup. For explicit calculation, we incorporated the option of GTR + I (proportion of invariable sites) + Γ (four discrete gamma rates) and allowed the ML parameters to be estimated.
Amino Acid Compositions
We used Mega 5 (Tamura et al. 2011) for computing the mean amino acid compositions for the six major groups of seed plants: angiosperms, Pinaceae, gnetophytes, cupressophytes, Ginkgo, and cycads.
Results
Cephalotaxus Retains a More Primitive CpDNA Organization Than Taiwania and Cryptomeria
The circular cpDNAs of Cephalotaxus (AP012265) and Taiwania (AP012266) are 136,196 and 132,588 bp, with GC contents of 35.1% and 34.6%, respectively (supplementary fig. 1, Supplementary Material online). They have atypical organizations that cannot be divided into four parts: two IRs, a large single-copy (LSC) region, and a small single-copy region. To reveal the evolution of these two atypical cpDNAs, we performed a dot-plot comparison, with the Cycas cpDNA used as the reference because of its ancestral organization (Wu et al. 2007). Supplementary figure 2, Supplementary Material online, shows fewer discontinuous fragments perpendicular to the diagonal lines in Cephalotaxus (7 major lines) than in Taiwania (13 major lines) and Cryptomeria (16 major lines). Therefore, in cpDNA organizations, Cephalotaxus experienced fewer rearrangements than Taiwania and Cryptomeria, which suggests that the cpDNA organization of Cephalotaxus is more primitive than those of Taiwania and Cryptomeria. Moreover, the retained IRB copy and its two adjacent regions of Cephalotaxus are syntenic with those of Cycas, so the cpDNA organization of Cephalotaxus is helpful in tracing the mechanism for the loss of an IR copy from cupressophytes.
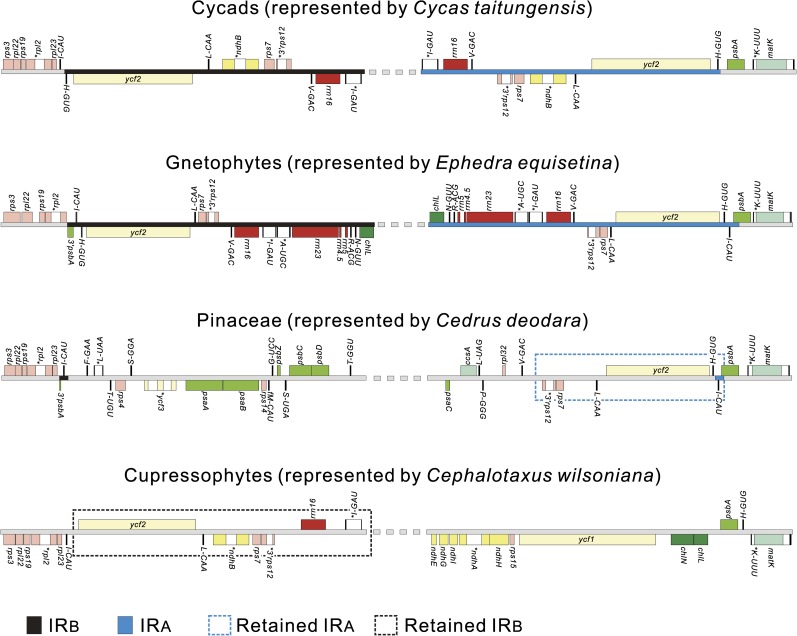
Pinaceae and Cupressophytes Retain Different Residual IR Copies
Figure 1 (for comparisons among more taxa, see supplementary fig. 3, Supplementary Material online) depicts comparisons of detailed LSC-IR junctions among elucidated cpDNA representatives from cycads (represented by Cycas), gnetophytes (represented by Ephedra), Pinaceae (represented by Cedrus), and cupressophytes (represented by Cephalotaxus because its cpDNA organization has fewer rearrangements as mentioned previously). We excluded the cpDNA of Ginkgo because it is not available in the GenBank. In cycads and gnetophytes, the two LSC-IR junctions have conserved gene orders, that is, the retained IRA and IRB are upstream of psbA genes and downstream of the rpl23–rps3 gene cluster (the cpDNAs of gnetophytes have lost the rpl23 gene), respectively. As compared with the two IRs of cycads, those of gnetophytes have expanded further to encompass the trnI-CAU and the 3′ region of psbA genes. However, the major components of IRs (the ycf2 gene and the rRNA operon) and the genes of LSC that flank the two IR regions of gnetophytes are apparently colinear with those of cycads. Therefore, gene orders of the two LSC-IR junctions are syntenic between cycads and gnetophytes, which provide an informative clue to clarify the evolution of IR dynamics in extant gymnosperms.
FIG. 1.
Comparisons of LSC-IR–adjoined regions among representative cpDNAs of four major gymnosperm groups revealed different IR copies retained in cupressophyte and Pinaceae cpDNAs.
From this conserved gene order, we could conclude that in the cpDNAs of Pinaceae and Cephalotaxus, the regions encompassing the whole ycf2 gene and the adjoined psbA or rpl23 gene should be the retained ancestral IRs. Most significantly, the flanking regions of the retained IR copy in the cpDNAs of Pinaceae and Cephalotaxus are colinear with those of the respective IRA and IRB in both cycads and gnetophytes. This observation suggests that the cpDNAs of Pinaceae and cupressophytes retain different IR copies. In other words, the IRB and IRA were lost from the cpDNAs of Pinaceae and cupressophytes, respectively. In summary, conifer evolution exhibited two independent losses of an IR copy. These two losses did not occur in the common ancestor of extant conifers, but rather, after Pinaceae and cupressophytes separated from each other, about 225 Ma (Miller 1999).
Incongruent Chloroplast Phylogenomics and Heterotachy between Gnetophytes and Pinaceae
We extracted 53 common cpDNA protein-coding genes (table 1) from 23 complete cpDNAs of 2 ferns, 5 angiosperms, and 16 gymnosperms, including the two first elucidated here. Furthermore, we determined 53 genes common to three cupressophyte representative families (i.e., Araucariaceae: Agathis, Podocarpaceae: Nageia, and Sciadopityaceae: Sciadopitys) and one cycad (Stangeriaceae: Bowenia) and incorporated them into our data set to increase sampling diversity, specifically the cupressophytes and cycads, and to improve phylogenetic estimates. Supplementary figure 4, Supplementary Material online, shows the ML (GTR + CAT model) and MP trees inferred from the concatenated 53 cpDNA genes (12,241 amino acids) with Angiopteris and Psilotum used as outgroups. Of note, the two trees are incongruent in topology. The GTR + CAT model was used for reconstruction of the ML tree because it could better describe sequence heterogeneity and efficiently replace the computation-intensive GTR + Γ model (Stamatakis 2006). The ML tree resolved gnetophytes to be sisterhood to cupressophytes (the gnecup hypothesis) and Pinaceae to be sister to the gnecup clade, whereas the MP tree resolved gnetophytes to the basal-most seed plants and conifers as a monophyletic clade (the gnetales-sister hypothesis). Nonetheless, ML and MP methods consistently generated the long-branched gnetophytes and the short-branched Pinaceae. This observation suggests extremely different rates in these two lineages and as such is a characteristic signal of heterotachy. In the next two sections, we examine where the heterotachy comes from and whether the pronounced heterotachy between gnetophytes and Pinaceae (HBGP) has a major influence on the incongruent tree topologies between ML and MP methods.
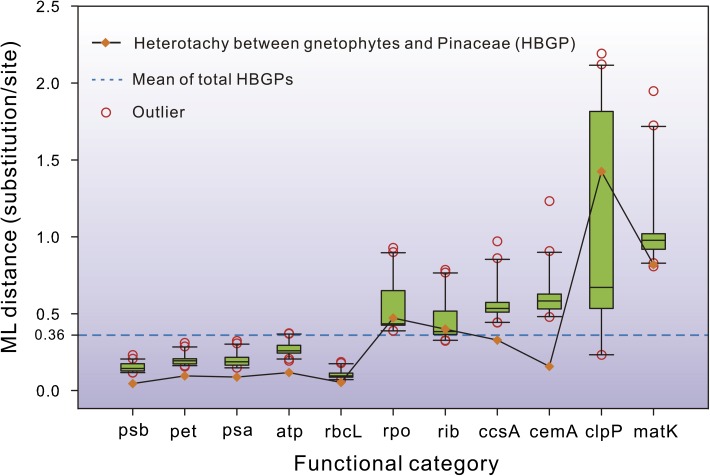
Levels of Heterotachy Are Associated with Gene Functions
To investigate which genes or functional categories contribute to HBGP, we divided the 53 genes into 11 categories (table 1) according to their functions (Race et al. 1999). We calculated the pairwise ML distances (hereafter referred as substitution rate) between each examined gymnosperm and Amborella (the basal-most angiosperm) from each category under a GTR + I + Γ model; the distribution of substitution rates for each category is shown in figure 2. Categories with lower substitution rates have narrow distributions (i.e., low heterotachy), whereas those with higher substitution rates tend to have broad distributions (high heterotachy). Significantly, low heterotachous categories have functions related to photosynthesis, and the high heterotachous categories are associated with gene expression or other functions. Because we are interested in the HBGP, we determined the difference in substitution rates between gnetophytes and Pinaceae (here defined as the mean substitution rate of gnetophytes minus the mean substitution rate of Pinaceae) for each category. Seven of the 11 functional categories (psb, pet, psa, atp, rbcL, ccsA, and cemA) with HBGPs lower than the mean of the total HBGPs (0.36 substitution/site) were concatenated to form the L-data set (7,315 amino acids) and the rest (rpo, rib, clpP, and matK) formed the H-data set (4,926 amino acids) for further analyses.
FIG. 2.
Box plots illustrating the distribution of pairwise ML distances between Amborella and each of the 20 sampled gymnosperm species and differences in heterotachous levels among 11 functional categories of 53 genes. The ML distances were calculated under a GTR + I + Γ model. The HBGP (defined as the mean substitution rate of gnetophytes minus the mean substitution rate of Pinaceae) of each category is indicated. Horizontal lines within boxes denote media.
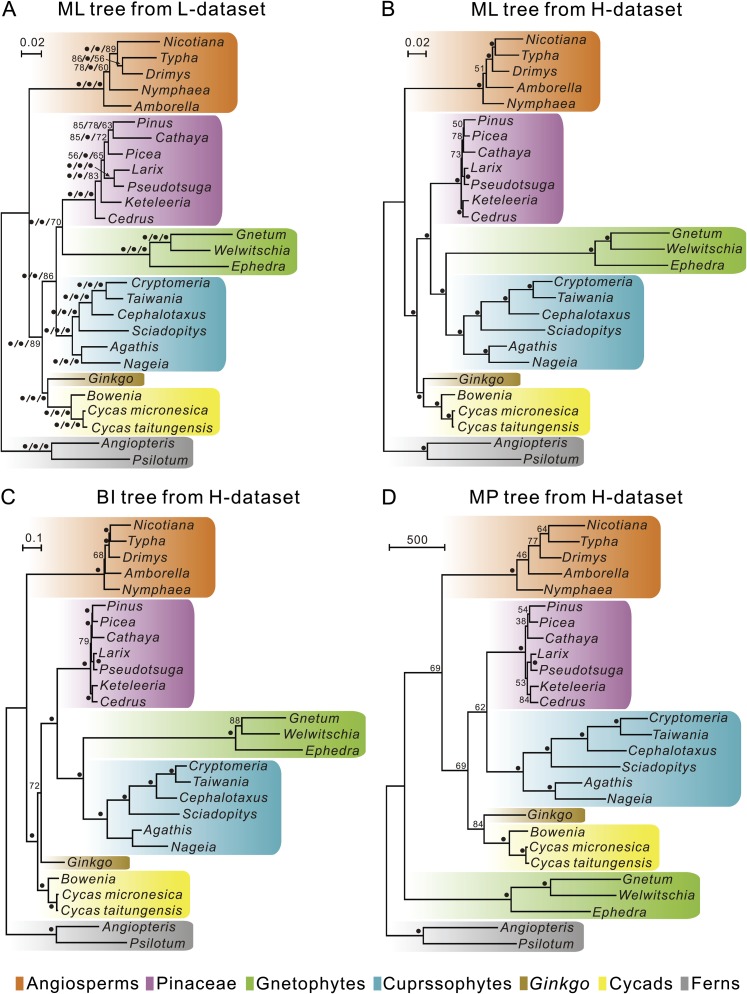
Topological Incongruence Depends on Data Sets Rather Than Tree-building Methods
Figure 3 shows that by using three different methods (ML, BI, and MP), the tree topologies inferred from L- and H-data sets differ in placement of Amborella, Cedrus, gnetophytes, and conifers. In the L-data set, all trees yielded an identical topology, regardless of the method used, and indicated that gnetophytes and Pinaceae formed a sister clade to cupressophytes (the gnepines hypothesis). In contrast, the trees inferred from the H-data set had incongruent topologies with different methods. For example, both ML and BI trees generated the gnecup topology, but the MP tree yielded the gnetales-sister topology. Moreover, the MP tree differs from both ML and BI trees in placement of the Larix–Pseudotsuga clade, and the ML and BI trees have almost identical topologies except for the position of Ginkgo.
FIG. 3.
Trees inferred from the low and high heterotachous data sets (L- and H-data set, respectively). (A) Trees inferred from the L-data set by use of the ML method with a GTR + CAT model, BI with an MBL + CAT + Γ model, and MP, respectively. The MP method generated a single most-parsimonious tree with consistency index (CI) = 0.65 and retention index (RI) = 0.69. Three different methods yielded an identical topology, and only the ML tree is presented. Bootstrap values for ML and MP and posterior probability for BI are arranged in ML/BI/MP. (B–D) show trees based on the H-data set by use of the ML method with a GTR + CAT model, BI with an MBL + CAT + Γ model, and MP, respectively. A single most-parsimonious tree was obtained with CI = 0.69 and RI = 0.73. Supported values estimated from 1,000 bootstrap replicates are shown along branches. Solid circles denote supports greater than 90%. Scales of branch lengths are indicated.
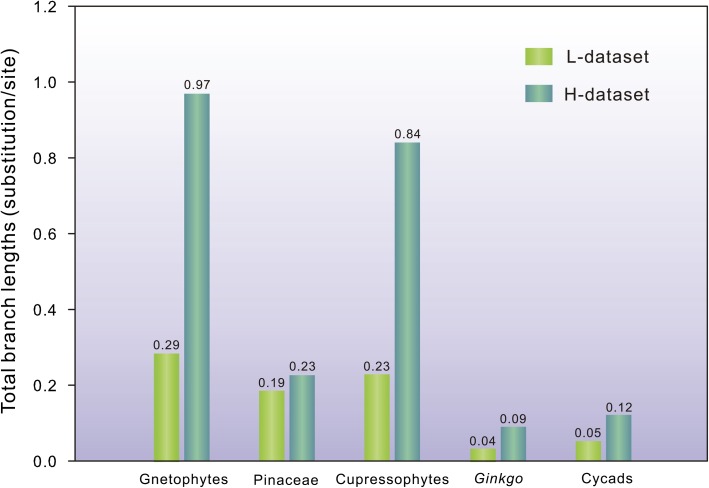
To evaluate the effects of branch lengths, we calculated the total branch length for each monophyletic clade of gymnosperms in the two ML trees. Figure 4 shows that the branch length of Pinaceae differs slightly between the L- and H-data sets (ratio between H- and L-data sets: 0.23/0.19 = 1.2), whereas for other gymnosperms, the branch lengths were longer in the H-data set than in L-data set (ratio 0.97/0.29 = 3.3 for gnetophytes, 0.84/0.23 = 3.7 for cupressophytes, 0.09/0.04 = 2.3 for Ginkgo, and 0.12/0.05 = 2.4 for cycads). Therefore, in the H-data set, the slight increase in branch length observed in the Pinaceae clade is abnormal as compared with those of other gymnosperms. This abnormality apparently elevates the level of heterotachy. Therefore, the H-data set has greater sensitivity than the L-data set to different tree-building methods, and this sensitivity might result from an asymmetric increase in branch lengths between Pinaceae and other gymnosperms.
FIG. 4.
Comparisons of total branch lengths estimated from the L- and H-data sets in each monophyletic group of gymnosperms. In the H-data set, the substitution rate of Pinaceae appears to be slightly elevated. The total branch lengths were calculated from the ML trees shown in figure 3. Numbers above bars denote the values of branch lengths.
Amino Acid Compositions of Phe, Lys, and Arg Are Extremely Biased in Gnetophyte cpDNAs
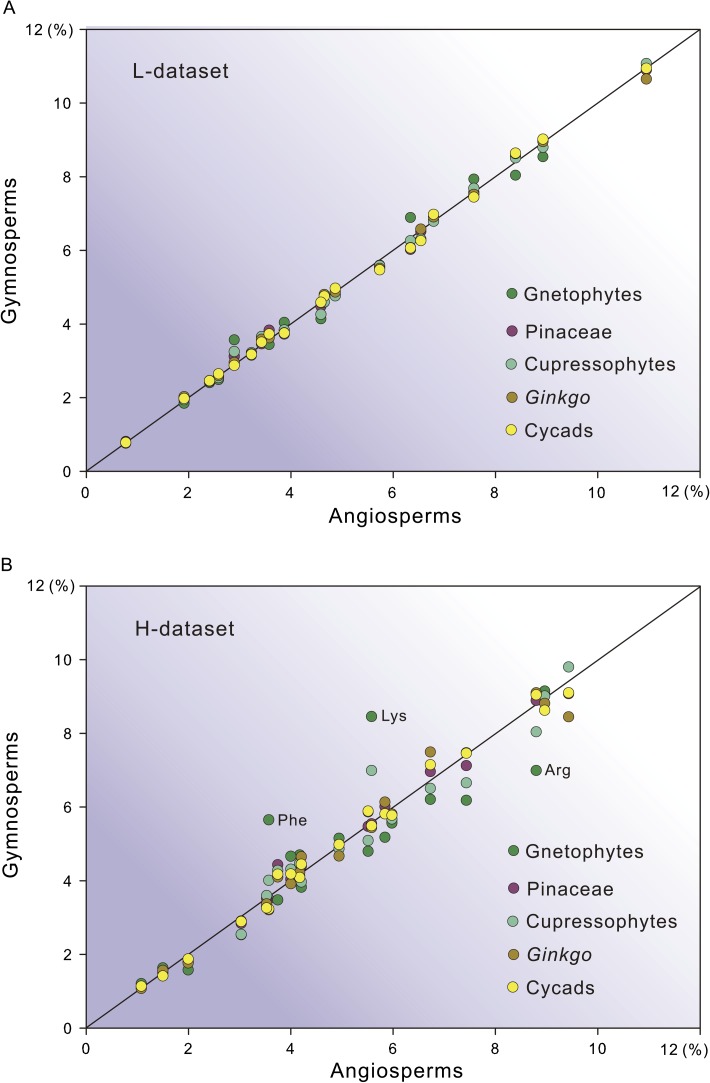
To better understand the sources of incongruence of topology, we compared amino acid compositions among the five gymnosperm lineages, with angiosperms used as the outgroup (fig. 5). This comparison could help determine which lineages have biased amino acid compositions because such compositional biases can cause systematic errors and mislead tree topologies (Philippe et al. 2005; Jeffroy et al. 2006). In the L-data set, most circles distribute along the diagonal line (fig. 5), which suggests that the amino acid compositions of the five gymnosperm groups are consistent with one another. In contrast, in the H-data set, many circles deviate from the diagonal line and are isolated from each other, with the Phe, Lys, and Arg of gnetophytes the most biased. These data indicate that the H-data set contains heterogeneous compositions of amino acids among the five gymnosperm groups, with gnetophytes being the most remarkable.
FIG. 5.
Comparisons of amino acid compositions among the five gymnosperm groups. The amino acid compositions (in percentage) appear less biased in the L-data set (A) than in the H-data set (B). The amino acid compositions of the five sampled angiosperms were used as outgroups. Circles along the diagonal line suggest that specific gymnosperms and angiosperms are similar in the compositions of amino acids, whereas circles deviating from the diagonal line indicate biased compositions between specific gymnosperms and angiosperms. Three species of amino acids with extreme biases are indicated.
Discussion
A Pitfall in Addressing Phylogeny from CpDNA Structural Mutations
Although extremely rearranged cpDNAs have been found in some lineages (e.g., Trifolium: Cai et al. 2008; Geraniaceae: Guisinger et al. 2011), cpDNA structural mutations are considered rare and therefore informative characters to address seed plant phylogeny (Kim et al. 2005; Jansen et al. 2007, 2008; Lee et al. 2007; Wu et al. 2007; Braukmann et al. 2009; Guisinger et al. 2010; Lin et al. 2010). However, caution is needed with phylogenetic inferences based on the cpDNA mutations (Raubeson and Jansen 2005). One of the most difficult questions is determining whether the shared mutations are homoplasious or synapomorphic. For instance, a 40–50 kb cpDNA inversion was used to distinguish Pseudotsuga menziesii from Pinus radiate (Strauss et al. 1988). However, a recent broad sampling of Pinaceae cpDNAs revealed that Pseudotsuga massoniana also possesses the above-mentioned 40- to 50-kb inversion and that the inversion is a homoplasy rather than a synapomorphy shared among Pinaceae genera and species (Wu et al. 2011). Previously, the absence of an IR copy was considered a derived character uniquely shared by all conifers (Raubeson and Jansen 1992), despite the possibility that Pinaceae and cupressophytes might have each lost a different copy of IRs.
In comparing the junctions near LSC regions and residual IR copies among gymnosperms, we discovered that Pinaceae and cupressophytes independently lost different IR copies (fig. 1). This finding suggests that loss of an IR copy is homoplasious between Pinaceae and cupressophytes, which contradicts the view of Raubeson and Jansen (1992, p. 20) that “the same copy has been lost throughout the conifers” and that “a single loss event defining the conifers as a monophyletic group.” In other words, there were two loss events in the conifer evolution. However, loss of different IR copies does not exclude the likelihood that conifers are monophyletic. To this end, this case indicates an apparent pitfall in the evaluation of cpDNA structural mutations for addressing phylogeny and highlights the need for caution in interpreting results when considering mutations of genomic structures.
LBA Artifact in the High Heterotachous Data Set
Zhong et al. (2010) claimed that the gnecup clade was a consequence of LBA artifact, although their ML tree inferred from 56 cpDNA-encoded genes highly supported this topology. Here, we demonstrated that high heterotachous genes contribute to the incongruent estimates of tree topologies (fig. 3). The high heterotachous data set (H-data set) showed coaccelerated substitution rates in both gnetophytes and cupressophytes as estimated by the ML method (fig. 4), which follows the “classic” LBA setting (Felsenstein 1978; Steel 2005), that is, an artificial grouping of two nonadjacent taxa with their substitution rates independently accelerated. As a result, our ML and BI trees inferred from the H-data set apparently generated the gnecup topology (fig. 3B and C). As well, the LBA artifact we observed could not be alleviated by our incorporating the heterotachy model—the MBL model (Zhou et al. 2007; Kolaczkowski and Thornton 2008) (fig. 3C).
On the other hand, the MP tree (fig. 3D) inferred from the H-data set placed the long-branched gnetophytes as the basal-most seed plants, which conforms to the topology of the gnetales-sister hypothesis. The gnetales-sister topology was repeatedly recovered in several MP trees of previous studies (e.g., Sanderson et al. 2000; Rydin et al. 2002; Rai et al. 2003), and those authors also noted that this misleading topology might result from the LBA artifact. We showed that in the H-data set, gnetophytes have biased amino acid compositions (fig. 5B), which greatly elevated the branch lengths of gnetophytes under the MP algorithm. Bergsten (2005) claimed that “outgroup taxa almost always represent long branches and are as such a hazard toward misplacing long-branched ingroup taxa.” Accordingly, the gnetales-sister topology generated by MP trees in this study is reasonably due to the artifact of LBA, which includes both long-branched gnetophytes and outgroups.
Gnepines Hypothesis Seems More Reliable
However, our use of the L-data set revealed that different tree-building methods yielded identical results with the gnepines topology in which cycads and Ginkgo formed a monophyletic clade. This finding implies that the L-data set is insensitive to methods used. The L-data set may contain significant phylogenetic signals for reconstructing a robust topology, which itself might alleviate the influences of inadequate models or methods (Kelchner and Thomas 2007). Of note, the gnepines topology is further reinforced by several structural mutations unique to the cpDNAs of gnetophytes and Pinaceae, such as losses of all ndh genes (Braukmann et al. 2009) and the rps16 gene (Wu et al. 2007), and expansion of IRs to 3′ psbA gene (Wu et al. 2007, 2009). In conclusion, the gnepines clade is strongly substantiated by the congruent phylogenetic estimates of L-data set and the above rare cpDNA structural mutations.
Signals of High Heterotachous Genes Are Dominant in Chloroplast Phylogenomic Estimates
With the growing increase in genomic data, phylogenomics was anticipated to eventually resolve incongruence in molecular phylogenetics (Gee 2003; Rokas et al. 2003). However, to date, a congruent phylogeny of the five groups of extant gymnosperms founded on phylogenomic analyses has not been reached. For instance, trees based on sequences of nucleotides (de la Torre-Bárcena et al. 2009) and amino acids (Cibrián-Jaramillo et al. 2010) from available expressed sequence tags (ESTs) consistently placed gnetophytes as the basal-most gymnosperms. However, the gnepines clade was also revealed from amino acids of available ESTs (Finet et al. 2010). Our ML and MP trees (supplementary fig. 3, Supplementary Material online), with amino acid sequences of 53 cpDNA-encoded genes, generated the gnecup and gnetales-sister trees, respectively, with strong supports (>90%). However, Rokas et al. (2003) argued that high supports do not guarantee a corrected phylogeny. For example, in yeast phylogenomics, highly supported clades were found incorrect because of incongruent topologies generated by different methods (Jeffroy et al. 2006).
In the H-data set, the resulting incongruent topologies suggested specific interpretations of informative signals among different methods. Of note, the H-data set contains both high heterotachous signals and biased amino acid compositions. These two kinds of signals were classified as “nonphylogenetic signals” that contributed to systematic errors (Philippe et al. 2005; Jeffroy et al. 2006). Our analyses showed that with the same tree-building methods, both the 53-gene data set and H-data set generated identical tree topologies. Therefore, in the 53-gene data set, nonphylogenetic signals of the H-data set may be dominant to produce misleading trees, even though the L-data set (7,315 amino acids) is larger than the H-data set (4,926 amino acids). This asymmetric power in phylogenetic estimates is also reflected in the more abundant variable sites in the H-data set (proportion of variable sites = 73.1%) than in the L-data set (proportion of variable sites = 35.0%).
Conclusions
Efforts to understand the seed plant phylogeny have been greatly improved in the beginning of the phylogenomic era. However, concatenation of multiple genes to a huge data set might not always lead to correct phylogenetic estimates. The extremely divergent signals among genes and lineages might be insufficiently described by current methods or models and also cause biases to mislead results. If the biases predominate over true phylogenetic signals, a misleading topology is suggested, with high supports (Phillips et al. 2004). We demonstrated that high heterotachous genes are the major source of incongruent and misleading topologies in the chloroplast phylogenomics of gymnosperms. The elevated heterotachy resulted from the abnormally low and high rates in the Pinaceae and gnetophytes, respectively (fig. 4). Whether these genes of Pinaceae and gnetophytes have undergone any specific selection is worthy of investigation.
In this study, the gnepines hypothesis is robustly supported by congruent phylogenetic estimates from the low heterotachous genes. In addition, the gnepines topology is reinforced by evidence from cpDNA structural mutations. These findings suggest that gnetophytes and Pinaceae form a monophyletic “gnepinophytes” clade separate from the “cupressophytes” clade.
Almost all of the low heterotachous genes function in photosynthesis. Here, we showed the power of concatenated multiple photosynthetic genes in addressing the deep phylogeny of seed plants. However, phylogenetic analyses based on a single or few photosynthetic genes may be problematic because of poor variable sites in these genes. We also demonstrated that adding more cpDNA data of non-Pinaceae conifers (cupressophytes) could not overcome the misleading phylogenetic estimates, which disagrees with the assumption of Zhong et al. (2010). We do not oppose the power of chloroplast phylogenomics, but we stress that determining “adequate” or “inadequate” genes is prerequisite to reduce the controversy over the tree topology. Removal of high heterotachous genes from data sets is a simple but powerful strategy for this purpose.
Supplementary Material
Supplementary figures S1–S4 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by research grants from the National Science Council, Taiwan (NSC972621B001003MY3) and the Biodiversity Research Center, Academia Sinica (to S-.M.C.). We greatly appreciate the two anonymous reviewers’ critical reading and suggestions that helped improve the manuscript.
References
- Albert VA, et al. Functional constraints and rbcL evidence for land plant phylogeny. Ann Mo Bot Gard. 1994;81:534–567. [Google Scholar]
- Bergsten J. A review of long-branch attraction. Cladistics. 2005;21:163–193. doi: 10.1111/j.1096-0031.2005.00059.x. [DOI] [PubMed] [Google Scholar]
- Bleidorn C, et al. On the phylogenetic position of Myzostomida: can 77 genes get it wrong? BMC Evol Biol. 2009;9:150. doi: 10.1186/1471-2148-9-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowe LM, Coat G, dePamphilis CW. Phylogeny of seed plants based on all three genomic compartments: extant gymnosperms are monophyletic and Gnetales’ closest relatives are conifers. Proc Natl Acad Sci U S A. 2000;97:4092–4097. doi: 10.1073/pnas.97.8.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braukmann TW, Kuzmina M, Stefanović S. Loss of all plastid ndh genes in Gnetales and conifers: extent and evolutionary significance for the seed plant phylogeny. Curr Genet. 2009;55:323–337. doi: 10.1007/s00294-009-0249-7. [DOI] [PubMed] [Google Scholar]
- Brinkmann H, et al. An empirical assessment of long-branch attraction artefacts in deep eukaryotic phylogenomics. Syst Biol. 2005;54:743–757. doi: 10.1080/10635150500234609. [DOI] [PubMed] [Google Scholar]
- Cai Z, et al. Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) is associated with numerous repeated sequences and novel DNA insertions. J Mol Evol. 2008;67:696–704. doi: 10.1007/s00239-008-9180-7. [DOI] [PubMed] [Google Scholar]
- Cannarozzi G, Schneider A, Gonnet G. A phylogenomic study of human, dog, and mouse. PLoS Comput Biol. 2007;3:e2. doi: 10.1371/journal.pcbi.0030002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaw SM, Aharkikh A, Sung HM, Lau TC, Li WH. Molecular phylogeny of extant gymnosperms and seed plant evolution: analysis of nuclear 18S rRNA sequences. Mol Biol Evol. 1997;14:56–68. doi: 10.1093/oxfordjournals.molbev.a025702. [DOI] [PubMed] [Google Scholar]
- Chaw SM, Parkinson CL, Cheng Y, Vincent TM, Palmer JD. Seed plant phylogeny inferred from all three plant genomes: monophyly of extant gymnosperms and origin of Gnetales from conifers. Proc Natl Acad Sci U S A. 2000;97:4086–4091. doi: 10.1073/pnas.97.8.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibrián-Jaramillo A, et al. Using phylogenomic patterns and gene ontology to identify proteins of importance in plant evolution. Genome Biol Evol. 2010;2:225–239. doi: 10.1093/gbe/evq012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre-Bárcena JE, et al. The impact of outgroup choice and missing data on major seed plant phylogenetics using genome-wide EST data. PLoS One. 2009;4:e5764. doi: 10.1371/journal.pone.0005764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet. 2005;6:361–375. doi: 10.1038/nrg1603. [DOI] [PubMed] [Google Scholar]
- Dunn CW, et al. Broad phylogenetic sampling improves the resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- Duvall MR, Bricker Ervin A. 18S gene trees are positively misleading for monocot/dicot phylogenetics. Mol Phylogenet Evol. 2004;30:97–106. doi: 10.1016/s1055-7903(03)00187-8. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Cases in which parsimony or compatibility methods will be positively misleading. Syst Zool. 1978;27:401–410. [Google Scholar]
- Finet C, Timme RE, Delwiche CF, Marlétaz F. Multigene phylogeny of the green lineage reveals the origin and diversification of land plants. Curr Biol. 2010;20:2217–2222. doi: 10.1016/j.cub.2010.11.035. [DOI] [PubMed] [Google Scholar]
- Gee H. Evolution: ending incongruence. Nature. 2003;425:782. doi: 10.1038/425782a. [DOI] [PubMed] [Google Scholar]
- Goremykin VV, Viola R, Hellwig FH. Removal of noisy characters from chloroplast genome-scale data suggests revision of phylogenetic placements of Amborella and Ceratophyllum. J Mol Evol. 2009;68:197–204. doi: 10.1007/s00239-009-9206-9. [DOI] [PubMed] [Google Scholar]
- Guisinger MM, Chumley TW, Kuehl JV, Boore JL, Jansen RK. Implications of the plastid genome sequence of Typha (Typhaceae, Poales) for understanding genome evolution in Poaceae. J Mol Evol. 2010;70:149–166. doi: 10.1007/s00239-009-9317-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisinger MM, Kuehl JV, Boore JL, Jansen RK. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol Biol Evol. 2011;28:583–600. doi: 10.1093/molbev/msq229. [DOI] [PubMed] [Google Scholar]
- Hajibabaei M, Xia J, Drouin G. Seed plant phylogeny: gnetophytes are derived conifers and a sister group to Pinaceae. Mol Phylogenet Evol. 2006;40:208–217. doi: 10.1016/j.ympev.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Hamby RK, Zimmer EA. Ribosomal RNA as a phylogenetic tool. In: Soltis PE, Soltis DE, Doyle JJ, editors. Molecular systematics of plants. London: Chapman & Hall; 1992. pp. 50–91. [Google Scholar]
- Hampl V, et al. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups”. Proc Natl Acad Sci U S A. 2009;106:3859–3864. doi: 10.1073/pnas.0807880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath TA, Hedtke SM, Hillis DM. Taxon sampling and the accuracy of phylogenetic analyses. J Syst Evol. 2008;46:239–257. [Google Scholar]
- Hedtke SM, Townsend TM, Hillis DM. Resolution of phylogenetic conflict in large data sets by increased taxon sampling. Syst Biol. 2006;55:522–529. doi: 10.1080/10635150600697358. [DOI] [PubMed] [Google Scholar]
- Hendy MD, Penny D. A framework for the quantitative study of evolutionary trees. Syst Zool. 1989;38:297–309. [Google Scholar]
- Hirao T, Watanabe A, Kurita M, Kondo T, Takata K. Complete nucleotide sequence of the Cryptomeria japonica D. Don. chloroplast genome and comparative chloroplast genomics: diversified genomic structure of coniferous species. BMC Plant Biol. 2008;8:70. doi: 10.1186/1471-2229-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki Y, Nakajima Y, Sato M, Sakaguchi M, Hashimoto T. Gene sampling can bias multi-gene phylogenetic inferences: the relationship between red algae and green plants as a case study. Mol Biol Evol. 2009;26:1171–1178. doi: 10.1093/molbev/msp036. [DOI] [PubMed] [Google Scholar]
- Inagaki Y, Simpson A, Dacks J, Roger A. Phylogenetic artifacts can be caused by leucine, serine, and arginine codon usage heterogeneity: dinoflagellate plastid origins as a case study. Syst Biol. 2004;53:582–593. doi: 10.1080/10635150490468756. [DOI] [PubMed] [Google Scholar]
- Jansen RK, Wojciechowski MF, Sanniyasi E, Lee SB, Daniell H. Complete plastid genome sequence of the chickpea (Cicer arietinum) and the phylogenetic distribution of rps12 and clpP intron losses among legumes (Leguminosae) Mol Phylogenet Evol. 2008;48:1204–1217. doi: 10.1016/j.ympev.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci U S A. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffroy O, Brinkmann H, Delsuc F, Philippe H. Phylogenomics: the beginning of incongruence? Trends Genet. 2006;22:225–231. doi: 10.1016/j.tig.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Kelchner SA, Thomas MA. Model use in phylogenetics: nine key questions. Trends Ecol Evol. 2007;22:87–94. doi: 10.1016/j.tree.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Kim KJ, Choi KS, Jansen RK. Two chloroplast DNA inversions originated simultaneously during early evolution in the sunflower family. Mol Biol Evol. 2005;22:1783–1792. doi: 10.1093/molbev/msi174. [DOI] [PubMed] [Google Scholar]
- Kolaczkowski B, Thornton JW. A mixed branch length model of heterotachy improves phylogenetic accuracy. Mol Biol Evol. 2008;25:1054–1066. doi: 10.1093/molbev/msn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25:2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- Lee HL, Jansen RK, Chumley TW, Kim KJ. Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Mol Biol Evol. 2007;24:1161–1180. doi: 10.1093/molbev/msm036. [DOI] [PubMed] [Google Scholar]
- Lin CP, Huang JP, Wu CS, Hsu CY, Chaw SM. Comparative chloroplast genomics reveals the evolution of Pinaceae genera and subfamilies. Genome Biol Evol. 2010;2:504–517. doi: 10.1093/gbe/evq036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart P, et al. Heterotachy and tree building: a case study with plastids and eubacteria. Mol Biol Evol. 2006;23:40–45. doi: 10.1093/molbev/msj005. [DOI] [PubMed] [Google Scholar]
- Mathews S, Clements MD, Beilstein MA. A duplicate gene rooting of seed plants and the phylogenetic position of flowering plants. Philos Trans R Soc Lond B Biol Sci. 2010;365:383–395. doi: 10.1098/rstb.2009.0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CN. Implications of fossil conifers for the phylogenetic relationships of living families. Bot Rev. 1999;65:239–277. [Google Scholar]
- Naver H, Boudreau E, Rochaix JD. Functional studies of Ycf3: its role in assembly of photosystem I and interactions with some of its subunits. Plant Cell. 2001;13:2731–2745. doi: 10.1105/tpc.010253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa S, et al. Biochemical and structural studies of the large Ycf4-photosystem I assembly complex of the green alga Chlamydomonas reinhardtii. Plant Cell. 2009;21:2424–2442. doi: 10.1105/tpc.108.063313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick KS, et al. Improved phylogenomic taxon sampling noticeably affects nonbilaterian relationships. Mol Biol Evol. 2010;27:1983–1987. doi: 10.1093/molbev/msq089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Delsuc F, Brinkmann H, Lartillot N. Phylogenomics. Annu Rev Ecol Evol Syst. 2005;36:541–562. [Google Scholar]
- Phillips MJ, Delsuc F, Penny D. Genome-scale phylogeny and the detection of systematic biases. Mol Biol Evol. 2004;21:1455–1458. doi: 10.1093/molbev/msh137. [DOI] [PubMed] [Google Scholar]
- Race HL, Herrmann RG, Martin W. Why have organelles retained genomes? Trends Genet. 1999;15:364–370. doi: 10.1016/s0168-9525(99)01766-7. [DOI] [PubMed] [Google Scholar]
- Rai HS, O'Brien HE, Reeves PA, Olmstead RG, Graham SW. Inference of higher-order relationships in the cycads from a large chloroplast data set. Mol Phylogenet Evol. 2003;29:350–359. doi: 10.1016/s1055-7903(03)00131-3. [DOI] [PubMed] [Google Scholar]
- Raubeson LA, Jansen RK. A rare chloroplast DNA structure mutation is shared by all conifers. Biochem Syst Ecol. 1992;20:17–24. [Google Scholar]
- Raubeson LA, Jansen RK. Chloroplast genomes of plants. In: Henry RJ, editor. Plant diversity and evolution: genotypic and phenotypic variation in higher plants. Cambridge (MA): CABI Publishing; 2005. pp. 45–68. [Google Scholar]
- Renner R. Gymnosperms. In: Hedges SB, Kumar S, editors. The timetree of life. New York: Oxford University Press; 2009. pp. 157–160. [Google Scholar]
- Rodríguez-Ezpeleta N, et al. Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr Biol. 2005;15:1325–1330. doi: 10.1016/j.cub.2005.06.040. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Ezpeleta N, et al. Detecting and overcoming systematic errors in genome-scale phylogenies. Syst Biol. 2007;56:389–399. doi: 10.1080/10635150701397643. [DOI] [PubMed] [Google Scholar]
- Rokas A, Williams BL, King N, Carroll SB. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature. 2003;425:798–804. doi: 10.1038/nature02053. [DOI] [PubMed] [Google Scholar]
- Rydin C, Källersjö M, Friis EM. Seed plant relationships and the systematic position of Gnetales based on nuclear and chloroplast DNA: conflicting data, rooting problems, and the monophyly of conifers. Int J Plant Sci. 2002;163:197–214. [Google Scholar]
- Sanderson MJ, Wojciechowski MF, Hu JM, Khan T, Brady SG. Error, bias, and long-branch attraction in data for two chloroplast photosystem genes in seed plants. Mol Biol Evol. 2000;17:782–797. doi: 10.1093/oxfordjournals.molbev.a026357. [DOI] [PubMed] [Google Scholar]
- Soltis PS, et al. The phylogeny of land plants inferred from 18S rDNA sequences: pushing the limits of rDNA signal? Mol Biol Evol. 1999;16:1774–1784. doi: 10.1093/oxfordjournals.molbev.a026089. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood–based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Steel M. Should phylogenetic models be trying to “fit an elephant”? Trends Genet. 2005;21:307–309. doi: 10.1016/j.tig.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Stewart CN, Jr, Via LE. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques. 1993;14:748–751. [PubMed] [Google Scholar]
- Strauss SH, Palmer JD, Howe GT, Doerksen AH. Chloroplast genomes of two conifers lack a large inverted repeat and are extensively rearranged. Proc Natl Acad Sci U S A. 1988;85:3898–3902. doi: 10.1073/pnas.85.11.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. Forthcoming 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Lai YT, Lin CP, Wang YN, Chaw SM. Evolution of reduced and compact chloroplast genomes (cpDNAs) in gnetophytes: selection towards a lower cost strategy. Mol Phylogent Evol. 2009;52:115–124. doi: 10.1016/j.ympev.2008.12.026. [DOI] [PubMed] [Google Scholar]
- Wu CS, Lin CP, Hsu CY, Wang RJ, Chaw SM. Comparative chloroplast genomes of Pinaceae: insights into the mechanism of diversified genomic organizations. Genome Biol Evol. 2011;3:309–319. doi: 10.1093/gbe/evr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Wang YN, Liu SM, Chaw SM. Chloroplast genome (cpDNA) of Cycas taitungensis and 56 cp protein-coding genes of Gnetum parvifolium: insights into cpDNA evolution and phylogeny of extant seed plants. Mol Biol Evol. 2007;24:1366–1379. doi: 10.1093/molbev/msm059. [DOI] [PubMed] [Google Scholar]
- Wu J, Susko E. General heterotachy and distance method adjustments. Mol Biol Evol. 2009;26:2689–2697. doi: 10.1093/molbev/msp184. [DOI] [PubMed] [Google Scholar]
- Zhong B, Yonezawa T, Zhong Y, Hasegawa M. The position of gnetales among seed plants: overcoming pitfalls of chloroplast phylogenomics. Mol Biol Evol. 2010;27:2855–2863. doi: 10.1093/molbev/msq170. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Rodrigue N, Lartillot N, Philippe H. Evaluation of the models handling heterotachy in phylogenetic inference. BMC Evol Biol. 2007;7:206. doi: 10.1186/1471-2148-7-206. [DOI] [PMC free article] [PubMed] [Google Scholar]