Abstract
Cancer progression involves multiple complex and interdependent steps, including progressive proliferation, angiogenesis and metastases. The complexity of these processes requires a comprehensive elucidation of the integrated signaling networks for better understanding. EAPII interacts with multiple cancer-related proteins, but its biological significance in cancer development remains unknown. In this report we identified the elevated level of EAPII protein in non-small-cell lung carcinoma (NSCLC) patients and NSCLC cell lines in culture. The oncogenic role of EAPII in lung cancer development was demonstrated using NSCLC cells with genetic manipulations that influence EAPII expression: EAPII overexpression increases proliferation of NSCLC cells with an accelerated transition of cell cycle and facilitates xenograft tumor growth in vivo; EAPII knockdown results in apoptosis of NSCLC cells and reduces xenograft tumor formation. To further explore the mechanism of EAPII's oncogenic role in lung cancer development and to elucidate the potential signaling pathway(s) that EAPII may impact, we employed antibody array to investigate the alternation of the major signaling pathways in NSCLC cells with altered EAPII level. We found that EAPII overexpression significantly activated Raf1 and ERK1/2, but not c-Jun N-terminal kinase and p38 pathways. Consistently, the protein and mRNA levels of MYC and cyclin D1, which are targets of the mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK–ERK) pathway, are significantly increased by EAPII overexpression. Taken together, we demonstrated that EAPII is an oncogenic factor and the activation of MAPK–ERK signaling pathway by EAPII may contribute to lung cancer development.
Keywords: EAPII, MAPK–ERK pathway, NSCLC
Introduction
We previously identified EAPII (ETS1-Associated Protein II) as a new protein interacting with ETS1, a transcription factor involved in tumorigenesis and metastasis, and this association modulates the transcriptional activity of ETS1 (Pei et al., 2003). EAPII was also independently identified and designated as TTRAP (TRAF and TNF receptor-associated protein) through association with the cytoplasmic domain of CD40, tumor necrosis factor (TNF) receptor-75, and TNF receptor-associated factors (TRAFs); this association inhibits nuclear factor κB activation (Pype et al., 2000). EAPII protects neuroblastoma cells from apoptosis induced by MG132, a proteasome inhibitor (Zucchelli et al., 2009), but the mutant of DJ-1, an oncogene in cooperation with Ras (Nagakubo et al., 1997), promotes apoptosis through c-Jun N-terminal kinase (JNK) and p38 mitogen-activated protein kinase (MAPK) pathways in an EAPII-dependent manner (Zucchelli et al., 2009). EAPII expression also promotes apoptosis of HL-60 cells induced by hydroquinone, a cytotoxic agent (Zhang et al., 2007). Furthermore, EAPII is upregulated in FOXO3a-induced apoptosis, in which FOXO3a turns TNF receptor signaling to a proapoptotic JNK-dependent pathway (Lee et al., 2008). EAPII also interacts with promyelocytic leukemia protein (Xu et al., 2008), which controls key pathways for growth suppression, induction of apoptosis and cellular senescence (Salomoni, 2009; Lallemand-Breitenbach and de The, 2010). These observations suggest that EAPII may protect cells from apoptosis or promote apoptosis in a cell-specific manner. However, the biological significance of EAPII in cancer development remains unknown. In this report we examined expression patterns of EAPII in lung cancer tissues and non-small-cell lung carcinoma (NSCLC) cell lines. The data from this regard suggest that EAPII may be involved in the development of NSCLC. We further showed that re-expression of EAPII in NSCLC cells promotes cell proliferation and enhances xenograft tumor growth, whereas EAPII knockdown using the short hairpin RNA (shRNA) technique induces apoptosis and reduces xenograft tumor growth. Results of the major signaling pathways analysis using antibody array showed that EAPII upregulates oncogenic protein MYC and cell cycle machinery such as cyclin D1. Significantly, extracellular signal-regulated kinases (ERK1/2), but not JNK or p38 MAPK pathways, are activated in these models, suggesting that EAPII is an upstream event of the Raf–ERK signaling pathway and has an oncogenic role in lung cancer development.
Results
Increased expression of EAPII in lung carcinomas
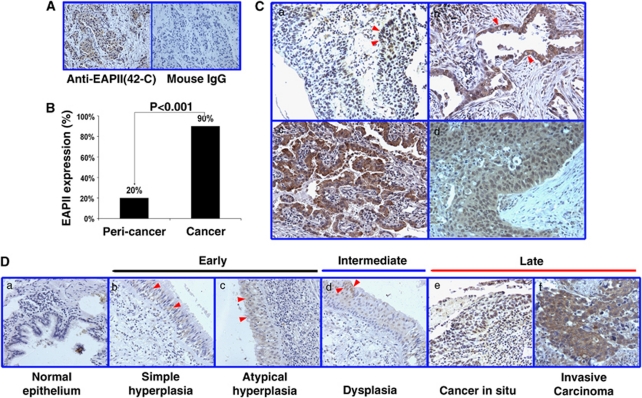
To understand the role of EAPII in lung cancer progression, we carried out immunohistochemistry in a tissue array (AccuMax, ISU ABXIS Co., Ltd, Korea) containing 103 cases of lung carcinoma and 44 adjacent non-neoplastic tissues. We had previously developed a mouse monoclonal anti-EAPII antibody (42C), and this antibody recognizes EAPII specifically in western blot (Pei et al., 2003) and produces specific signals in human tissue immunohistochemical staining, whereas normal mouse IgG produces no signal (Figure 1A). EAPII expression is significantly elevated in lung carcinomas: Compared with 20% (9/44) in adjacent non-neoplastic tissue, 90% (93/103) of lung carcinomas showed positive staining (Figure 1B). In lung carcinomas, elevated EAPII expression is observed in the cytoplasm, nucleus or both (Figure 1C). The expression level and localization of EAPII change along with the course of lung cancer development. In Figure 1D, we have listed the representative images of each stage of lung cancer development. No EAPII staining was observed in normal bronchial epithelium (Figure 1Da). With increased cell proliferation, weak staining of EAPII in the nucleus occurs, and EAPII expression appears in hyperplastic epithelium (Figure 1Db and c) and dysplastic epithelium (Figure 1Dd). Furthermore, significantly elevated EAPII expression is observed in lung carcinomas (Figure 1De and f). Overall, cytoplasmic staining was observed in 84% (78/93) and nuclear staining in 56% (52/93) of cases analyzed. Whereas nuclear staining of EAPII can be seen in the highly proliferative tissue, the majority of lung carcinomas showed cytoplasmic staining or cytoplasmic and nuclear staining. This result suggests that EAPII may contribute to both early and advanced stages of lung cancer development.
Figure 1.
Increased expression of EAPII in lung carcinomas. (A) The representative staining of anti-EAPII antibody 42C and normal mouse IgG. (B) Percentages of EAPII-positive staining from the array containing 103 lung carcinomas and 44 matched peri-tumor tissues. (C) Representative images of a tissue microarray stained with 42C: EAPII expression is observed in the nucleus (arrows in a), cytoplasma (arrows in b) or both the nucleus and cytoplasm (c, d) of NSCLC tissues. (D) EAPII expression in the development of lung cancer: no EAPII staining is observed in normal bronchial epithelium (a); EAPII is expressed in the nucleus of the hyperplastic epithelium (arrows in b and c), dysplastic epithelium (arrows in d) and lung carcinomas (e, f).
EAPII is highly expressed in most of the lung cancer cell lines
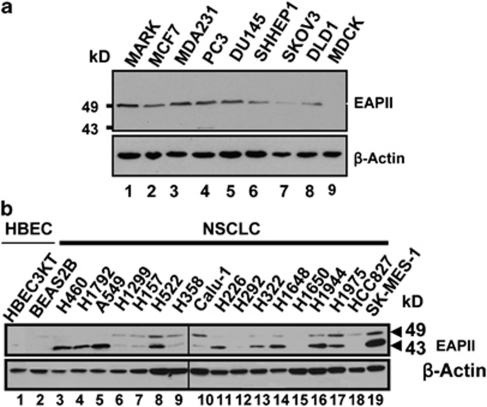
Antibody 42C was further used in western blot to determine the EAPII expression level in cancer cell lines. A single band, migrated at 49 kDa, was observed in multiple cancer cell lines including breast, prostate, liver, ovarian and colon cancers, but not in the Madin–Darby canine kidney epithelium, further showing the specificity of the antibody. Consistent with the immunohistochemistry study, a high level of EAPII expression is observed in nearly all lung carcinoma cell lines tested: In these NSCLC cells, two bands, 49 and 43, were recognized by anti-EAPII antibody, and 15 out of 17 lung cancer cell lines (88%) showed detectable EAPII protein levels, whereas barely detectable EAPII protein levels were observed in the immortalized bronchial epithelial cell lines HBEC3KT and BEAS2B (Figure 2b). Both 43 and 49 kDa bands are specifically recognized by multiple EAPII antibodies, suggesting that the 43-kDa band is likely a variant form of EAPII lacking the N-terminus (see Supplementary Figure S1). Consistently, these double bands of 43 and 49 kD EAPII protein can be seen in other types of cancer such as ovarian cancer (see Supplementary Figure S2).
Figure 2.
The expression of EAPII protein from various cancer cell lines was examined by western blot analyses. (a) EAPII is expressed in multiple cancer cell lines. The cell lines used included MCF7 (breast adenocarcinoma), MDA-MB-231 (breast adenocarcinoma), PC-3 (prostate adenocarcinoma), DU145 (prostate carcinoma), SK-HEP-1 (liver adenocarcinoma), SKVO3 (ovarian adenocarcinoma), DLD-1 (colorectal adenocarcinoma) and MDCK (Madin–Darby canine kidney) cell lines. Lysates extracted from cells overexpressing EAPII were used as EAPII marker (MARK). Protein marker is indicated, and β-actin was used as a loading control. (b) EAPII expression is determined in immortalized human bronchial epithelial cells (HBEC): (1) HBEC3KT, (2) BEAS2B and NSCLC cells (NSCLC), (3) H460, (4) H1792, (5) A549, (6) H1299, (7) H157, (8) H522, (9) H358, (10) Calu-1, (11) H226, (12) H292, (13) H322, (14) H1648, (15) H1650, (16) H1944, (17) H1975, (18) HCC827 and (19) SK-MES-1.
Overexpression of EAPII promotes cell proliferation in NSCLC cells
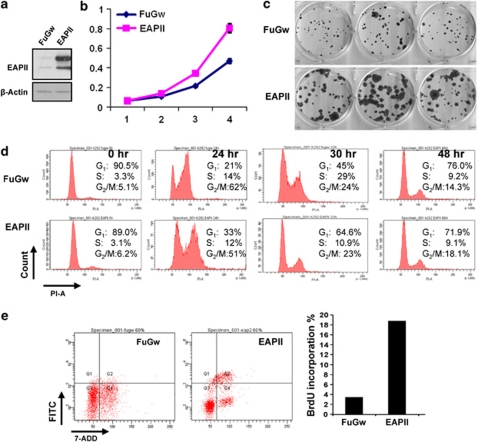
To demonstrate the oncogenic role of EAPII in lung cancer development, we enforced expression of exogenous EAPII in H292 cells, which have barely detectable endogenous EAPII protein. H292 cells were infected with lentiviral EAPII or a control lentiviral vector FuGw. Like the endogenous EAPII protein, the exogenous EAPII protein appears as two bands at 49 and 43 kDa, respectively (Figure 3a). Cell growth was analyzed by the sulforhodamine B method and by colony formation assays on plates in 10 days as described in Materials and methods. Compared with the cells containing the control vector (FuGw), EAPII-expressing cells grow significantly faster (Figure 3b) and form much larger clones (Figure 3c). To better understand the impact of EAPII overexpression on cell growth, we carried out the cell cycle analysis using flow cytometry. Ninety percent of cells were synchronized in the G1 phase after 72 h of serum starvation. After addition of serum, control cells (FuGw) sequentially proceed through S and G2/M phase, approaching normal distribution of the cell phase at 48 h, but EAPII-expressing cells return to normal distribution of the cell cycle at 30 h, when 29% of control cells are still in the S phase, suggesting that EAPII promotes G1/S transition, leading to a faster restoration of the normal distribution of cell cycle (Figure 3d). Because pulse labeling of cells by brief BrdU exposures is a sensitive measurement of cell proliferation, the BrdU assay was applied in our model. Compared with FuGw control, EAPII overexpression significantly enhanced the incorporation of BrdU into H292 cells from 4% to 19% after 40 min of BrdU incubation (Figure 3e), demonstrating that EAPII expression promotes cell proliferation.
Figure 3.
Overexpression of EAPII promotes proliferation of NSCLC cells and accelerates cell cycle progression. H292 cells were infected with lentiviral EAPII or lentiviral empty vector FuGw, and cell growth was analyzed by sulforhodamine B (SRB) method (b) and by colony formation assays on 12-well plates in 10 days (c). The protein level of EAPII in H292 clones was analyzed by western blot with antibody 42C. β-Actin was used as a loading control (a). EAPII-expressing and control (FuGw) H292 cells were deprived of serum for 72 h, and cells were released from G1 arrest by adding serum, then subjected to flow-cytometric analysis at the indicated time points (d). EAPII-expressing and control (FuGw) H292 cells were pulse-labeled with 10 μ BrdU for 40 min, and cells were stained with FITC-conjugated anti-BrdU and DNA-content marker 7-AAD, then subjected to flow-cytometric analysis. FACS histogram and the percentage of BrdU incorporation are shown (e).
EAPII knockdown induces apoptosis of NSCLC cells
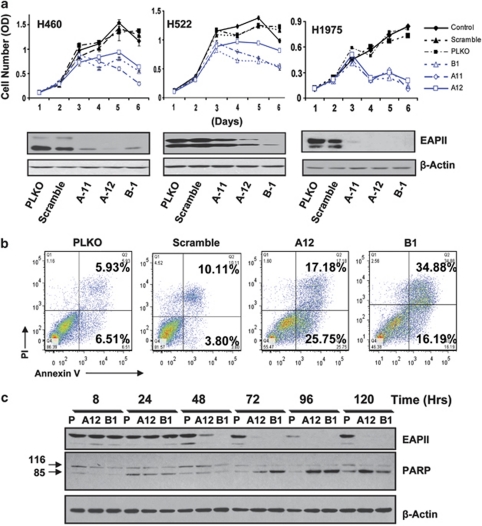
To determine whether the elevated EAPII protein in NSCLC cells is essential for proliferation and tumor growth, shRNA was employed to silence EAPII gene expression. Lentivirus-delivered EAPII shRNA was used to knock down EAPII expression in NSCLC cells. Three EAPII shRNAs were selected to target three regions of human EAPII sequences (accession no. AF201687) corresponding to the coding regions 300–320 (clone B1), 791–811 (clone A-11) and 992–1012 (clone A-12) relative to the first nucleotide of the start codon. All three shRNAs showed variable downregulation of EAPII proteins (both 49 and 43 kDa bands) in H460, H522 and H1975 lung cancer cells, whereas EAPII levels did not change in the vector (PLKO) or scramble control. Reduced cell growth was observed in cells with lower expressions of EAPII protein compared with cells infected with vector control or scramble shRNA (Figure 4a). To determine whether the reduced cell proliferation is due to apoptotic induction, ApoAlert Annexin V kit was employed to characterize the death feature of H1975 cells with EAPII knockdown. Clearly, a significant population of early and late apoptosis (40%) was observed in cells with EAPII knockdown compared with vector or scramble controls (10%) 72 h after virus infection (Figure 4b), demonstrating that EAPII knockdown results in apoptosis of the lung cancer cells. Furthermore, the time course of EAPII knockdown was determined by western blot. EAPII protein levels were significantly reduced at 48 h, with maximum reduction at 72 h. Consistent with the expected apoptosis induction by EAPII knockdown, detection of the cleaved form of poly(ADP-ribose) polymerase (PARP) protein (85 kDa), an early marker of apoptotic cell death (Kaufmann et al., 1993), tracked closely along with the time course of EAPII knockdown (Figure 4c) as significant difference of PARP cleavages between EAPII knockdown and the control group occurred in 72 h although trivial PARP cleavages were seen after 24 h in both control and EAPII knockdown groups due to cell spontaneous apoptosis or apoptosis initiated by the lentivirus infection.
Figure 4.
EAPII knockdown induces NSCLC cell apoptosis. H460, H522 and H1975 lung cancer cells were infected with lentivirus-shRNAEAPII (B1, A11, and A12), control vector (PLKO) or scramble-shRNA and cell growth curves were analyzed by sulforhodamine B (SRB) method (a). The efficiency of EAPII protein knockdown at 48 h after infection with shRNA was determined by western blot (shown below each growth curve). Apoptotic cell death was determined by flow-cytometric analysis with annexin V and PI staining. H1975 cells were infected with lentivirus-shRNAEAPII (B1 and A12), control vector (PLKO) or scramble-shRNA, harvested 72 h postinfection, and then subjected to apoptosis assay. The percentage of apoptosis, including early and late stage of apoptotic cell death in each group, is shown in the right panel (b). Time courses of EAPII knockdown and PARP induction after lentivirus infection were determined in H1975 cells by western blots in vector controls (P) and EAPII knockdown groups (A12 and B1). Protein marker is indicated on the left, and β-actin was used as a loading control (c).
Altered EAPII expression in NSCLC cells regulates growth of tumor xenografts in nude mice
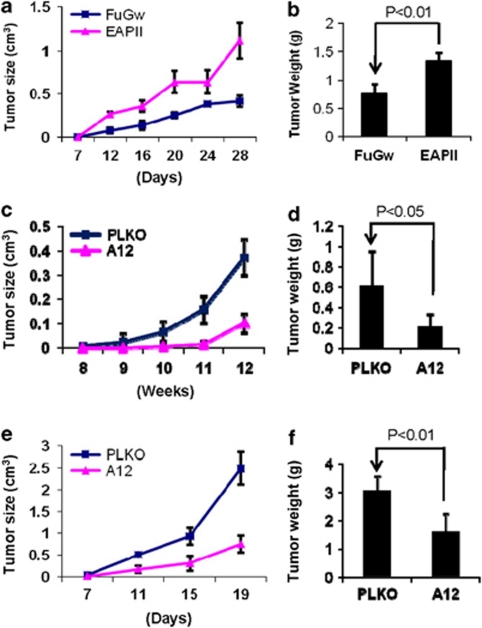
To examine the effect of the altered EAPII expression on the development and growth of lung cancer, EAPII-expressing and EAPII-knockdown cell models were employed in xenograft experiments in nude mice. EAPII-expressing and FuGw-control H292 cells were inoculated in nude mice, and their tumorigenicities were compared. Consistently, tumor growth was strongly enhanced in EAPII-expressing cells. At 4 weeks post-inoculation the average tumor volume was 0.3 cm3 in controls versus 1 cm3 in the EAPII-expressing group (Figure 5a). Similarly, harvested tumor weight was significantly increased in the EAPII-expressing group compared with the control group (Figure 5b). The effect of EAPII knockdown on tumor growth was determined in H1975 and H460 cells. The cells were infected with lentivirus-shRNAEAPII (A12) or control vector (PLKO). After 24 h, the cells were subcutaneously injected into the right flanks of the nude mice, and tumor formation was measured twice a week. As significant apoptotic induction in vitro occurred 3 days after lentivirus infection (Figure 4a), inoculation of the cells into mice within 24 h should provide the equivalent live cells for both EAPII knockdown and control groups at the starting point of the experiment. Xenograft H1975 tumors grew very slowly and became palpable in 8 weeks. Compared with control, EAPII knockdown significantly reduces tumor growth in both H1975 (Figure 5c) and H460 (Figure 5e) regardless of the tumor growth rate. A significant difference was further confirmed with the comparison of the average of the tumor weights in both xenograft tumors (Figures 5d and f). In addition, EAPII protein levels in the xenograft tumors were checked at the end of the experiments, and the result clearly showed that the EAPII expression is correlated with tumor growth (see Supplementary Figure S3). These results further demonstrated that EAPII is essential for lung cancer tumor growth and that the elevated expression of EAPII contributes to lung cancer development.
Figure 5.
Modulation of NSCLC xenograft growth by altered EAPII expression. H292 cells infected with control (FuGw) or EAPII (a, b), H1975 cells with control (PLKO) or shRNAEAPII (A12) (c, d), or H460 cells with control (PLKO) or shRNAEAPII (A12) (e, f) grew in nude mice. Tumors were measured twice a week unless otherwise indicated, and the mean tumor volume growth curves were compared (a, c, e). At the end of the experiments, the tumors were removed and weighed. The comparisons of final tumor weights were analyzed by the Student t-test. Results represent means±s.d. (n=6) (b, d, f).
Altered EAPII level regulates the activation of the MAPK–ERK pathway in lung cancer cells
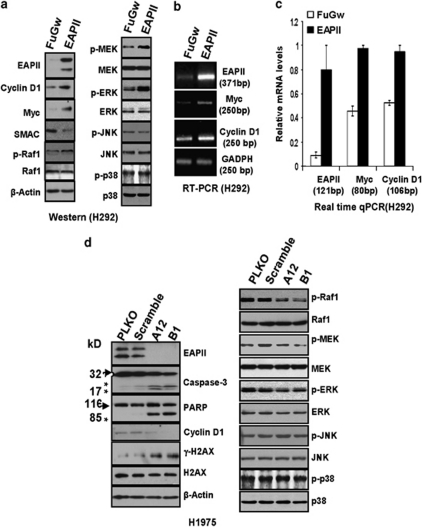
To determine the major molecular events induced by the altered EAPII expression, an array (cat #CSAA1, Sigma, St Louis, MO, USA) comprising 224 antibodies selected from a variety of pathways, including apoptotic and major cell signaling pathways, was used in the EAPII-expressing H292 and EAPII-knockdown H1975 cell models. The H292 cells with lentiviral EAPII or control vector FuGw serve as an oncogenic model and the H1975 cells with lentiviral shRNAEAPII or vector control (PLKO) serve as an apoptotic model, as H292 does not express endogenous EAPII and H1975 expresses robust levels of both the 43 and 49-kDa forms (Figure 2b). Cell lysates from each model were collected 48 h post-infection, and fluorescent dye labeling and hybridization were carried out according to the manufacturer's instruction (see Supplementary Materials and Methods). After normalization within and between arrays, a linear model was used to fit the data, and the log odds for each protein were calculated. The differentially expressed proteins or posttranslational modified forms were those with p-values less than 0.05: Upregulation of phospho-Raf1, phospho-ERK1, ERK5, MYC and cyclin D1 was identified in H292 cells with EAPII overexpression; downregulation of Bcl-xl, Bcl10, cyclin A, p16ink4a and p19ink4d and upregulation of caspase-3 active form and SMAC/DIABlO were identified in H1975 cells with EAPII knockdown (see Supplementary Figure S5, Table S1 and Table S2). To generally evaluate the array results, we further validated protein changes among a broad panel of altered proteins using western blot. In the EAPII-expression model, phospho-Raf1, phospho-MEK1/2 and phospho-ERK1/2 were increased, although the total protein level of each of these proteins remained the same. In contrast, we did not detect any change for the JNK and p38 MAPK pathways (phosphorylation state and total proteins) (Figure 6a), suggesting that the MAPK–ERK pathway is one of the downstream targets of EAPII-mediated signaling. Furthermore, MYC and cyclin D1 were upregulated and SMAC was downregulated in this model (Figure 6a). MYC and cyclin D1 are the downstream events in the MAPK–ERK pathway. This prompted us to further determine whether MYC and cyclin D1 are regulated at the mRNA level. To this end, reverse transcription–polymerase chain reaction (RT–PCR) was used to amplify specific target genes, and indeed, results showed the elevated MYC and cyclin D1 at transcript level (Figure 6b). Furthermore, real-time qRT–PCR was performed and the results showed that EAPII overexpression increased Myc and cyclin D1 expression levels by 2.13 and 1.68-fold, respectively (Figure 6c).
Figure 6.
Protein regulation in EAPII overexpression and EAPII knockdown models. Cell lysates were collected from H292 cells with control or EAPII lentivirus after 72 h of infection and subjected to western blots for the indicated proteins (a). Total RNA, extracted from H292 cells with control or EAPII lentivirus after 72 h of infection, was used for RT–PCR. PCR analyses of EAPII, MYC, cyclin D1 and GADPH were conducted under the following conditions: EAPII, 52 °C, 28 cycles; MYC, 52 °C, 28 cycles; cyclin D1, 52 °C, 28 cycles; and GADPH, 55 °C, 28 cycles. Each of these genes and the size of the products are indicated (b). Real-time qRT–PCR was used to measure the relative gene expression of EAPII, MYC and cyclin D1 in H292 cells infected with lentiviral EAPII and the FuGw control. Relative gene expression for each gene was obtained by dividing the expression of each gene by the expression of GADPH. The size of each amplicon is indicated. These results represent at least two independent experiments (c). Cell lysates were collected from H1975 cells with control (PLKO, Scramble) or EAPII (A12 and B1) shRNA-lentivirus after 72 h of infection and then subjected to western blot of the indicated proteins (d). Protein marker is indicated on the left, and the asterisks indicate the cleaved forms of caspase-3 or PARP proteins. β-Actin was used as a loading control.
In the EAPII knockdown-induced apoptosis model we validated the upregulation of caspase-3 active form alone with the downregulation of both 49 and 43 kDa bands of EAPII. Downregulation of cyclin D1 was also observed (Figure 6d). PARP cleavage was also observed along with cell death. Consistent with the observed effect of EAPII overexpression, reduction of the phospho-Raf1, phospho-MEK1/2, and phospho-ERK1/2 occurred in this EAPII-knockdown model although total protein level remained unchanged (Figure 6d). To determine whether ERK activity is important for EAPII's function, we further tested the effects of MEK inhibitor PD98059 on the proliferative role of EAPII. The result showed that treatment with MEK inhibitor PD98059 significantly reduced the phosphorylation of ERK1/2 and abolished the cell growth advantage induced by ectopic expression of EAPII (see Supplementary Figure S4).
In addition, we determined the phosphorylated form of H2A histone family member X (γ-H2AX), which is a marker of DNA double-strand break (Fernandez-Capetillo et al., 2004). With EAPII knockdown, accumulation of γ-H2AX was increased although total H2AX remained unchanged (Figure 6d), supporting the recent discovery that EAPII participates in DNA repair processes for double-strand breaks (Cortes Ledesma et al., 2009).
Discussion
In this study, we identified elevated expression of EAPII protein in lung cancer tissues and NSCLC cell lines, which suggests the biological significance of EAPII expression during lung cancer development. We further demonstrated that forced alteration of EAPII expression modulated cell proliferation in vitro and xenograft lung cancer in mice. These observations support our hypothesis that EAPII has an oncogenic role in lung cancer development. Additionally, using antibody array and western blots we identified the activation of the Raf1–MEK1/2–ERK1/2 cascade, as evidenced by the phosphorylation of Raf1, MEK1/2 and ERK1/2, demonstrating the role of EAPII in the regulation of MAPK–ERK pathway. Consistently, MAPK–ERK activation leads to transcriptional regulation of MYC and cyclin D1, resulting in increased cell proliferation, accelerated G1/S transition and tumor formation, suggesting that the ERK–MYC–cyclin D1 axis can be, at least partly, an oncogenic mechanism by which EAPII contributes to lung cancer development.
NSCLC constitutes around 85% of all lung tumors (Molina et al., 2008). A diverse range of genetic abnormalities is observed in lung cancer cells, which may include most common alterations such as inactivation of the p16/pRb pathway, expression of hTERT and TP53 mutations, and less common genetic changes in NSCLC (Singhal et al., 2008; Blanco et al., 2009). These genetic changes may have a direct role in lung cancer etiology or an indirect role in regulating cancer-related signal networks in the context of gene–environment interactions (Borczuk et al., 2009; Sanchez-Cespedes, 2009). The complexity of cancer demands a comprehensive understanding of the integrated signaling networks. Overexpression of EAPII occurs in over 80% of NSCLCs, suggesting that EAPII may have a role in lung cancer etiology. Among the EAPII-expressing tissues, 84% (78/93) of lung carcinomas showed cytoplasmic staining of EAPII. In particular, we observed that nuclear staining of EAPII occurs in the highly proliferative tissue, and the majority of lung carcinomas showed cytoplasmic staining or cytoplasmic and nuclear staining (Figure 1d). This observation suggests that cytoplasmic EAPII may be more closely related to its potential oncogenic role. It has recently been shown that EAPII functions as a 5′-Tyr-DNA phosphodiesterase and promotes DNA topological conformational changes during DNA synthesis (Cortes Ledesma et al., 2009). Therefore, it is possible that an elevated nuclear expression of EAPII favors enhanced proliferative activity in both normal bronchial epithelium and tumor cells, implying that EAPII may have different roles in the cytoplasm or nucleus. In addition, two bands (49 and 43 kDa) of EAPII protein were detected for both endogenous (Figure 2) and exogenous EAPII (Figure 3a), suggesting that EAPII variant forms may exist. It is currently unknown how the function and subcellular distribution are associated with the specific form. However, forced expression of the exogenous EAPII provided both forms of EAPII and EAPII knockdown silenced both of them (Figures 3a and 4a), which allows us to analyze the oncogenic role of EAPII as a whole. Undoubtedly, these observations also raised critical questions such as: (1) Is the short form the cleaved product or a variant form? (2) Does the short form function differently? An independent study is warranted to fully address these questions. The oncogenic role of EAPII was first suggested by its pro-proliferation and pro-tumor activity in H292 and H1650 cells with EAPII overexpression (Figures 3 and 5) and further validated by the indispensable role of EAPII in the survival of lung cancer cells using EAPII knockdown technique (Figures 4 and 5). Protein sequence alignment of EAPII indicates that EAPII contains an Exo_endo_phos domain (88–332 aa). Exo_endo_phos is a conserved domain retaining all six motifs that are hallmarks for an endonuclease/exonuclease/phosphatase superfamily (Exo_endo_phos, PF03372) (Rodrigues-Lima et al., 2001). Many lines of evidence indicate that EAPII may be a component of the signal transduction pathway with functions other than the removal of 5′-phosphate termini at double-strand DNA break: (1) EAPII is associated with multiple signal transduction pathways, including the TNF (CD40) (Pype et al., 2000) and TGF-β receptor (ALK4) (Esguerra et al., 2007). EAPII modulates the transcriptional activity of multiple transcription factors. Besides ETS1, EAPII directly interacts with SMAD3, which is a transcription factor that mediates TGF-β1 signaling and represses its activity (Esguerra et al., 2007). (2) EAPII also responds to extracellular stress. The expression of EAPII could be induced by IFN-γ (Xu et al., 2008). (3) EAPII is differentially localized in the nucleus and/or the cytoplasm. EAPII has a role in DJ-1 mutant-mediated apoptosis through activation of JNK1/2 and p38 MAPK pathways in neuroblastoma cells (Zucchelli et al., 2009). In the present study, however, overexpression of EAPII in lung cancer cells activates the Raf–MEK–ERK cascade, but not the JNK or p38 MAPK pathway (Figure 6). These results not only reveal the involvement of EAPII in signaling transduction but also provide a potential mechanism of the oncogenic role of EAPII: activation of MAPK–ERK pathways. It is well known that the Ras–MAPK signaling pathway regulates cell proliferation (Karnoub and Weinberg, 2008) and the activation of Ras oncogene causes lung carcinoma in mice (Johnson et al., 2001). We speculated that elevated EAPII in lung cancer is one of the genetic alterations that provide constitutive activation of Ras–MAPK signaling and subsequently drive cell proliferation and tumor formation. In support of this hypothesis, upregulation of MYC and cyclin D1 is observed in EAPII-expressing cells. This is a logical outcome of the activation of the MAPK–ERK pathway, as both MYC and cyclin D1 are oncogenic factors implicated in lung cancer development (Little et al., 1983; Schauer et al., 1994; Mitani et al., 2001; Gautschi et al., 2007; Rapp et al., 2009). MYC promotes cell proliferation by transcriptionally regulating a broad spectrum of genes (Eilers and Eisenman, 2008), and cyclin D1 interacts with cell cycle machinery by promoting G1/S transition (Aktas et al., 1997). In contrast to the proapoptosis activity of EAPII in neuroblastoma cells, EAPII provides a promalignant activity through the MAPK–ERK pathway.
EAPII knockdown induced severe apoptosis of H1975 cells and moderate apoptosis in H522 and H460 (Figure 4). Apoptotic induction by EAPII knockdown was confirmed by PARP cleavage and activation of caspase-3 (Figure 4c). It is likely that these lung cancer cells are highly dependent on the activity of EAPII in continued cell proliferation. This phenomenon is called ‘oncogene addiction' (Hubner et al., 2006). Although the mechanism of ‘oncogene addiction' is not fully understood (Weinstein and Joe, 2008), results from the antibody array and western blot suggested that apoptotic induction by silencing EAPII could be due to (1) blockage of the survival signal such as MAPK–ERK1/2 or Bcl-xl and (2) upregulation of pro-apoptosis proteins such as SMAC/DIABLO. Elevated EAPII in lung cancer may provide pro-survival signals necessary for the aberrant growth of lung cancer cells through a constitutive activation of RAS–ERK–MAPK pathway, which is known to modulate survival factors such as nuclear factor κB. In addition, both MYC and cyclin D1 are downstream targets of ERK–MAPK pathway (Chang et al., 2003), and many cancer cells have been shown to have ‘oncogene addiction' to these genes (Weinstein and Joe, 2008; Lee et al., 2010).
Contrary to previous reports showing that EAPII represses the activation of transcription factors including ETS1 (Pei et al., 2003) and nuclear factor κB (Pype et al., 2000), EAPII provides a different scenario in the current setting with lung cancer. The possible causes for the discrepancy are (1) cell-context specificity and (2) the limit of analysis methods, in which the read-out was mainly based on the luciferase reporter assays. It is noteworthy that accumulation of γ-H2AX, a marker for DNA double-strand breaks, occurs in lung cancer cells following EAPII knockdown, supporting the claim of the enzymatic activity of 5′-Tyr-DNA phosphodiesterase of EAPII (Cortes Ledesma et al., 2009). Therefore, we had speculated that accumulation of double-strand breaks by EAPII knockdown might lead to apoptosis. However, our results showed that EAPII knockdown induces apoptosis in both cells with wild-type p53 (H1975, H460) and with mutant p53 (H522), and neither antibody array nor western blot analysis showed a significant augmentation in p53 level or its phosphorylation status (data not shown). These results suggest that lack of EAPII is not sufficient to trigger p53-dependent apoptosis in this case, although it is possible that EAPII knockdown-induced apoptosis can be boosted by p53-dependent or -independent pathways in cells exposed to genomic insults or DNA-damaging agents. Further exploring the biochemical role of EAPII in the regulation of signaling networks and the downstream target(s) will be critical to understanding the biological significance of EAPII.
In conclusion, our results showed that EAPII exerts proproliferative and antiapoptotic activities in lung cancer development, and EAPII is involved in the modulation of MAPK–ERK signaling. Lung cancer cell growth and survival can be impaired by the inactivation of EAPII, suggesting the oncogenic role of EAPII and potentially a novel target in chemointervention.
Materials and methods
Cell lines, transfections, western blots and antibodies
The NSCLC cell lines H460, H1792, A549, H1299, H157, H522, H358, Calu-1, H226, H292, H322, H1648, H1650, H1944, H1975, HCC827 and SK-MES-1 were purchased from ATCC and cultured as previously described (Li et al., 2010). The immortalized human bronchial epithelial cell line HBEC3KT was kindly provided by Dr JD Minna (University of Texas Southwestern Medical Center, Dallas, TX, USA) and BEAS2B by Dr R Lotan (University of Texas MD Anderson Cancer Center, Houston, TX, USA). Western blots were developed by enhanced chemiluminescence. Anti-EAPII rabbit polyclonal antibody (220) and monoclonal antibody (42C) were previously described (Pei et al., 2003). Antibodies against activated caspase-3 (no. 9662), PARP (no. 9542), phospho-Raf1 (no. 9421), phospho-MEK/1/2 (no. 9154), phospho-ERK1/2 (no. 9101) and phospho-p38MAPK (no. 9215) were purchased from Cell Signaling, Inc. (Danvers, MA, USA); phospho-JNK1/2/3(sc-6254), MYC(sc-764) and γ-H2AX(sc-101696) from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA); and cyclin D1(#MS-210-P0) from Lab Vision Products (Fremont, CA, USA).
Immunohistochemistry study and tissue array
Paraffin-embedded lung carcinoma tissue sections were obtained from the Tumor Bank at the Winship Cancer Institute of Emory University with approval from Emory IRB. Tissue array was purchased from AccuMax (ISU ABXIS Co., Ltd.), which contains 103 cases of lung carcinoma and 44 adjacent non-neoplastic tissues. All samples were anonymous, and the diagnosis of normal or tumor tissue was reconfirmed by one of the authors (Fan, pathologist). Immunohistochemical staining was done as follows: Sections were deparaffinized and rehydrated; microwave-heated antigen retrieval was performed in 10 m citrate buffer, pH 6.0, for 3 × 5 min followed by incubation in proteinase K (DAKO, Carpinteria, CA, USA) for 10 min. Sections were treated with 3% H2O2 for 10 min to quench endogenous peroxidase activity and blocked with 1.5% normal serum for 20 min. 42C (a monoclonal EAPII antibody) was incubated at a dilution of 1:50 at room temperature for 1 h. Secondary biotinylated anti-mouse antibody was used at a dilution of 1:200 (Vector Laboratories, Burlingame, CA, USA) for 30 min at room temperature. The sections were counterstained with hematoxylin. A mouse negative IgG (DAKO) was used as a negative control in each staining process.
Generation of EAPII-expressing or shRNA lentiviruses
EAPII-expressing lentiviruses (EAPII-FuGw) were generated in the FuGw vector, a self-inactivating and replication-incompetent lentiviral green fluorescent protein-expressing vector (Lois et al., 2002), by inserting the PCR fragment of EAPII with BamHI/EcoRI sites into a FuGw to replace green fluorescent protein DNA. FuGw was used as a control. Lentiviral shRNAs against human EAPII (AF201687), empty vector control (pLKO.1 empty vector control) and the control lentiviral scrambled shRNAs were purchased from The RNA Consortium lentiviral shRNA library (TRC-Hs1.0) (Open Biosystems, Huntsville, AL, USA). 293FT cells were seeded in a 10-cm culture dish in DMEM medium supplemented with 10% fetal bovine serum (Sigma), -glutamine (10 ml/l), penicillin and streptomycin (100 units/ml) the night before transfection. 293FT cells were transfected at a confluence of 40–50% with 8.5 μg each of the lentiviral vectors along with 4.2 μg of pVSVG and 6.25 μg of Δ8.91 plasmids using Fugene VI (Qiagen, Valencia, CA, USA) as per the manufacturer's directions, and 20% FBS was added 24 h post-transfection. Cell supernatant was collected 48 h post-transfection and then used for further infections.
Cell proliferation, colony formation, cell cycle and BrdU assays
Cell proliferation assay
Cells were seeded at densities of approximately 2 × 103 cells per well in 96-well tissue culture plates. Surviving cell numbers were estimated by sulforhodamine B assay as previously described (Li et al., 1999). Colony formation assays were carried out as described previously (Wang et al., 2008). Briefly, cells (single-cell suspension) were plated in six-well plates at a density of 100 per well. The medium was replaced every 3 days, and cell colonies were stained for 14 days with crystal violet (0.1% in 20% methanol). Pictures were taken using a digital camera to record the result. Cell cycle analysis was performed by propidium iodide staining: 5 × 105 cells were harvested and fixed in 70% ethanol overnight; cells were washed twice in cold phosphate-buffered saline and resuspended in 1 ml of phosphate-buffered saline containing propidium iodide (20 mg/ml) and 50 μl RNase A (10 mg/ml). Cells were subjected to fluorescence-activated cell sorting (FACS) analysis using BD FACSCanto II (32) (BD Biosciences, San Jose, CA, USA). Annexin V-PE apoptosis detection kit (BD Biosciences, San Jose, CA, USA) was used to detect apoptotic cells as previously described (Pei et al., 2005). The rate of DNA synthesis was determined by the BrdU Flow Kit (BD Biosciences, San Diego, CA, USA) according to the manufacturer's protocol. Briefly, 1 × 106 cells were seeded in 10-cm dishes. After overnight culture, cells (∼50% confluent) were labeled with 10 μ BrdU for 40 min, harvested by trypsinization, and then fixed and permeabilized with BD Cytofix/Cytoperm buffer (BD Biosciences, San Jose, CA, USA). The incorporated BrdU was immunostained with FITC-conjugated anti-BrdU, and 7-AAD was used to stain DNA contents. Cells were subjected to FACS analysis. FLOWJO software (Ashland, OR, USA) was used for flow cytometry analysis.
Reverse transcription–PCR and real-time qRT–PCR analysis
Reverse transcription–PCR assays were performed as previously described (Li et al., 1999). First-strand cDNA was produced from total RNA, extracted from H292 cells with control or EAPII lentivirus after 48 h of infection using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA), and were PCR-amplified with primers specific to EAPII, MYC, cyclin D1 or glyceraldehyde-3-phosphate dehydrogenase (GADPH): MYC-F, 5′-CCTCAACGTTAGCTTCACCAA-3′ MYC-R, 5′-TTTGATGAAGGTCTCGTCGTC-3′ cyclin D1–F, 5′-CCCTCGGTGTCCTACTTCAAA-3′ cyclin D1–R, 5′-CCAGGTTCCACTTGAGCTTGT-3′ EAPII-F, 5′-GAATGTGTCAGGAAATGAGC-3′ EAPII-R, 5′-AATAATGTGTCCCTCTTCTGC-3′ GADPH-F, 5′-CACAGTCAAGGCTGAGAATGGGAA-3′ and GADPH-R, 5′-GTGGTTCACACCCATCACAAACAT-3′. Real-time amplification was performed in a final volume of 25 μl, containing 30 ng of cDNA from the reversed transcribed reaction, primer mixture (0.3 μ each of sense and antisense primers) and 12.5 μl of 2 × SYBR Green Master Mix (A&B Applied Biosystems, Foster City, CA, USA) using the Sequence Detector System (7000 Sequence Detection System and software; A&B Applied Biosystems). The following primers were used: MYC-F, 5′-TCAAGAGGTGCCACGTCTCC-3′ MYC-R, 5′-TCTTGGCAGCAGGATAGTCCTT-3′ GAPDH-F, 5′-TTCAACAGCGACACCCACTC-3′ GAPDH-R, 5′-GTGGTCCAGGGGTCTTACTC-3′ EAPII-F, 5′-CTGCTGCTTGTAAACTTCGT-3′ EAPII-R, 5′-AGGAAATCTACCACAGTCCAG-3′ cyclin D1-F, 5′-CCTCGGTGTCCTACTTCAAA-3′ cyclin D1-R, 5′-CTCCTCGCACTTCTGTTCCT-3′. The standard amplification program, comprising 40 cycles of 15 s at 95 °C and 30 s at 60 °C, was used. Fluorescent product was detected at the last step of each cycle. The relative level of each amplicon was normalized by GADPH mRNA level using the comparative CT method (Schmittgen and Livak, 2008).
Xenograft tumor model in nude mice
Under an IACUC-approval protocol, 2 × 106 cells in 100 μl of phosphate-buffered saline of each cell line with altered EAPII expression, and controls were injected subcutaneously into the right anterior flank of 6- to 8-week-old athymic nude mice (Charles River Laboratories, Wilmington, DE, USA). Tumors were measured twice weekly, unless otherwise indicated, with a calipers, and tumor volumes were calculated using the formula [0.52(L × S2)], where L represents the largest tumor diameter and S represents the smallest tumor diameter as previously described (Pei et al., 2005).
Acknowledgments
We thank Drs JD Minna (University of Texas Southwestern Medical Center, Dallas) and R Lotan (University of Texas MD Anderson Cancer Center, Houston) for providing HBEC3KT and BEAS2B cell lines and Biomolecular Computing Resource (BIMCORE) at the Emory University School of Medicine for assistance with antibody array analysis. This work was supported in part by National Institutes of Health grants K22CA109577 (RL) and RO1CA118450 (SYS), a start-up fund from the Department of Hematology and Medical Oncology, Emory University (RL).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Aktas H, Cai H, Cooper GM. Ras links growth factor signaling to the cell cycle machinery via regulation of cyclin D1 and the Cdk inhibitor p27KIP1. Mol Cell Biol. 1997;17:3850–3857. doi: 10.1128/mcb.17.7.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco R, Iwakawa R, Tang M, Kohno T, Angulo B, Pio R, et al. A gene-alteration profile of human lung cancer cell lines. Hum Mutat. 2009;30:1199–1206. doi: 10.1002/humu.21028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borczuk AC, Toonkel RL, Powell CA. Genomics of lung cancer. Proc Am Thorac Soc. 2009;6:152–158. doi: 10.1513/pats.200807-076LC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Steelman LS, Lee JT, Shelton JG, Navolanic PM, Blalock WL, et al. Signal transduction mediated by the Ras/Raf/MEK/ERK pathway from cytokine receptors to transcription factors: potential targeting for therapeutic intervention. Leukemia. 2003;17:1263–1293. doi: 10.1038/sj.leu.2402945. [DOI] [PubMed] [Google Scholar]
- Cortes Ledesma F, El Khamisy SF, Zuma MC, Osborn K, Caldecott KW. A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 2009;461:674–678. doi: 10.1038/nature08444. [DOI] [PubMed] [Google Scholar]
- Eilers M, Eisenman RN. Myc′s broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esguerra CV, Nelles L, Vermeire L, Ibrahimi A, Crawford AD, Derua R, et al. Ttrap is an essential modulator of Smad3-dependent Nodal signaling during zebrafish gastrulation and left-right axis determination. Development. 2007;134:4381–4393. doi: 10.1242/dev.000026. [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. H2AX: the histone guardian of the genome. DNA Repair (Amst) 2004;3:959–967. doi: 10.1016/j.dnarep.2004.03.024. [DOI] [PubMed] [Google Scholar]
- Gautschi O, Ratschiller D, Gugger M, Betticher DC, Heighway J. Cyclin D1 in non-small cell lung cancer: a key driver of malignant transformation. Lung Cancer. 2007;55:1–14. doi: 10.1016/j.lungcan.2006.09.024. [DOI] [PubMed] [Google Scholar]
- Hubner A, Jaeschke A, Davis RJ. Oncogene addiction: role of signal attenuation. Dev Cell. 2006;11:752–754. doi: 10.1016/j.devcel.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann SH, Desnoyers S, Ottaviano Y, Davidson NE, Poirier GG. Specific proteolytic cleavage of poly(ADP-ribose) polymerase: an early marker of chemotherapy-induced apoptosis. Cancer Res. 1993;53:3976–3985. [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, de The H. PML nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2:a000661. doi: 10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Youn SW, Kim JY, Park KW, Hwang CI, Park WY, et al. FOXO3a turns the tumor necrosis factor receptor signaling towards apoptosis through reciprocal regulation of c-Jun N-terminal kinase and NF-kappaB. Arterioscler Thromb Vasc Biol. 2008;28:112–120. doi: 10.1161/ATVBAHA.107.153304. [DOI] [PubMed] [Google Scholar]
- Lee JT, Shan J, Gu W. Targeting the degradation of cyclin D1 will help to eliminate oncogene addiction. Cell Cycle. 2010;9:857–858. doi: 10.4161/cc.9.5.11175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Chen S, Yue P, Deng X, Lonial S, Khuri FR, et al. Proteasome inhibitor PS-341 (bortezomib) induces calpain-dependent IkappaB(alpha) degradation. J Biol Chem. 2010;285:16096–16104. doi: 10.1074/jbc.M109.072694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Pei H, Papas T. The p42 variant of ETS1 protein rescues defective Fas-induced apoptosis in colon carcinoma cells. Proc Natl Acad Sci USA. 1999;96:3876–3881. doi: 10.1073/pnas.96.7.3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little CD, Nau MM, Carney DN, Gazdar AF, Minna JD. Amplification and expression of the c-myc oncogene in human lung cancer cell lines. Nature. 1983;306:194–196. doi: 10.1038/306194a0. [DOI] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Mitani S, Kamata H, Fujiwara M, Aoki N, Tango T, Fukuchi K, et al. Analysis of c-myc DNA amplification in non-small cell lung carcinoma in comparison with small cell lung carcinoma using polymerase chain reaction. Clin Exp Med. 2001;1:105–111. doi: 10.1007/s10238-001-8020-5. [DOI] [PubMed] [Google Scholar]
- Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83:584–594. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagakubo D, Taira T, Kitaura H, Ikeda M, Tamai K, Iguchi-Ariga SM, et al. DJ-1, a novel oncogene which transforms mouse NIH3T3 cells in cooperation with ras. Biochem Biophys Res Commun. 1997;231:509–513. doi: 10.1006/bbrc.1997.6132. [DOI] [PubMed] [Google Scholar]
- Pei H, Yordy JS, Leng Q, Zhao Q, Watson DK, Li R. EAPII interacts with ETS1 and modulates its transcriptional function. Oncogene. 2003;22:2699–2709. doi: 10.1038/sj.onc.1206374. [DOI] [PubMed] [Google Scholar]
- Pei H, Li C, Adereth Y, Hsu T, Watson DK, Li R. Caspase-1 is a direct target gene of ETS1 and plays a role in ETS1-induced apoptosis. Cancer Res. 2005;65:7205–7213. doi: 10.1158/0008-5472.CAN-04-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pype S, Declercq W, Ibrahimi A, Michiels C, Van Rietschoten JG, Dewulf N, et al. TTRAP, a novel protein that associates with CD40, tumor necrosis factor (TNF) receptor-75 and TNF receptor-associated factors (TRAFs), and that inhibits nuclear factor-kappa B activation. J Biol Chem. 2000;275:18586–18593. doi: 10.1074/jbc.M000531200. [DOI] [PubMed] [Google Scholar]
- Rapp UR, Korn C, Ceteci F, Karreman C, Luetkenhaus K, Serafin V, et al. MYC is a metastasis gene for non-small-cell lung cancer. PLoS One. 2009;4:e6029. doi: 10.1371/journal.pone.0006029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Lima F, Josephs M, Katan M, Cassinat B. Sequence analysis identifies TTRAP, a protein that associates with CD40 and TNF receptor-associated factors, as a member of a superfamily of divalent cation-dependent phosphodiesterases. Biochem Biophys Res Commun. 2001;285:1274–1279. doi: 10.1006/bbrc.2001.5328. [DOI] [PubMed] [Google Scholar]
- Salomoni P. Stemming out of a new PML era. Cell Death Differ. 2009;16:1083–1092. doi: 10.1038/cdd.2009.63. [DOI] [PubMed] [Google Scholar]
- Sanchez-Cespedes M. Lung cancer biology: a genetic and genomic perspective. Clin Transl Oncol. 2009;11:263–269. doi: 10.1007/s12094-009-0353-7. [DOI] [PubMed] [Google Scholar]
- Schauer IE, Siriwardana S, Langan TA, Sclafani RA. Cyclin D1 overexpression vs. retinoblastoma inactivation: implications for growth control evasion in non-small cell and small cell lung cancer. Proc Natl Acad Sci USA. 1994;91:7827–7831. doi: 10.1073/pnas.91.16.7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Singhal S, Miller D, Ramalingam S, Sun SY. Gene expression profiling of non-small cell lung cancer. Lung Cancer. 2008;60:313–324. doi: 10.1016/j.lungcan.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Yue P, Kim YA, Fu H, Khuri FR, Sun SY. Enhancing mammalian target of rapamycin (mTOR)-targeted cancer therapy by preventing mTOR/raptor inhibition-initiated, mTOR/rictor-independent Akt activation. Cancer Res. 2008;68:7409–7418. doi: 10.1158/0008-5472.CAN-08-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–3080. doi: 10.1158/0008-5472.CAN-07-3293. [DOI] [PubMed] [Google Scholar]
- Xu GL, Pan YK, Wang BY, Huang L, Tian L, Xue JL, et al. TTRAP is a novel PML nuclear bodies-associated protein. Biochem Biophys Res Commun. 2008;375:395–398. doi: 10.1016/j.bbrc.2008.08.023. [DOI] [PubMed] [Google Scholar]
- Zhang ZB, Hu J, Bi YY, Zhao ZW, Tao N, Yan H, et al. Effect of TTRAP expression on apoptosis induced by hydroquinone in HL-60 cells in vitro. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi. 2007;25:654–656. [PubMed] [Google Scholar]
- Zucchelli S, Vilotti S, Calligaris R, Lavina ZS, Biagioli M, Foti R, et al. Aggresome-forming TTRAP mediates pro-apoptotic properties of Parkinson's disease-associated DJ-1 missense mutations. Cell Death Differ. 2009;16:428–438. doi: 10.1038/cdd.2008.169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.