Abstract
The hepatitis delta virus (HDV) ribozyme and related RNAs are widely dispersed in nature. This RNA is a small nucleolytic ribozyme that self-cleaves to generate products with a 2’3’-cyclic phosphate and a free 5’-hydroxyl. Although small ribozymes are dependent on divalent metal ions under biologically-relevant buffer conditions, they function in the absence of divalent metal ions at high ionic strength. This characteristic suggests that a functional group within the covalent structure of small ribozymes is facilitating catalysis. Structural and mechanistic analyses have demonstrated that the HDV ribozyme active site contains a cytosine with a perturbed pKa which serves as a general acid to protonate the leaving group. The reaction of the HDV ribozyme in monovalent cations alone never approaches the velocity of the Mg2+-dependent reaction and there is significant biochemical evidence that a Mg2+ ion participates directly in catalysis. A recent crystal structure of the HDV ribozyme revealed that there is a metal binding pocket in the HDV ribozyme active site. Modeling of the cleavage site into the structure suggested that this metal ion can interact directly with the scissile phosphate and the nucleophile. In this manner, the Mg2+ ion can serve as a Lewis acid, facilitating deprotonation of the nucleophile and stabilizing the conformation of the cleavage site for in-line attack of the nucleophile at the scissile phosphate. This catalytic strategy had previously only been observed in much larger ribozymes. Thus, in contrast to most large and small ribozymes, the HDV ribozyme uses two distinct catalytic strategies in its cleavage reaction.
Ribozymes, like their protein enzyme counterparts, achieve biological catalysis by folding into stable, compact tertiary structures (1-3). Ribozymes function in many fundamental biological processes, including mRNA splicing, protein translation, gene regulation, and critical RNA processing events. RNA possesses a negatively charged backbone and thus folding of ribozymes is often dependent on the presence of divalent cations, typically Mg2+. In addition to playing structural roles, Mg2+ ions can be directly involved catalysis (2, 4).
While proteins are composed of a diverse group of amino acids, strands of RNA are synthesized from only four similar nucleosides: adenosine, guanosine, cytidine and uridine. The naturally occurring nucleobases are remarkably stable. They have pKa's that are outside biologically relevant buffer conditions and they do not readily tautomerize. RNA chains lack sequence diversity and therefore they are prone to misfolding into non-native conformations. RNAs also lack the extended hydrophobic core of proteins that is so effective at shifting the pKa's of buried charged amino acids (5, 6).
To counteract the fundamental inertness of RNA, ribozymes rely on a few common mechanistic strategies: metal-mediated catalysis and general acid/general base catalysis (2, 3, 7, 8). The phosphate groups in the RNA backbone are capable of chelating divalent metal ions and positioning them accurately to participate in catalysis. Metal-mediated catalysis is observed in a wide variety of naturally-occurring and artificially-evolved ribozymes (2, 9-11). Surprisingly, RNA molecules are also known to use general acid/general base catalysis without the participation of divalent cations as a catalytic strategy (1, 3, 8). In many small ribozymes, nucleobases, including adenine, guanine, and cytosine, and small molecule co-factors are observed in position to participate in proton transfer reactions. These functional groups have pKa's far from neutrality when free in solution. To make most efficient use of these general acids/general bases at biologicaly relevant pH, the pKa's of the groups would have to shift >1 pH unit toward neutrality.
The hepatitis delta virus (HDV1) ribozyme (Figure 1) was one of the first two ribozymes whose crystal structure was solved (12). Yet, key questions still remain about its structure and catalytic mechanism. Although there was significant evidence that a Mg2+ ion lurked in the active site of this ribozyme, the location and the role of this ion remained undefined (13-15). Was a major conformational switch required over the course of the catalytic reaction as previously suggested (16-18)? Here, I will present a recent model for the cleavage reaction of the HDV ribozyme which invokes both of the two key mechanisms by which ribozymes catalyze reactions, metal-mediated catalysis and general acid/base catalysis.
Figure 1. The structure of the HDV ribozyme.
A. Secondary structure of the HDV ribozyme. The HDV ribozyme folds into a pseudoknot containing five core paired regions (P1-P4, P1.1). In the trans-acting version of the ribozyme, a short substrate, U(-1) to A8 here, base pairs to the ribozyme to form the P1 helix. The cleavage site is indicated by an asterisk. The junctions J1.1/3 and J4/2 contribute key nucleobases to the active site of the ribozyme. B. Tertiary structure of the HDV ribozyme (3NKB). The active site is located at the junction of helices P1, P1.1 and P3. Key residues are labeled. A largely hydrated active site Mg2+ ion is shown as spheres. Figure adapted from (33).
Small self-cleaving ribozymes and large ribozymes exhibit fundamental differences
Ribozymes can be divided into two families. Large ribozymes, including group I introns, group II introns and RNase P, are typically > 200 nucleotides in length and have a multidomain structure. Their active site pockets are created by the junctions of multiple double helices and multiple ‘single-stranded’ regions of RNA (2, 19-23). These complex active sites bring together clusters of 2-3 negatively charged phosphate groups from the ribozyme core that serve to chelate and orient catalytic metal ions. The large ribozymes are critically dependent on Mg2+ (or sometimes Mn2+) ion for catalytic activity and are thus obligatory metalloenzymes. Large ribozymes activate water or a distant exogenous or endogenous nucleotide for nucleophilic attack at a scissile phosphate to generate products with a 5’-phosphate and a free 3’ hydroxyl. The active sites of these molecules are large enough to bind and position small substrates, such as guanosine or water, and can bind and position helical substrates, such as tRNA, in trans and position them for cleavage.
Small nucleolytic ribozymes, including the hepatitis delta virus (HDV), hammerhead, hairpin Varkud satellite (VS) and glucosamine-6-phosphate-responsive (glmS) ribozymes, catalyze related RNA self-cleavage reactions (3, 24). Small ribozymes perform a reaction that is distinct from that performed by the large ribozymes. The 2’-OH of the upstream nucleotide is activated for nucleophilic attack at the scissile phosphate to cleave their RNA substrate and generate 2’,3’-cyclic phosphate and 5’-OH termini on the cleavage products (1) (Figure 2). All have evolved as self-cleaving, cis-acting elements and all can be engineered to base pair and, subsequently cleave, a single stranded RNA substrate in trans. The active sites of small ribozymes are usually formed by the juxtaposition of two base-paired helices, and typically lack the phosphate clusters observed in large ribozymes. All can react in the absence of Mg2+, albeit slower, (13, 25) if sufficiently large concentrations of monovalent cation are present to allow the RNA to fold. It was therefore anticipated that these ribozymes would function by Mg2+-free mechanisms. The ability to function in the absence of Mg2+ ion suggests that functional groups within the covalent structure of RNA itself accelerates the chemical reaction. Indeed, all of these ribozymes appear to use general acid/general base mechanisms to cleave their RNA substrates.
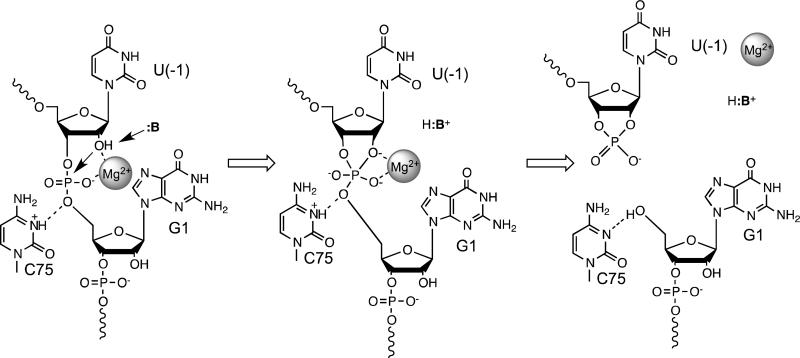
Figure 2. The catalytic mechanism of the HDV ribozyme.
In the substrate bound state (left), C75 is protonated and donates a hydrogen bond to the 5’-O of G1. The active site metal ion is directly coordinated to the pro-RP oxygen of G1 and the 2’-hydroxyl of U(-1). Metal binding to the nucleophile facilitates deprotonation by a yet unknown base (:B). C75+ and the active site Mg2+ ion have the potential to stabilize a phosphorane intermediate (middle) generated by attack of the 2’-O on the scissile phosphate. This intermediate is resolved to form the 2’,3’-cyclic phosphate on U(-1) and a free 5’-hydroxyl on G1 (right).
The hepatitis delta virus ribozyme
The HDV ribozyme (~85 nt) was originally identified in the both genomic and antigenomic strands of the HDV RNA (1.7 kb) (26). This ribozyme plays an integral part in viral replication. Both genomic and antigenomic RNAs are synthesized by a rolling circle mechanism from a circular RNA template, and both contain closely related ribozymes. The RNA produced in this manner contains tandem repeats of the genome. The genomic and antigenomic ribozymes self cleave to resolve the long concatemers (27) into single, unit-sized pieces that can then be ligated to create circular single-stranded RNA. While this RNA evolved as a self-cleaving, cis-acting ribozyme, it can be engineered to cleave an RNA substrate in trans as shown in Figure 1. Folding of such trans-acting ribozymes requires the presence of the substrate RNA. Thus while we make the distinction between the ‘active-site’ of the ribozyme and the ‘cleavage-site’ of the substrate for convenience, the structure of each these elements is critically dependent on the other.
For many years, the HDV ribozyme was believed to be an orphan, found only in a rare human virus. Recently, however, HDV-like ribozymes have been discovered to be wide-spread (28-30). These catalytic RNAs and their cleavage sites appear to have defined the 5’-end of a mobile genetic element that was broadcast throughout nature (29, 30). What might the HDV-like ribozymes be doing in modern biology? Outside of the ribozymes found within the HDV, it is not yet clear what celluar functions these ribozymes are playing. The catalytic activity of ribozymes from Anopheles gambiae are developmentally regulated, suggesting that these molecules may play roles in gene expression (29).
Structural biology of the HDV ribozyme
The three-dimensional structure of the HDV ribozyme has been extensively studied by X-ray crystallography (Figure 1). In 1998, Ferré-D’Amaré et al. determined the structure of the ribozyme postcleavage (12). This crystal structure revealed the overall fold of the self-cleaving ribozyme and defined the nucleotides that were well positioned to participate in catalysis. The N3 of C75 was observed to serve as a hydrogen bond acceptor for the 5’-hydroxyl of G1, the leaving group in the cleavage reaction (12) (Figure 2). Thus, this structure suggests that C75 participates in the cleavage reaction. Mutation of C75 to a U results in a catalytically inactive ribozyme that can be rescued by imidazole, further implicating this nucleotide as a catalytic nucleobase (31). There are only a few interactions between the ribozyme active site and the nucleotides upstream of the cleavage site. Therefore, dissociation of the upstream fragment containing the 2’,3’-cyclic phosphate is rapid relative to the reverse, ligation, reaction (32). The upstream cleavage product, including the scissile phosphate, is therefore not visible in this crystal structure. To fully understand how the ribozyme activates the 2’-hydroxyl nucleophile, another crystal structure was needed.
Recently, we determined the crystal structure of the HDV ribozyme trapped in a pre-cleavage state (33) (Figure 1). Ribozyme cleavage was inhibited by replacing the 2’-hydroxyl of U(-1) with a hydrogen. The electron density for the RNA at the cleavage site, including the scissile phosphate and the upstream nucleotide U(-1), is poor in this crystal form. Fortunately, the crystal structure provides sufficient constraints to generate a model for the cleavage site. There are steric constraints: the ribozyme and the cleavage site dinucleotide cannot occupy the same space. There are basepairing constraints: downstream nucleotide, G1, is basepaired to U37 and G1 is resolved in the electron density map. There are chemical constraints: the 2’-hydroxyl of U(-1) must be positioned for in-line attack at the scissile phosphate. There are crystal structures available for several small ribozymes and these provide models for cleavage site dinucleotides in conformations consistent with in-line attack (27, 34-37).
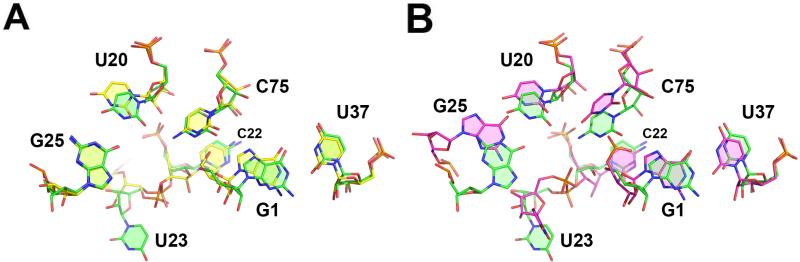
To create a picture of the RNA substrate precleavage, we superposed the downstream nucleotide of a ribozyme's cleavage site with G1 from the HDV ribozyme. The cleavage site from the hammerhead ribozyme (27) provided the best fit to the HDV ribozyme active site (Figure 3). The conformation of the dinucleotide in the HDV ribozyme is unchanged from that observed in the hammerhead ribozyme, except for rotations about the glycosidic bonds to maximize stacking interactions in the HDV ribozyme active site. Inspection of the electron density in this region of the molecule indicates that the crystallographic data are consistent with this model, even if they are insufficient to uniquely define it. The conformation of the cleavage site dinucleotide is satisfying in that there is complementarity between the ribozyme active site and the cleavage site. Key atoms surrounding the scissile phosphate are held in place by two hydrogen bonds or metal ion interactions within the active site and there is extensive shape complementarity between the ribozyme active site and the cleavage site (Figure 3). The resulting model is consistent with the wealth of biochemical data available for the HDV ribozyme (described below).
Figure 3. The cleavage site of the hammerhead ribozyme docked into the active site of the HDV ribozyme.
The HDV ribozyme active site is shown in green and the cleavage site is shown in pink. A. Key atoms in the cleavage site, including the 2’-hydroxyl of U(-1), the pro-R P oxygen of G1, and the 5’-hydroxyl of G1, are held in place by at least two hydrogen-bonds, or metal mediated interactions. G25 is positioned directly under G27, and it is not labeled. B. There is extensive shape complementarity between the cleavage site and the active site. Molecular surfaces for the cleavage site (pink) and the active site (green) were generated independently. The active site metal ion and its hydration shell are shown as spheres.
Comparison of the conformation of the HDV ribozyme in the precleavage and postcleavage states reveals very little change in the overall structure (Figure 4A). There are minor differences at nucleotides U20 and U23. However a reexamination of the diffraction data from the postcleavage, product state suggests that the structure of the active site of the precleavage ribozyme is compatible with the crystallographic data of the cleaved ribozyme determined by Ferré-D’Amaré et al (12). The differences between the two models are likely to be the result of ambiguous electron density in the structure of the ribozyme product. Thus, no major conformational switching is necessary to get from the precleavage state to the product form of the ribozyme as had been previously suggested (16-18).
Figure 4. The active site of the product-bound ribozyme is similar to the active site of the inhibitor-bound ribozyme.
A. The active site of the product bound ribozyme (yellow), 1CXO, was superposed on the active site of the inhibitor-bound ribozyme (green), 3NKB. Only minor differences were observed between the two structures. The structure of the inhibited ribozyme is consistent with the electron density of the product-bound ribozyme. Thus the observed differences are likely due to the weak electron density in this region of the 1CXO structure. B. The active site of the C75U mutant (red), 1SJ3, is significantly different than the active site of the inhibitor-bound ribozyme (green), 3NKB. Mutation of C75 to a U disrupts a critical hydrogen bond network that organizes the active site. The active site of the C75U mutant is therefore significantly less compact. Figure adapted from (33).
C75 does not only participate in the cleavage reaction, but it also contributes to the architecture of the active site pocket. This is demonstrated by the structure of the catalytically inactive mutant, C75U (17). Uracil differs from protonated cytosine in that the exocyclic amine of C is replaced by a keto group, and N3, while protonated in both nucleobases, is positively charged only in cytosine. The exocyclic amine of C75 makes key interactions with the ribozyme active site, including hydrogen bonds to the pro-RP oxygens at the C22 and scissile phosphates. Furthermore, protonation of cytosine results in a significant positive dipole on the exocyclic amine (38, 39). When this functional group is replaced with a keto group, the active site misfolds. Comparison of the C75U mutant to the WT product ribozyme reveals that the mutant active site is significantly disrupted and is more open, consistent with a broken network of hydrogen bonds in the C75U mutant (Figure 4B). This result emphasizes that the functional groups on C75 also contribute to the stability of the active site structure. Although the nucleotides upstream of the substrate are visible in the C75U crystal structure (17), disruption of the active site likely prevented the substrate from binding in a functional conformation.
A cytosine with a perturbed pKa serves as a general acid
The 5’-hydroxyl leaving group donates a hydrogen bond to the nucleobase of C75 in the product state (12). Postcleavage, the 5’-O is protonated and can serve as a hydrogen bond donor. Precleavage, however, there is no proton on the 5’-O and this group can only serve as a hydrogen bond acceptor. If this hydrogen bond is present in the pre-cleaved state, then C75 must serve as the proton donor. Thus, the crystal structure of the cleaved ribozyme suggested that prior to cleavage C75 would be protonated at the N3 position and positively charged (Figure 5). It would transfer this proton to the 5’-hydroxyl leaving group during catalysis to help stabilize the leaving group. Thus, a nucleobase would serve as a general acid in the RNA cleavage reaction. This is reminiscent of the protein ribonuclease RNase A. However unlike the histidine side chains in the RNase A active site, cytosine typically has a pKa of ~ 4.
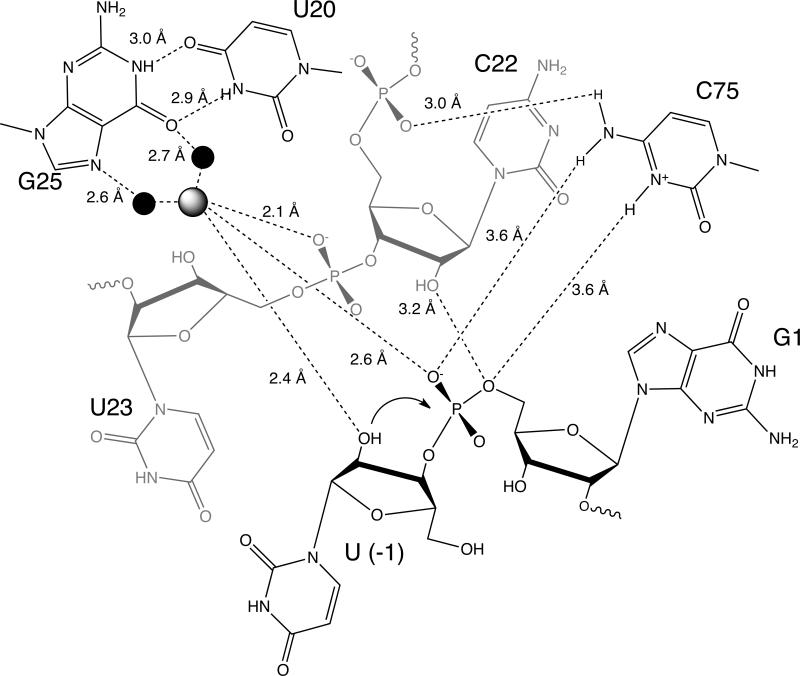
Figure 5. Hydrogen bonds and metal mediated interactions in the HDV ribozyme active site.
The cleavage site dinucleotide from the hammerhead ribozyme was docked into the active site of the HDV ribozyme as previously described. Distances for hydrogen bonds and metal mediated interactions in the active site are shown. Figure adapted from (33).
Although protonated cytosine would be an excellent proton donor, only a very small population of cytosine is protonated at neutral pH under biologically relevant conditions. In Mg2+ free conditions, the pH-rate profile of the HDV ribozyme reveals an apparent pKa near neutrality (13, 31). To directly measure the pKa of C75 and to determine if the apparent pKa observed in solution arose from a cytosine with a shifted pKa, we used Raman spectroscopy of single RNA crystals to probe the protonation state of C75 (40). A spectral feature corresponding to neutral cytosine can be observed in the Raman spectrum of HDV ribozyme crystals. As the pH of the surrounding buffer is raised, the intensity of that feature increases. By plotting the area of that peak as a function of pH, a cytosine with a pKa of ~6 is observed. When C75 is mutated to a uridine, this feature is lost. Thus, the active site of the HDV ribozyme appears to shift the pKa of C75 by > 2 pH units.
Mutagenesis experiments confirm a critical role for C75 in the cleavage reaction. The C75U mutation is completely inactive but it can be rescued in the presence of imidazole, in the reaction buffer. Adenosine is bulkier than cytosine but, like cytosine, can be protonated and has a pKa ~ 3.5. The C75A mutation of the HDV ribozyme is less active than the WT ribozyme, but it is functional and has a pKa shifted by the same amount as C75 (13, 41). These data suggest that C75 is involved in an essential proton transfer reaction.
Mutagenesis and pH-rate profiles alone cannot be used to determine if a functional group is serving as a general acid or a general base. To address the role of C75 in the cleavage reaction, Das and Piccirilli synthesized a modified substrate in which the 5’-O was substituted with a sulfur atom, which serves as a hyperactivated leaving group (42). This modified substrate was readily cleaved by the HDV ribozyme containing the C75U mutation. When the 5’-hydroxyl at the cleavage site is replaced with an activated leaving group, protonation of the leaving group is no longer essential for efficient cleavage, and the proton donor is not required for catalysis. These experiments strongly suggest that C75 serves as a general acid, donating a proton to the 5’-hydroxyl leaving group during the cleavage reaction.
How does a ribozyme active site accomplish pKa shifting? Insight into this issue has been provided by electrostatics calculations (39, 43). When the N3 of C75 is in position to hydrogen bond with the 5’-O of G1, it is juxtaposed with the negatively charged scissile phosphate. Protonation of cytosine results in significant positive charge on both the N3 and the exocyclic N4 nitrogens. The pro-RP oxygen of the scissile phosphate makes a hydrogen bond to the N4 of C75. The significant interactions between the nucleobase and the negatively charged scissile phosphate serve to stabilize the positive charge on the protonated cytosine base and thereby shift the pKa. Indeed, the pKa of C75 is not shifted in the postcleavage ribozyme, which lacks the scissile phosphate (44).
The active sites of the hairpin and VS ribozymes contain adenosine residues with shifted pKa's (24, 45, 46). There are two crystal structures of the hairpin ribozymes bound to vanadate-containing transition state mimics (47, 48). In both of these models, hydrogen bonds between the exocyclic amine of adenosine and the non-bridging vanadate oxygens are observed in addition to hydrogen bonds from the N1 of adenosine to the 5’-hydroxyl leaving groups. Likewise, the glmS ribozyme shifts the pKa of an amine group from its glucosamine-6-phosphate cofactor which then serves as a general acid in the reaction (49, 50). The theme emerges that the general acids used by the nucleolytic ribozymes are positively charged, as opposed to neutral, when protonated. The interaction of the general acid with the 5’-hydroxyl leaving group results in a juxtaposition of the general acid and the negatively-charged scissile phosphate. Proximity of the general acid and the anionic phosphate stabilizes the positive charge of the protonated general acid, and this serves to shift the pKa towards neutrality.
There is a metal binding pocket in the HDV ribozyme active site
The recent crystal structure of the HDV ribozyme bound to an inhibitor RNA revealed a Mg2+ ion in the active site in proximity to the 2’-hydroxyl of U(-1) (33). Ligands to this Mg2+ ion are the pro-SP oxygen of C22, the pro-RP oxygen of the scissile phosphate, the 2’-hydroxyl of (U-1), and 3 water molecules (Figure 5). In contrast to the catalytic Mg2+ ions observed in the large ribozymes where 3-4 phosphate oxygen atoms typically coordinate catalytic Mg2+ ions (2), there is only a single phosphate ligand from the HDV ribozyme core and a total of two phosphate oxygen ligands to this Mg2+ ion. The HDV ribozyme therefore uses an alternate strategy, a rare base pair with metal binding properties, to create an active site metal-binding pocket.
Divalent metal ions are often observed to interact with canonical G•U wobble base pairs through the ion's hydration shells (51, 52) (Figure 6). In contrast to Watson-Crick base pairs, canonical G•U wobble pairs provide a concentration of negative dipoles in the major groove that attracts cations. The major groove of A-form RNA, however, is deep and the metal ions bound there are inaccessible for the long-range tertiary structure formation needed to build ribozyme active sites. When a reverse G•U wobble is formed with a syn G base, an electron-rich surface patch is also formed, but it is found on the more accessible minor groove face of the helix (Figure 6) (39). Thus, reverse wobbles represent a strategy to create minor-groove metal binding motifs, which could be used to facilitate metal-mediated tertiary contacts or the binding of catalytic metal ions.
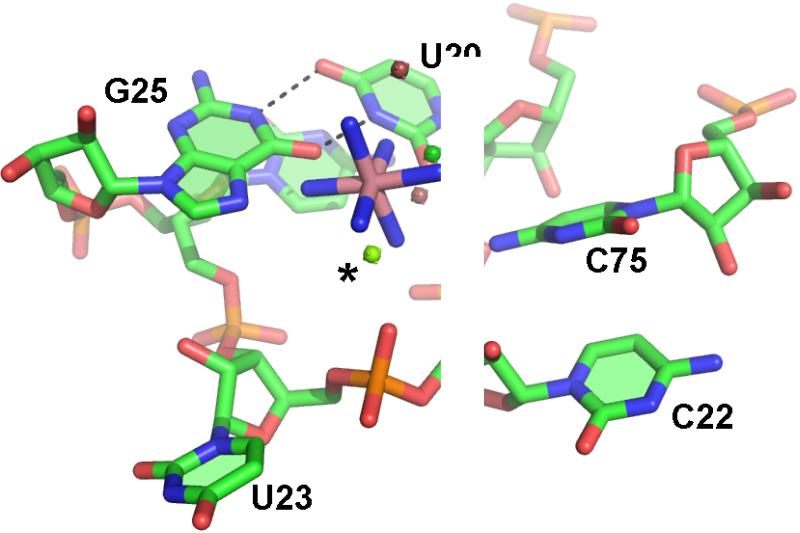
Figure 6. Comparison of metal binding by a standard G•U and a reverse G•U wobble.
Standard G•U wobble base pairs (left) are often associated with major groove Mg(H2O)62+ binding sites (PDBID 1HR2). G25 and U20 base pair in the reverse G•U wobble geometry (right). This helps to create a binding site for a largely hydrated Mg2+ ion in the minor groove. The minor groove is on the left in both panels.
In the HDV ribozyme, G25 and U20 form this rare, reverse G•U wobble base pair (Figure 5). These nucleotides are universally conserved in all of the known HDV-like ribozymes (although the significance of this is unknown as the identity of these nucleotides was used in the screening process) (28, 29). G25 is a second shell ligand to the catalytic metal ion, interacting with this ion through two water molecules. The reverse G•U wobble, the scissile phosphate, and the phosphate at position U23 create a highly negatively charged binding pocket in the active site. This negatively charged pocket not only serves to bind the catalytic Mg2+ ion, but also interacts with C75 through its exocyclic amine (33, 39), a feature that may contribute to the anti-cooperative binding of the proton on C75 and the catalytic Mg2+.
A related metal binding site was previously observed in the crystal structure of the C75U variant ribozyme although the metal binding pocket is significantly distorted due to the mutation (17). Ligands to this metal ion include keto oxygens from U20(O2) and U75(O4), phosphate oxygens from C22, U23 and U(-1), and the 5’-oxygen leaving group from G1. It is important to note that the O4 of U75 is not, however, present in the wild-type ribozyme, as position 75 is an invariant C nucleotide. In addition the 5’-oxygen of G1 is hydrogen bound to C75 in the WT ribozyme and therefore unavailable to bind the active site metal. All of the distances between the metal ion and ligands from the C75U ribozyme are 2.7 Å or longer, consistent with water-mediated interactions. Divalent cations (Mg2+, Mn2+, Sr2+, Ba2+), trivalent cations, Co(NH3) 3+6, and monovalent cation (Tl+) are all observed to bind in similar positions (Figure 7) (17, 53).
Figure 7. The active site metal binding pocket can accommodate a variety of cations.
The active site Mg2+ (yellow, marked with an asterisk) observed in the inhibitor-bound ribozyme (3NKB) overlaps with the binding sites for Mg2+ (yellow), Mn2+ (violet), Ba2+ (green), Tl+ (red) and Co(NH3) 3+6 (blue and pink) observed in the structures of the C75U mutant (1SJ3, 1SJF, 2OIH, 1VBZ, 1VBY).
There are two classes of small ribozymes
Small ribozymes have distinct responses to variations in cation identity and these responses suggest that while some small ribozymes are metal-independent, others, like the HDV ribozyme, are likely metalloenzymes.
We can sort ribozymes into two major classes (referred to here as Class 1 and Class 2) on the basis of their response to the trivalent ion complex [Co(NH3)6]3+. [Co(NH3)6]3+ mimics a fully hydrated magnesium ion, but differs in two manners (54): First, the ligands are exchange inert; thus, there can be no direct coordination between ligands on the ribozyme and the cobalt center. Second, the pKa of the amine groups of [Co(NH3)6]3+ is much greater than that of water bound to magnesium ions; therefore, they do not participate in proton transfer reactions (54).
Class 1 ribozymes include the hairpin, VS and the glmS ribozymes. These RNAs function in [Co(NH3)6]3+ alone, and their Mg2+ dependent reactions are not inhibited by [Co(NH3)6]3+ (55-59). These data suggest that, even in biologically relevant buffer conditions, Mg2+ does not participate directly in their chemical reactions. Mechanistic studies have suggested that the active site of the hairpin and VS ribozymes contains two critical bases, an adenine and a guanine, that interact with the 5’-oxygen leaving group and the 2’-hydroxyl nucleophile, respectively, to facilitate catalysis through general acid/general base catalysis (34, 60, 61). The glucosamine-6-phosphate (glmS) responsive ribozyme, which uses a guanosine nucleobase and a substrate amine to perform general acid/base catalysis, is probably also in this category. Binding of the glucosamine-6-phosphate ligand is metal ion dependent however and this complicates this analysis (58, 59).
The class II ribozymes include the hammerhead and HDV ribozymes. These RNAs exhibit a distinctly different response to [Co(NH3)6]3+ than class I ribozymes (62-64). In physiological buffers (low to moderate ionic strength and in the presence of Mg2+), [Co(NH3)6]3+ is strongly inhibitory to the HDV and hammerhead ribozymes. In the HDV ribozyme [Co(NH3)6]3+ has been shown to inhibit in a manner that is competitive with Mg2+ ion (13). In addition, Raman spectroscopy of the HDV ribozyme reveals that a [Co(NH3)6]3+ ion binds in the active site near C75 and can be readily displaced by a Mg2+ ion (65). These data suggest that [Co(NH3)6]3+ is capable of displacing catalytic Mg2+ ions and thereby inhibiting the reaction. In high ionic strength buffers (1 M NaCl), the requirement for a catalytic Mg2+ can be bypassed by an alternate mechanism, and [Co(NH3)6]3+ is not inhibitory (62). In the hammerhead ribozyme, functional groups required for the Mg2+-dependent catalysis are not required for reaction under high ionic strength (66). This provides evidence that under biologically-relevant conditions, the hammerhead ribozyme functions as a metalloenzyme. Under Mg2+-free, high-ionic strength conditions, both the HDV and hammerhead ribozymes cannot work at maximal rates, and lose at least 25-fold in activity due to loss of catalytic metal ions (14, 25, 66, 67).
These data indicate a significant role for Mg2+ in the mechanisms of the HDV and hammerhead ribozymes. Indeed, crystal structures of the HDV and hammerhead, but not the hairpin or glmS ribozymes reveal active site metal ions positioned to participate in catalysis. The metal ion observed in the recent crystal structures of the hammerhead ribozyme is not close enough to the cleavage site to participate in catalysis (68). However, spectroscopic analysis (69) and computational simulations (70, 71) suggest that the active site metal ion moves closer to the scissile phosphate prior to catalysis, possibly to interact with the 5’-hydroxyl leaving group. Thus, both the hammerhead and HDV ribozymes are likely to be metal-dependent under biologically relevant conditions.
There is significant biochemical evidence for participation of a divalent cation in HDV ribozyme catalysis
The importance of the active site metal ion in the HDV ribozyme is underscored by the deleterious effects of mutations and modifications to nucleotides surrounding this ion. The G25•U20 reverse wobble, which interacts with the active site metal ion through its hydration shell, is present in all known HDV-like ribozymes. Single mutations, such as G25A or U20C, which would allow formation of a Watson-Crick G-C or U-A base pair at this position reduce the rate constant by ~104-fold (72, 73). In addition, modifications that affect the positioning of U20, including phosphorothioate substitution at the pro-RP oxygen of C22 and deoxyribose substitution at U20, inhibit the reaction (74, 75).
In the crystal structure of the HDV ribozyme precleavage, the pro-RP oxygen at the scissile phosphate is an inner sphere ligand to the active site Mg2+ (33) (Figure 5). Substitution of this oxygen atom with a sulfur atom has an interesting effect: 80-90% of the modified substrate is not cleaved by the ribozyme while 10-20% is cleaved with the same rate constant as unmodified substrate or a substrate with a sulfur substitution at the pro-SP oxygen (76). It is not clear why 10-20% of the modified substrate is reactive while 80-90% is unreactive. It is possible that the modified substrate was contaminated with residual unmodified substrate or the alternate stereoisomer, or it is possible that minor fraction of the modified substrate was being cleaved by an alternate, metal-independent reaction pathway (14). This result suggests that the pro-RP oxygen at the scissile phosphate is playing a critical role although it does not address whether that role is metal binding. Sulfur substitution at the pro-RP oxygen could not be rescued by thiophilic metal ions and therefore a direct interaction between the pro-RP oxygen of G1 and a metal ion cannot be inferred from this experiment (76). However, in addition to its interaction with the active site Mg2+ ion, the pro-RP oxygen of G1 is within 3.6 Å of the N4 of C75. Substitution of the pro-RP oxygen of the scissile phosphate with the bulkier sulfur atom may disrupt interactions with the N4 of C75, and thereby misposition the general acid within the active site (39) or unfavorably modulate the pKa of C75 (40). These multiple roles may explain why sulfur substitution at the pro-RP oxygen of the scissile phosphate negatively influences catalysis but cannot be rescued by thiophilic metal ions.
Lack of specificity in metal preference does not mean lack of active site metal
There are only two direct contacts between the ribozyme and the catalytic metal ion and the rest are solvent mediated. This characteristic might be the reason that the HDV ribozyme active site is able to accommodate metal ions with a wide variety of atomic radii and other chemical characteristics. Large ribozymes, including the group I and II self splicing introns and RNase P use phosphate clusters to bind and position metal ions and are exquisitely sensitive to the identity of the divalent cation. All will work in the presence of Mg2+ ion and many will work in the presence of Mn2+ ion. Ca2+ ion, however, is often found to support the formation of tertiary structure but does not support catalytic activity. In contrast, the HDV and hammerhead ribozymes bind their active site metal through solvent mediated interactions (33, 68) and are more promiscuous with respect to metal ion specificity (62, 77, 78). This lack of specificity is likely the result of the nature of the small ribozyme catalytic metal binding site. This is important to note because lack of specificity could be interpreted as lack of metal ions in the catalytic mechanism, but this does not appear to be true.
A change in the catalytic mechanism in the absence of Mg2+
The cleavage reaction of the HDV ribozyme is multichannel (62), occuring in the presence or absence of the catalytic Mg2+ (13, 14, 79). Thus, in buffers containing only monovalent cations, the reaction proceeds through an alternative but related mechanism to that catalyzed in the presence of the catalytic Mg2+. The overall rate of reaction is ~3,000-fold slower in 1 M NaCl than in saturating Mg2+. Mg2+ contributes ~125-fold by to facilitating folding and ~25-fold by facilitating chemistry.
Evidence for the participation of Mg2+ in the cleavage reaction is provided by analysis of the pH-rate profile of the reaction. In the presence of Mg2+ the pH-rate profile increases log-linearly until ~pH 9.0, where it plateaus. In contrast, in the absence of Mg2+ the catalytic rate constant decreases log-linearly with increasing pH, revealing a pKa near neutrality (13, 41, 64). The pKa observed in the absence of Mg2+ is consistent with the pKa of C75, and the value of this pKa is consistent with direct measurement of the pKa of C75 by Raman spectroscopy in low Mg2+ (40). As the general acid is deprotonated, the rate of reaction decreases. However this pH dependence of the general acid is obscured in the presence of Mg2+ by a second, Mg2+-dependent pKa, that is ≥ 9. This pKa is too high to measure because RNA unfolds and hydrolyzes non-specifically under these conditions. This suggests that, in the presence of Mg2+, there is a second proton transfer associated with a Mg2+ ion involved in the catalytic mechanism that is not present in the Mg2+-free reaction.
Is this Mg2+-dependent pKa associated with the active site, or the result of a long-range structural effect? The pKa of C75 is sensitive to the concentration of Mg2+ ion, suggesting that there is at least one Mg2+ ion bound in the active site in the vicinity of C75. When the Mg2+-binding site is occupied, protonation of C75 is more difficult and the pKa of C75 drops. Conversely, as the Mg2+ concentration is dropped and the Mg2+ binding site is less occupied, the pKa of C75 is raised. Anti-cooperative binding of the proton on C75 and a Mg2+ ion is directly observable by Raman spectroscopy (65) suggests that the two positive charges are in close enough proximity to interact with each other electrostatically.
Proton inventory experiments also suggest a Mg2+ ion is involved in the catalytic mechanism. The proton inventory of the HDV cleavage reaction is two when the concentration of Mg2+ is low or zero, implying two proton transfers in the rate limiting step. However, proton inventories approach 1 at high Mg2+ concentrations (80, 81). These data are consistent with a mechanism in which an active site Mg2+ binds to the U(–1) 2’-O and lowers the pKa of the 2’-O to facilitate deprotonation the nucleophile. This leaves only a single proton transfer, from C75H+ to the O5’ of G1, in the rate limiting step. Na+ is observed to bind within the active site pocket in a position that partially overlaps the catalytic Mg2+ ion (82). Na+ may be able to facilitate catalysis by electrostatic stabilization, but it does not serve as a Lewis acid. Thus, under these conditions, there are two proton transfers in the rate limiting step. This is supported by solvent isotope effects under Mg2+-free conditions which suggest that the 2’-OH of U(-1) is deprotonated by a hydroxide ion from solution (64). These results are thus generally consistent with the catalytic Mg2+ ion serving as a Lewis acid in the Mg2+-containing cleavage reaction.
The HDV ribozyme has a hybrid engine
Although the HDV ribozyme is capable of catalysis in the absence of Mg2+ ion, this RNA is a metalloenzyme under biologically relevant conditions. The recent crystal structure of this RNA suggests that the active site Mg2+ ion interacts directly with the 2’-hydroxyl nucleophile and the scissile phosphate to facilitate catalysis. This type of interaction between a divalent cation and the cleavage site had previously been observed only in large ribozymes, such as group I and II introns and RNAse P. The active site Mg2+ likely helps to activate and to stabilize developing negative charge on the nucleophile, to orient the cleavage site for in-line attack, and to stabilize the transition state (Figure 2). In this manner, the HDV ribozyme uses metal ion catalysis characteristic of the large ribozymes.
The HDV ribozyme is capable of functioning in the absence of Mg2+ ion because much of its catalytic power comes from a catalytic nucleobase, C75, with a pKa dramatically shifted towards neutrality and capable of participating in general acid catalysis (Figure 2). The hairpin and VS ribozymes both use an adenosine with a pKa shifted towards neutrality as the general acid in the cleavage reaction. The glmS ribozyme appears to use an amine group, also with a shifted pKa, from its cofactor glucosamine-6-phosphate as a general acid. In its use of a general acid, the HDV ribozyme also uses nucleobase catalysis typical of the small ribozymes. Thus we see that two distinct catalytic strategies, those typified by large ribozymes and by small ribozymes, are found under the hood of the HDV ribozyme.
Acknowledgements
I very grateful to my collaborators Phil Bevilacqua, Paul Carey and Sharon Hammes-Schiffer and to Jui-Hui Chen, Elaine Chase, Durga Chadalavada, Bo Yang, and Rieko Yajima for their contributions to the HDV ribozyme project. I would like to thank Phil Bevilaqua and Mark Hermodson for helpful suggestions on this manuscript.
Footnotes
This project was supported by NIH grant R01GM095923, the Purdue University Department of Biochemistry, the Markey Center for Structural Biology and the Purdue University Center for Cancer Research.
Abbreviations used: glmS, glucosamine-6-phosphate; HDV, hepatitis delta virus; VS, Varkud satellite; WT, wild-type.
Literature cited
- 1.Fedor MJ. Comparative enzymology and structural biology of RNA self-cleavage. Ann. Rev. Biophys. 2009;38:271–299. doi: 10.1146/annurev.biophys.050708.133710. [DOI] [PubMed] [Google Scholar]
- 2.Reiter NJ, Chan CW, Mondragon A. Emerging structural themes in large RNA molecules. Curr. Opin. Struct. Biol. 2011;21:319–326. doi: 10.1016/j.sbi.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liberman JA, Wedekind JE. Base ionization and ligand binding: how small ribozymes and riboswitches gain a foothold in a protein world. Curr. Opin. Struct. Biol. 2011;21:327–334. doi: 10.1016/j.sbi.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeRose VJ. Metal ion binding to catalytic RNA molecules. Curr. Opin. Struct. Biol. 2003;13:317–324. doi: 10.1016/s0959-440x(03)00077-0. [DOI] [PubMed] [Google Scholar]
- 5.Isom DG, Cannon BR, Castaneda CA, Robinson A, Garcia-Moreno B. High tolerance for ionizable residues in the hydrophobic interior of proteins. Proc. Natl. Acad. Sci. U. S. A. 2008;105:17784–17788. doi: 10.1073/pnas.0805113105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Isom DG, Castaneda CA, Cannon BR, Garcia-Moreno B. Large shifts in pKa values of lysine residues buried inside a protein. Proc. Natl. Acad. Sci. U. S. A. 2011;108:5260–5265. doi: 10.1073/pnas.1010750108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fedor MJ, Williamson JR. The catalytic diversity of RNAs. Nat. Rev. Mol. Cell. Biol. 2005;6:399–412. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- 8.Wilcox JL, Ahluwalia AK, Bevilacqua PC. Charged Nucleobases and Their Potential for RNA Catalysis. Acc. Chem. Res. 2011 doi: 10.1021/ar2000452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan T, Uhlenbeck OC. A small metalloribozyme with a two-step mechanism. Nature. 1992;358:560–563. doi: 10.1038/358560a0. [DOI] [PubMed] [Google Scholar]
- 10.Glasner ME, Bergman NH, Bartel DP. Metal ion requirements for structure and catalysis of an RNA ligase ribozyme. Biochemistry. 2002;41:8103–8112. doi: 10.1021/bi012179b. [DOI] [PubMed] [Google Scholar]
- 11.Shan S, Kravchuk AV, Piccirilli JA, Herschlag D. Defining the catalytic metal ion interactions in the Tetrahymena ribozyme reaction. Biochemistry. 2001;40:5161–5171. doi: 10.1021/bi002887h. [DOI] [PubMed] [Google Scholar]
- 12.Ferre-D'Amare AR, Zhou KH, Doudna JA. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 13.Nakano S, Chadalavada DM, Bevilacqua PC. General acid-base catalysis in the mechanism of a hepatitis delta virus ribozyme. Science. 2000;287:1493–1497. doi: 10.1126/science.287.5457.1493. [DOI] [PubMed] [Google Scholar]
- 14.Nakano S, Proctor DJ, Bevilacqua PC. Mechanistic characterization of the HDV genomic ribozyme: assessing the catalytic and structural contributions of divalent metal ions within a multichannel reaction mechanism. Biochemistry. 2001;40:12022–12038. doi: 10.1021/bi011253n. [DOI] [PubMed] [Google Scholar]
- 15.Chen JH, Gong B, Bevilacqua PC, Carey PR, Golden BL. A catalytic metal ion interacts with the cleavage Site G•U wobble in the HDV ribozyme. Biochemistry. 2009;48:1498–1507. doi: 10.1021/bi8020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pereira MJ, Harris DA, Rueda D, Walter NG. Reaction pathway of the trans-acting hepatitis delta virus ribozyme: a conformational change accompanies catalysis. Biochemistry. 2002;41:730–740. doi: 10.1021/bi011963t. [DOI] [PubMed] [Google Scholar]
- 17.Ke A, Zhou K, Ding F, Cate JH, Doudna JA. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature. 2004;429:201–205. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- 18.Krasovska MV, Sefcikova J, Spackova N, Sponer J, Walter NG. Structural dynamics of precursor and product of the RNA enzyme from the hepatitis delta virus as revealed by molecular dynamics simulations. J. Mol. Biol. 2005;351:731–748. doi: 10.1016/j.jmb.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 19.Guo F, Gooding A, Cech TR. Structure of the tetrahymena ribozyme: Base triple sandwhich and metal ion at the active site. Mol. Cell. 2004;16:351–362. doi: 10.1016/j.molcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004;430:45–50. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- 21.Golden BL, Kim H, Chase E. Crystal structure of a phage Twort group I ribozyme-product complex. Nat. Struct. Mol. Biol. 2005;12:82–89. doi: 10.1038/nsmb868. [DOI] [PubMed] [Google Scholar]
- 22.Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reiter NJ, Osterman A, Torres-Larios A, Swinger KK, Pan T, Mondragon A. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature. 2010;468:784–789. doi: 10.1038/nature09516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson TJ, Lilley DM. Do the hairpin and VS ribozymes share a common catalytic mechanism based on general acid-base catalysis? A critical assessment of available experimental data. RNA. 2011;17:213–221. doi: 10.1261/rna.2473711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murray JB, Seyhan AA, Walter NG, Burke JM, Scott WG. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem. Biol. 1998;5:587–595. doi: 10.1016/s1074-5521(98)90116-8. [DOI] [PubMed] [Google Scholar]
- 26.Kuo MY, Sharmeen L, Dinter-Gottlieb G, Taylor J. Characterization of self-cleaving RNA sequences on the genome and antigenome of human hepatitis delta virus. J. Virol. 1988;62:4439–4444. doi: 10.1128/jvi.62.12.4439-4444.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salehi-Ashtiani K, Luptak A, Litovchick A, Szostak JW. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science. 2006;313:1788–1792. doi: 10.1126/science.1129308. [DOI] [PubMed] [Google Scholar]
- 29.Webb CH, Riccitelli NJ, Ruminski DJ, Luptak A. Widespread occurrence of self-cleaving ribozymes. Science. 2009;326:953. doi: 10.1126/science.1178084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eickbush DG, Eickbush TH. R2 retrotransposons encode a self-cleaving ribozyme for processing from an rRNA cotranscript. Mol. Cell. Biol. 2010;30:3142–3150. doi: 10.1128/MCB.00300-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perrotta AT, Shih I, Been MD. Imidazole rescue of a cytosine mutation in a self-cleaving ribozyme. Science. 1999;286:123–126. doi: 10.1126/science.286.5437.123. [DOI] [PubMed] [Google Scholar]
- 32.Wadkins TS, Been MD. Ribozyme activity in the genomic and antigenomic RNA strands of hepatitis delta virus. Cell. Mol. Life Sci. 2002;59:112–125. doi: 10.1007/s00018-002-8409-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen JH, Yajima R, Chadalavada DM, Chase E, Bevilacqua PC, Golden BL. A 1.9 Å crystal structure of the HDV ribozyme precleavage suggests both Lewis acid and general acid mechanisms contribute to phosphodiester cleavage. Biochemistry. 2010;49:6508–6518. doi: 10.1021/bi100670p. [DOI] [PubMed] [Google Scholar]
- 34.Rupert PB, Ferre-D'Amare AR. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- 35.Alam S, Grum-Tokars V, Krucinska J, Kundracik ML, Wedekind JE. Conformational heterogeneity at position U37 of an all-RNA hairpin ribozyme with implications for metal binding and the catalytic structure of the S-turn. Biochemistry. 2005;44:14396–14408. doi: 10.1021/bi051550i. [DOI] [PubMed] [Google Scholar]
- 36.Klein DJ, Ferre-D'Amare AR. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- 37.Cochrane JC, Lipchock SV, Strobel SA. Structural investigation of the GlmS ribozyme bound to Its catalytic cofactor. Chem. Biol. 2007;14:97–105. doi: 10.1016/j.chembiol.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Veeraraghavan N, Bevilacqua PC, Hammes-Schiffer S. Long-distance communication in the HDV ribozyme: insights from molecular dynamics and experiments. J. Mol. Biol. 2010;402:278–291. doi: 10.1016/j.jmb.2010.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Veeraraghavan N, Ganguly A, Chen JH, Bevilacqua PC, Hammes-Schiffer S, Golden BL. Metal binding motif in the active site of the HDV ribozyme binds divalent and monovalent ions. Biochemistry. 2011;50:2672–2682. doi: 10.1021/bi2000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gong B, Chen JH, Chase E, Chadalavada DM, Yajima R, Golden BL, Bevilacqua PC, Carey PR. Direct measurement of a pKa near neutrality for the catalytic cytosine in the genomic HDV ribozyme using Raman crystallography. J. Amer. Chem. Soc. 2007;129:13335–13342. doi: 10.1021/ja0743893. [DOI] [PubMed] [Google Scholar]
- 41.Nakano S, Bevilacqua PC. Mechanistic characterization of the HDV genomic ribozyme: a mutant of the C41 motif provides insight into the positioning and thermodynamic linkage of metal ions and protons. Biochemistry. 2007;46:3001–3012. doi: 10.1021/bi061732s. [DOI] [PubMed] [Google Scholar]
- 42.Das SR, Piccirilli JA. General acid catalysis by the hepatitis delta virus ribozyme. Nature Chem. Biol. 2005;1:45–52. doi: 10.1038/nchembio703. [DOI] [PubMed] [Google Scholar]
- 43.Tang CL, Alexov E, Pyle AM, Honig B. Calculation of pKas in RNA: on the structural origins and functional roles of protonated nucleotides. J. Mol. Biol. 2007;366:1475–1496. doi: 10.1016/j.jmb.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 44.Luptak A, Ferre-D'Amare AR, Zhou K, Zilm KW, Doudna JA. Direct pKa measurement of the active-site cytosine in a genomic hepatitis delta virus ribozyme. J. Amer. Chem. Soc. 2001;123:8447–8452. doi: 10.1021/ja016091x. [DOI] [PubMed] [Google Scholar]
- 45.Guo M, Spitale RC, Volpini R, Krucinska J, Cristalli G, Carey PR, Wedekind JE. Direct Raman measurement of an elevated base pKa in the active site of a small ribozyme in a precatalytic conformation. J. Amer. Chem. Soc. 2009;131:12908–12909. doi: 10.1021/ja9060883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cottrell JW, Scott LG, Fedor MJ. The pH dependence of hairpin ribozyme catalysis reflects ionization of an active site adenine. J. Biol. Chem. 2011;286:17658–17664. doi: 10.1074/jbc.M111.234906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rupert PB, Massey AP, Sigurdsson ST, Ferre-D'Amare AR. Transition state stabilization by a catalytic RNA. Science. 2002;298:1421–1424. doi: 10.1126/science.1076093. [DOI] [PubMed] [Google Scholar]
- 48.Torelli AT, Krucinska J, Wedekind JE. A comparison of vanadate to a 2'-5' linkage at the active site of a small ribozyme suggests a role for water in transition-state stabilization. RNA. 2007;13:1052–1070. doi: 10.1261/rna.510807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davis JH, Dunican BF, Strobel SA. glmS riboswitch binding to the glucosamine-6-phosphate alpha-anomer shifts the pKa toward neutrality. Biochemistry. 2011 doi: 10.1021/bi200471c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gong B, Klein DJ, Ferre-D'Amare AR, Carey PR. The glmS Ribozyme Tunes the Catalytically Critical pKa of Its Coenzyme Glucosamine-6-phosphate. J. Amer. Chem. Soc. 2011;133:14188–14191. doi: 10.1021/ja205185g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Juneau K, Podell E, Harrington DJ, Cech TR. Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA--solvent interactions. Structure. 2001;9:221–231. doi: 10.1016/s0969-2126(01)00579-2. [DOI] [PubMed] [Google Scholar]
- 52.Keel AY, Rambo RP, Batey RT, Kieft JS. A general strategy to solve the phase problem in RNA crystallography. Structure. 2007;15:761–772. doi: 10.1016/j.str.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ke A, Ding F, Batchelor JD, Doudna JA. Structural roles of monovalent cations in the HDV ribozyme. Structure. 2007;15:281–287. doi: 10.1016/j.str.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 54.Suga H, Cowan JA, Szostak JW. Unusual metal ion catalysis in an acyl-transferase ribozyme. Biochemistry. 1998;37:10118–10125. doi: 10.1021/bi980432a. [DOI] [PubMed] [Google Scholar]
- 55.Hampel A, Cowan JA. A unique mechanism for RNA catalysis: the role of metal cofactors in hairpin ribozyme cleavage. Chem. Biol. 1997;4:513–517. doi: 10.1016/s1074-5521(97)90323-9. [DOI] [PubMed] [Google Scholar]
- 56.Nesbitt S, Hegg LA, Fedor MJ. An unusual pH-independent and metal-ion-independent mechanism for hairpin ribozyme catalysis. Chem. Biol. 1997;4:619–630. doi: 10.1016/s1074-5521(97)90247-7. [DOI] [PubMed] [Google Scholar]
- 57.DeYoung MB, Siwkowski A, Hampel A. Determination of catalytic parameters for hairpin ribozymes. Methods Mol. Biol. 1997;74:209–220. doi: 10.1385/0-89603-389-9:209. [DOI] [PubMed] [Google Scholar]
- 58.Klawuhn K, Jansen JA, Souchek J, Soukup GA, Soukup JK. Analysis of metal ion dependence in glmS ribozyme self-cleavage and coenzyme binding. Chem. Biochem. 2010;11:2567–2571. doi: 10.1002/cbic.201000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brooks KM, Hampel KJ. Rapid steps in the glmS ribozyme catalytic pathway: cation and ligand requirements. Biochemistry. 2011;50:2424–2433. doi: 10.1021/bi101842u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lafontaine DA, Wilson TJ, Norman DG, Lilley DM. The A730 loop is an important component of the active site of the VS ribozyme. J. Mol. Biol. 2001;312:663–674. doi: 10.1006/jmbi.2001.4996. [DOI] [PubMed] [Google Scholar]
- 61.Wilson TJ, McLeod AC, Lilley DM. A guanine nucleobase important for catalysis by the VS ribozyme. EMBO J. 2007;26:2489–2500. doi: 10.1038/sj.emboj.7601698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakano S, Cerrone AL, Bevilacqua PC. Mechanistic characterization of the HDV genomic ribozyme: classifying the catalytic and structural metal ion sites within a multichannel reaction mechanism. Biochemistry. 2003;42:2982–2994. doi: 10.1021/bi026815x. [DOI] [PubMed] [Google Scholar]
- 63.Roychowdhury-Saha M, Burke DH. Distinct reaction pathway promoted by non-divalent-metal cations in a tertiary stabilized hammerhead ribozyme. RNA. 2007;13:841–848. doi: 10.1261/rna.339207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cerrone-Szakal AL, Chadalavada DM, Golden BL, Bevilacqua PC. Mechanistic characterization of the HDV genomic ribozyme: the cleavage site base pair plays a structural role in facilitating catalysis. RNA. 2008;14:1746–1760. doi: 10.1261/rna.1140308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gong B, Chen JH, Bevilacqua PC, Golden BL, Carey PR. Competition between Co(NH3)63+ and inner sphere Mg2+ ions in the HDV ribozyme. Biochemistry. 2009;48:11961–11970. doi: 10.1021/bi901091v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O'Rear JL, Wang S, Feig AL, Beigelman L, Uhlenbeck OC, Herschlag D. Comparison of the hammerhead cleavage reactions stimulated by monovalent and divalent cations. RNA. 2001;7:537–545. doi: 10.1017/s1355838201002461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Curtis EA, Bartel DP. The hammerhead cleavage reaction in monovalent cations. RNA. 2001;7:546–552. doi: 10.1017/s1355838201002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martick M, Lee TS, York DM, Scott WG. Solvent structure and hammerhead ribozyme catalysis. Chem. Biol. 2008;15:332–342. doi: 10.1016/j.chembiol.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Osborne EM, Ward WL, Ruehle MZ, DeRose VJ. The identity of the nucleophile substitution may influence metal interactions with the cleavage site of the minimal hammerhead ribozyme. Biochemistry. 2009;48:10654–10664. doi: 10.1021/bi900614v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee TS, Silva Lopez C, Giambasu GM, Martick M, Scott WG, York DM. Role of Mg2+ in hammerhead ribozyme catalysis from molecular simulation. J. Amer. Chem. Soc. 2008;130:3053–3064. doi: 10.1021/ja076529e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wong KY, Lee TS, York DM. Active participation of Mg ion in the reaction coordinate of RNA self-cleavage catalyzed by the hammerhead ribozyme. J. Chem. Theory Comput. 2011;7:1–3. doi: 10.1021/ct100467t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Perrotta AT, Been MD. Core sequences and a cleavage site wobble pair required for HDV antigenomic ribozyme self-cleavage. Nucleic Acids Res. 1996;24:1314–1321. doi: 10.1093/nar/24.7.1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tanner NK, Schaff S, Thill G, Petit-Koskas E, Crain-Denoyelle AM, Westhof E. A three-dimensional model of hepatitis delta virus ribozyme based on biochemical and mutational analyses. Curr. Biol. 1994;4:488–498. doi: 10.1016/s0960-9822(00)00109-3. [DOI] [PubMed] [Google Scholar]
- 74.Jeoung YH, Kumar PK, Suh YA, Taira K, Nishikawa S. Identification of phosphate oxygens that are important for self-cleavage activity of the HDV ribozyme by phosphorothioate substitution interference analysis. Nucleic Acids Res. 1994;22:3722–3727. doi: 10.1093/nar/22.18.3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nishikawa F, Shirai M, Nishikawa S. Site-specific modification of functional groups in genomic hepatitis delta virus (HDV) ribozyme. Eur. J. Biochem. 2002;269:5792–5803. doi: 10.1046/j.1432-1033.2002.03280.x. [DOI] [PubMed] [Google Scholar]
- 76.Fauzi H, Kawakami J, Nishikawa F, Nishikawa S. Analysis of the cleavage reaction of a trans-acting human hepatitis delta virus ribozyme. Nucleic Acids Res. 1997;25:3124–3130. doi: 10.1093/nar/25.15.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Boots JL, Canny MD, Azimi E, Pardi A. Metal ion specificities for folding and cleavage activity in the Schistosoma hammerhead ribozyme. RNA. 2008;14:2212–2222. doi: 10.1261/rna.1010808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Suh YA, Kumar PK, Taira K, Nishikawa S. Self-cleavage activity of the genomic HDV ribozyme in the presence of various divalent metal ions. Nucleic Acids Res. 1993;21:3277–3280. doi: 10.1093/nar/21.14.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wadkins TS, Shih I, Perrotta AT, Been MD. A pH-sensitive RNA tertiary interaction affects self-cleavage activity of the HDV ribozymes in the absence of added divalent metal ion. J. Mol. Biol. 2001;305:1045–1055. doi: 10.1006/jmbi.2000.4368. [DOI] [PubMed] [Google Scholar]
- 80.Nakano S, Bevilacqua PC. Proton inventory of the genomic HDV ribozyme in Mg2+-containing solutions. J. Amer. Chem. Soc. 2001;123:11333–11334. doi: 10.1021/ja0166850. [DOI] [PubMed] [Google Scholar]
- 81.Shih IH, Been MD. Involvement of a cytosine side chain in proton transfer in the rate-determining step of ribozyme self-cleavage. Proc. Natl. Acad. Sci. U. S. A. 2001;98:1489–1494. doi: 10.1073/pnas.98.4.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Veeraraghavan N, Ganguly A, Golden BL, Bevilacqua PC, Hammes-Schiffer S. Mechanistic Strategies in the HDV Ribozyme: Chelated and Diffuse Metal Ion Interactions and Active Site Protonation. J Phys Chem B. 2011 doi: 10.1021/jp203202e. [DOI] [PMC free article] [PubMed] [Google Scholar]