Abstract
Overexpression of S100A4, a member of the S100 family of Ca2+-binding proteins, is associated with a number of human pathologies, including fibrosis, inflammatory disorders, and metastatic disease. The identification of small molecules that disrupt S100A4/target interactions provides a mechanism for inhibiting S100A4-mediated cellular activities and their associated pathologies. Using an anisotropy assay that monitors the Ca2+-dependent binding of myosin-IIA to S100A4, NSC 95397 was identified as an inhibitor that disrupts the S100A4/myosin-IIA interaction and inhibits S100A4-mediated depolymerization of myosin-IIA filaments. Mass spectrometry demonstrated that NSC 95397 forms covalent adducts with Cys81 and Cys86, which are located in the canonical target binding cleft. Mutagenesis studies showed that covalent modification of just Cys81 is suffcient to inhibit S100A4 function with respect to myosin-IIA binding and depolymerization. Remarkably, substitution of Cys81 with serine or alanine significantly impaired the ability of S100A4 to promote myosin-IIA filament disassembly. As reversible covalent cysteine modifications have been observed for several S100 proteins, we propose that modification of Cys81 may provide an additional regulatory mechanism for mediating the binding of S100A4 to myosin-IIA.
The S100 proteins, of which there are 21 family members in humans, are small acidic proteins characterized by two EF-hand Ca2+-binding loops: an N-terminal pseudo EF-hand comprised of 14 residues (EF1) and a C-terminal canonical EF-hand (EF2) comprised of 12 residues.1,2 The majority of S100 proteins exist as either symmetric, antiparallel homodimers, or antiparallel heterodimers, in which the N- and C-terminal helices (helices 1 and 4) from each subunit form a tight four helix bundle that constitutes the dimer interface. S100-target protein interactions are mediated by Ca2+-binding to the C-terminal EF-hand, which is flanked by helices 3 and 4. Calcium-binding to EF2 induces a conformational rearrangement that alters the angle between helices 3 and 4 and exposes a hydrophobic cleft that provides a binding surface for target proteins.3−5 As a consequence, S100 proteins function as calcium-activated switches that bind and modulate the activity of their biological targets.
S100 proteins are distributed in a cell- and tissue-specific manner.6 Given the diversity of S100 protein targets, these proteins modulate multiple cellular processes, including the regulation of protein phosphorylation and enzyme activity, cytoskeletal dynamics, cell growth and differentiation, and inflammatory responses.6,7 In addition to the regulation of normal physiology, increased expression of specific S100 family members is associated with human diseases such as cancer, cardiac disease, and neurodegenerative and inflammatory disorders.8 In particular, high S100A4 expression levels are associated with pathological conditions that involve fibrosis and inflammation, including kidney and liver fibrosis, rheumatoid arthritis, and cardiac hypertrophy.9,10 Moreover, S100A4 overexpression is a marker for poor prognosis in a number of human cancers and in mouse models of cancer S100A4 has been shown to promote the development of metastatic disease.11,12 A common feature of these pathologies is invasive cell motility; thus, inhibition of S100A4-mediated motile processes may afford a novel therapeutic strategy for many of these diseases.
S100A4 has a number of reported Ca2+-dependent protein targets, including tropomyosin,13 myosin-IIA,14 p53,15 and liprin β1.16 We have focused on the identification of compounds that inhibit the Ca2+-dependent binding of S100A4 to myosin-IIA, as this interaction is an established regulator of cell motility.17−19 Previously, we identified trifluoperazine as an inhibitor that disrupts the S100A4/myosin-IIA interaction by sequestering S100A4 via small molecule-induced oligomerization.20 We report here the identification of small molecules that inhibit S100A4 binding to myosin-IIA due to covalent modification of cysteine residues in the target binding cleft. Moreover, we demonstrate that the sulfhydryl functionality of Cys81 is critical for the S100A4/myosin-IIA interaction. These findings suggest that modification of Cys81 could function as an additional in vivo regulatory mechanism for mediating S100A4 binding to myosin-IIA and other S100A4 protein targets.
EXPERIMENTAL PROCEDURES
Purification of S100A4
For the production of the untagged protein, the codon-optimized human S100A4 sequence was subcloned into the Nde1/BamH1 sites of pET11b (Novagen). Mutation of C81 to alanine or serine was performed by QuikChange Site-Directed Mutagenesis (Stratagene). All amino acid substitutions were verified by DNA sequencing. BL21(DE3)* cells were transformed with the codon-optimized human WT S100A4 or C81 mutants, and cultures were grown in Luria broth at 37 °C to an OD600 of 0.8−1.0. Protein expression was induced with 0.7 mM IPTG, and the cultures were grown for 18−22 h at 27 °C. The cells were harvested at 8000 rpm for 10 min, and the cell pellets were resuspended in 15 mL of lysis buffer (50 mM Tris pH 7.5, 10% glycerol, 300 mM KCl, 5 mM DTT, 1 mM EDTA, 1 mM PMSF and 5 μg/mL each of chymostatin, leupeptin, and pepstatin) per cell pellet from 1.0 L of cell culture. The cell lysates were frozen at −80 °C, thawed on ice, and sonicated. Following centrifugation of the lysate at 30000g for 30 min, ammonium sulfate was added to the supernatant to 42% saturation; the sample was stirred for 20 min on ice and centrifuged at 30000g for 10 min. CaCl2 was added to the supernatant to a final concentration of 2 mM, and the sample was applied to a Phenyl-Sepharose column (GE Healthcare) equilibrated in buffer P (20 mM Tris pH 7.5, 2 mM CaCl2, 300 mM KCl, 5 mM DTT, 1 mM EDTA, and 0.02% NaN3) containing 25 g of ammonium sulfate/100 mL. The column was washed with 3 column volumes of buffer P containing 25 g of ammonium sulfate/100 mL and then with 3 column volumes of buffer P without ammonium sulfate, and S100A4 was eluted with 20 mM Tris pH 7.5, 5 mM EGTA, 2 mM DTT, 2 mM TCEP, 1 mM EDTA and 0.02% NaN3. Fractions containing S100A4 were pooled and dialyzed against 2.0 L of buffer Q (20 mM Tris pH 7.5, 2 mM DTT, 2 mM TCEP and 0.02% NaN3). The dialyzed pool was applied to a Hi Prep 16/10 QXL column (GE Healthcare), and the column was washed with 3 column volumes of buffer Q followed by a 200 mL of gradient of 0−0.5 M NaCl in buffer Q. S100A4 eluted at the beginning of the gradient. Typical protein yields were ~125 mg of S100A4 per liter of cells. All C81 mutant proteins were purified as described for the WT S100A4. The purified proteins were stored at −80 °C.
The C3R/C86S and C81S/C86S S100A4 cDNAs were cloned into the NdeI/HindIII sites of pET23a and were purified as described previously.21
S100A4 protein concentrations were determined using the Bradford protein assay (Bio-Rad) and a S100A4 standard of known concentration. The concentration of the S100A4 standard was determined by quantitative amino acid analysis (Keck Biotechnology Resource Laboratory at Yale University, New Haven, CT).
Myosin-IIA Peptide
The FITC-Ahx-DAMNR-EVSSLKNKLRR-OH peptide (FITC-MIIA1908−1923) was obtained from Bio-Synthesis Inc. A 5 mM stock solution of FITC-MIIA1908−1923 was prepared in 20 mM Tris pH 7.5, 150 mM KCl, 1 mM DTT, and 0.02% NaN3, aliquoted, and stored at −80 °C. The concentration of FITC-MIIA1908−1923 was determined using the extinction coeffcient for FITC.
Purification of Myosin-IIA1338−1960
Cloning and bacterial expression of the human myosin-IIA rod (residues 1338−1960) into pET28a is described in ref 22. The cell pellet from 1.0 L of bacterial culture was resuspended in 50 mL of buffer A (50 mM Tris-HCl pH 7.5, 50 mM NaCl, 1 mM DTT, 1 mM EDTA, 25% sucrose) containing 5 μg/mL each of chymostatin, leupeptin, and pepstatin, 1 mM PMSF, and 0.5 mg/mL lysozyme. The cell lysate was frozen at −80 °C. After thawing the cell pellet on ice, Triton X-100 was added to the lysate to a final concentration of 0.6% followed by sonication. The lysate was centrifuged at 30000g for 15 min, and the pellet was resuspended in 60 mL of buffer B (20 mM Tris-HCl pH 7.5, 50 mM NaCl, 1 mM DTT, 5 mM EDTA, 0.5% Triton X-100), stirred for 30 min on ice, and centrifuged as before. This wash step was repeated four to six times. Finally, the washed pellet was resuspended in 7 mL of 20 mM Tris-HCl pH 7.5, 0.6 M NaCl, 1 mM DTT, 2 mM EDTA, 0.02% NaN3. The resuspended pellet was stirred on ice for 1−2 h to extract myosin-IIA1338−1960. The sample was spun in a SS34 rotor at 15 000 rpm for 20 min and then applied to a HiPrep 2.6 × 60 cm Sephacryl S300HR column (GE Healthcare) equilibrated in the same high salt buffer. Fractions containing the myosin-IIA rod were assembled by dialysis against 5 mM PIPES pH 6.5, 20 mM NaCl, 10 mM MgCl2, 1 mM DTT, 0.02% NaN3 overnight. The assembled rods were centrifuged at 14400g for 20 min, and the pellet was resuspended and dialyzed against Storage Buffer (20 mM Tris-HCl pH 7.5, 0.6 M NaCl, 1 mM DTT, 0.02% NaN3). Following centrifugation at 11 000 rpm for 30 min in a SS34 rotor the protein was stored at −80 °C. Typical protein yields were 100 mg of myosin-IIA rods per liter of cells. The concentration of the myosin-IIA rod was determined using the Bradford protein assay (Bio-Rad) and a myosin-IIA rod standard of known concentration. The concentration of the myosin-IIA rod standard was determined by quantitative amino acid analysis (Keck Biotechnology Resource Laboratory at Yale University, New Haven, CT).
Small Molecule Screen
A high-throughput fluorescence polarization assay was developed to identify compounds capable of disrupting the S100A4/myosin-IIA interaction. Briefly, 98 μL of a buffer containing 20 mM Tris, 150 mM KCl, 1 mM DTT, 0.02% NaN3 was added to the wells of a 384-well dilution plate, using a Biomek FX liquid handling robot (Beckman Coulter). Two microliters of test compound was added from 384-well compound library plates to the wells of the dilution plate. The contents of the wells were mixed and 10 μL of the solution was transferred to wells on a 384-well assay plate (Corning Black Low Volume Catalog #3677). Ten microliters of a 2X stock of the S100A4/FITC-MIIA1908−1923 complex (final assay concentrations: 0.5 mM CaCl2, 50 nM FITC-MIIA1908−1923, 15 μM S100A4 dimer) was added to each well. Each experimental plate contained (1) an EGTA control (2 mM), which completely disrupted the S100A4/FITC-MIIA1908−1923 complex, (2) a background control, which contained all assay components except S100A4, (3) a no compound control, (4) a positive control containing 200 μM trifluoperazine, and (5) four concentrations of each experimental compound. The plates were incubated for 1 h at room temperature in the dark and then read on a Tecan Ultra plate reader in fluorescence polarization mode using excitation and emission wavelengths of 485 and 535 nm, respectively. The LOPAC 1280 compound library was screened in quantitative high-throughput mode using individual wells at final compound concentrations of 100, 10, 1, and 0.1 μM. Data were expressed as the percent of the DMSO control, and IC50 values were determined from the concentration response curves by linear interpolation. In confirmatory assays, 16 compound concentrations were tested in triplicate using 2-fold dilutions covering a range of 200−0.0061 μM. Replicate wells were averaged, and the IC50 values were calculated as described above.
Anisotropy Myosin-IIA Binding Assays
Fluorescence anisotropy measurements were performed at 22 °C using a Fluoromax-3-spectrofluorometer (Jobin Yvon). Individual reactions (200 μL) contained 100 nM FITC-MIIA1908−1923 and 0−600 μM S100A4 dimer in 20 mM Tris pH 7.5, 150 mM KCl, 1 mM DTT, 0.02% NaN3, and 0.5 mM CaCl2. For assays using S100A4 concentrations ≥100 μM, the CaCl2 concentration was adjusted to 2.5 mM. Anisotropy was measured with excitation at 490 nm and emission at 520 nm. Measurements were acquired at the magic angle of 55° between the vectors of polarization of the excitation and emission light using a G factor of 0.634 as determined previously for FITC on this instrument. Data were plotted using Graphpad Prism v4, and the dissociation constant was calculated by fitting to a single site saturation binding equation allowing for a floating Ymin value.21
For competitive binding assays, we used 2 μM S100A4 dimer as this S100A4 concentration yields a starting anisotropy value of ~50% of the maximal anisotropy change observed in saturation experiments. NSC 95397 (Tocris Bioscience) and NSC 672121 (NCI/DTP Open Chemical Repository) were stored as 50 mM stocks in DMSO at −20 °C. Final reactions contained 2 μM S100A4 dimer, 100 nM FITC-MIIA1908−1923, and 0−600 μM compound in 20 mM Tris pH 7.5, 150 mM KCl, 1 mM DTT, 0.02% NaN3, 0.5 mM CaCl2, and 2% DMSO. Controls included experiments representing maximum anisotropy (15 μM S100A4, 100 nM FITC-MIIA1908−1923, and calcium) and minimum anisotropy (100 nM FITC-MIIA1908−1923 and calcium). Experiments were performed in triplicate and fit to a sigmoidal dose response equation with a variable slope to obtain the IC50.
Promotion of Disassembly Assays
Assays comparing the ability of wild-type S100A4 and the C81 mutants to depolymerize assembled myosin-IIA rods were performed as described in Li et al.,22 using 1.5 μM S100A4 dimer and 1.5 μM myosin-IIA rod dimer in 20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM DTT, 2 mM MgCl2, 0.5 mM CaCl2, and 0.02% NaN3. For assays performed in the presence of NSC 95397 or NSC 672121 the final sample contained 2% DMSO.
Myosin-IIA Filament Assembly
Myosin-IIA rods (1.5 μM dimer) in 20 mM Tris-HCl pH 7.5, 1 mM DTT, 2 mM MgCl2, 0.5 mM CaCl2, and 0.02% NaN3 with varying NaCl concentrations of 150−500 mM were incubated for 2 h at 22 °C. The samples were centrifuged at 80K rpm (175000g) for 10 min at 25 °C in a TL-100 ultracentrifuge (Beckman). Samples of the mixtures and supernatants were separated on a 12% Tris-Tricine SDS−polyacrylamide gel. Coomassie-stained gels were scanned, and the extent of myosin-IIA polymerization was quantified by densitometry and analyzed with the ImageQuant version 5.2 software package.
Ca2+-Binding Measurements of the Wild-Type S100A4 and C81S Mutant
Ca2+-binding affnities were determined using a competition assay with the chromophoric Ca2+ chelator 5,5′Br2-BAPTA as described previously.21 Briefly, calcium was titrated into a solution containing 25 μM chelator and 25 μM wild-type or C81S S100A4 monomer in 20 mM Tris-HCl, pH 7.5, 150 mM KCl, 1 mM DTT, 0.02% NaN3 at 23 °C. The decrease in absorbance was monitored at 263 nm. The initial calcium concentration was less than 0.15 mol Ca2+/mol protein. Using the CaLigator software,23 data from two independent experiments were fit to two Ca2+ binding sites using a stepwise macroscopic binding equation in the presence of chelator. Since the low affnity site (EF1) does not compete well with 5,5′Br2-BAPTA, lower limits for this value were obtained by monitoring the χ2 value as a function of the low affnity constant.
LC-MS Analysis of S100A4
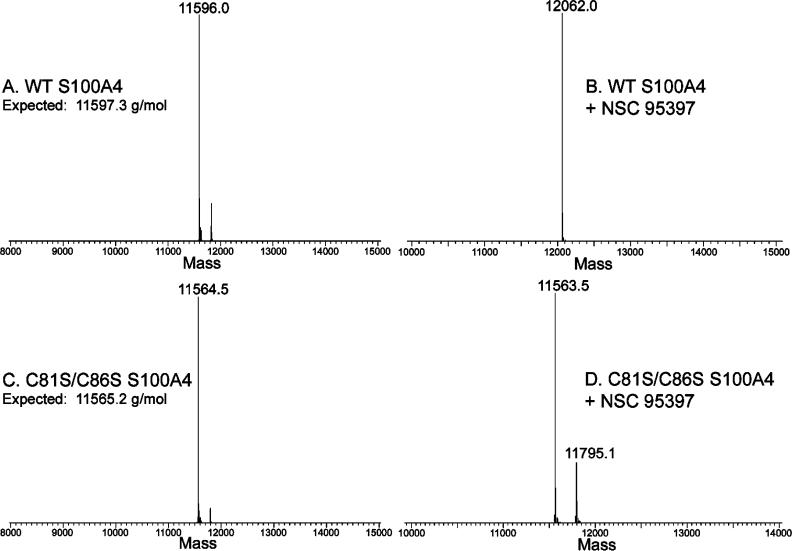
Covalent modification of S100A4 was examined by LC-ESI MS. S100A4-inhibitor complexes were prepared by mixing 174 μM S100A4 monomer with 350 μM inhibitor (1:2 molar ratio) in 20 mM Tris pH 7.5, 150 mM KCl, 1 mM DTT, 0.02% NaN3, 0.5 mM CaCl2, and 2% DMSO. The complex was incubated for 1 h at room temperature and then processed for LC-MS analysis. 500 ng of the S100A4-inhibitor complex was loaded onto a 1.0 × 50 mm C3 column (Micro Tech Scientific) attached to a Shimadzu HPLC with two LC-10AD pumps. The column was equilibrated in buffer A (5% acetonitrile, 0.1% formic acid). After a 5 min desalt, the complex was eluted using a 2 min gradient composed of 15−60% buffer B (95% acetonitrile, 0.1% formic acid) at a flow rate of 50 μL/min. The effuent was delivered directly into a LTQ mass spectrometer (Thermo) for mass analysis. WT and C81S/C86S S100A4 have predicted masses of 11 597.3 and 115 65.2 g/mol, respectively. NSC 95397 has a mass of 310.38 g/mol.
RESULTS
Identification of S100A4 Inhibitors
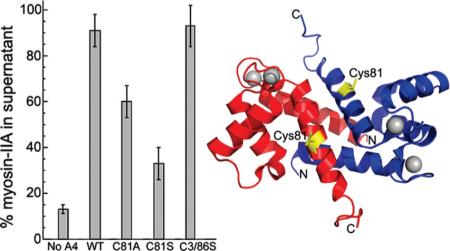
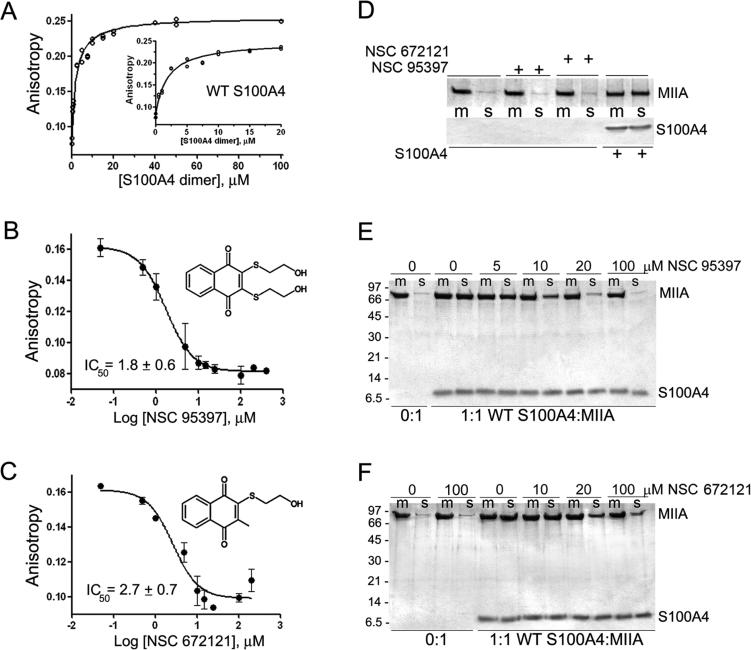
Using a fluorescence anisotropy assay that monitors the binding of S100A4 to the minimal binding site on myosin-IIA (MIIA1908−1923),21 we screened the Library of Pharmacologically Active Compounds (LOPAC) for small molecules that disrupt the S100A4/myosin-IIA interaction in a dose-dependent manner. The screen identified NSC 95397 (2,3-bis[2-hydroxyethylsulfanyl]-1,4-naphthoquinone) as a moderately potent S100A4 inhibitor, which disrupted the S100A4/myosin-IIA complex with an IC50 value in the low micromolar range (5.9 μM). Since the high-throughput screen was performed at relatively high S100A4 concentrations, which could lead to an underestimation of compound affnities, we confirmed disruption of the S100A4-FITC-MIIA1908−1923 complex in competitive binding assays using 2 μM S100A4 dimer since this S100A4 concentration yielded a starting anisotropy value of ~50% of the maximal anisotropy change observed in saturation experiments (Figure 1A). Titration of the S100A4-FITC-MIIA1908−1923 complex with NSC 95397 demonstrated that it competed for myosin-IIA binding with an IC50 of 1.8 ± 0.6 μM (Figure 1B). We also observed disruption of the S100A4-FITC-MIIA1908−1923 complex with NSC 672121 (2-(2-hydroxyethylsulfanyl)-3-methylnaphthalene-1,4-dione), a close congener of NSC 95397, which had an IC50 of 2.7 ± 0.7 μM (Figure 1C). We confirmed that NSC 95397 and NSC 672121 directly modulate the binding of S100A4 to myosin-IIA filaments in assays examining the ability of these compounds to block S100A4-mediated disassembly of myosin-IIA filaments. At a 1:1 molar ratio of S100A4 dimer per myosin-IIA rod, 94% of the myosin-IIA rods were in the supernatant. In the presence of 10 μM NSC 95397, only 52% of the myosin-IIA rods were found in the supernatant, demonstrating that NSC 95397 inhibits the ability of S100A4 to depolymerize myosin-IIA filaments (Figure 1E). Similarly, in the presence of 20 μM NSC 672121, only 61% of the myosin-IIA rods were in the supernatant (Figure 1F). Neither NSC 95397 nor NSC 672121 had any effect on the myosin-IIA filaments in the absence of S100A4 (Figure 1D).
Figure 1.
Napthoquinones disrupt S100A4 binding to myosin-IIA. Assays were performed with 2 μM S100A4 dimer, 100 nM FITC-MIIA1908−1923, and 0−600 μM compound in 20 mM Tris pH 7.5, 150 mM KCl, 1 mM DTT, 0.02% NaN3, and 0.5 mM CaCl2, 2% DMSO. (A) Fluorescence anisotropy measurements of S100A4 binding to FITC-MIIA1908−1923. Values represent two independent experiments. A Kd of 2.3 ± 0.3 μM was determined from the fit to a single site saturation binding curve. Inset: S100A4 binding to FITC-MIIA1908−1923 at S100A4 concentrations ≤20 μM. Competition assays with the napthoquinones NSC 95397 (B) and NSC 672121 (C). Values represent the mean ± standard deviation from 2 to 5 independent experiments. (D) Sedimentation of 1.5 μM myosin-IIA rods in the presence of 100 μM NSC 95397, 100 μM NSC 672121, or 1.5 μM S100A4 dimer. Neither NSC 95397 nor NSC 672121 affects the sedimentation of the myosin-IIA filaments (m = reaction mixture, s = supernatant). Myosin-IIA disassembly assays performed in the presence of NSC 95397 (E) and NSC 672121 (F). At a ratio of 1:1 WT S100A4 dimer:myosin-IIA rod, S100A4 depolymerizes the myosin-IIA filaments (m = reaction mixture, s = supernatant). Both NSC 95397 and NSC 672121 inhibit S100A4-mediated promotion of myosin-IIA filament disassembly. Representative gels from two independent experiments with each inhibitor.
NSC 95397 is a quinone with a p-naphthoquinone substructure and was identified previously as an inhibitor of Cdc25 phosphatase,24 which inactivates Cdc25 by sulfhydryl arylation of the active site cysteine.25,26 As S100A4 contains two solvent accessible cysteine residues (Cys81 and Cys86) in the predicted target binding cleft,27 we considered that NSC 95397 and associated analogues might disrupt the S100A4/myosin-IIA interaction due to a similar covalent modification of S100A4. To evaluate potential covalent modification of S100A4, we used mass spectrometry techniques. Using LC-ESI, we obtained a mass of 11 596.0 Da for the recombinant human Ca2+-S100A4 after incubation with DMSO (Figure 2A), which is consistent with a calculated mass of 11 597.3 Da after removal of the N-terminal methionine. Following incubation of Ca2+-S100A4 with NSC 95397, a peak with a mass of 12 062.0 Da was observed (Figure 2B), which is a mass difference of 466 Da as compared to the unmodified S100A4. This difference is consistent with covalent modification of each S100A4 subunit with 2 mol of NSC 95397 and the loss of one hydroxyethylsulfanyl side chain from each NSC 95397 molecule, which would have a predicted mass of 12 061.8 Da. Notably, we did not detect covalent modification of S100A4 in the absence of calcium (data not shown).
Figure 2.
Mass spectrometry of S100A4. Deconvolved mass spectra for wild-type S100A4 following treatment with vehicle control (A) or NSC 95397 (B). Spectra for C81S/C86S S100A4 following treatment with vehicle control (C) or NSC 95397 (D).
To determine if cysteine residues, and in particular Cys81 and Cys86, were the sites of modification, we performed mass spectrometry on vehicle and NSC 95397 treated S100A4 that contains serine substitutions at Cys81 and Cys86 (C81S/C86S S100A4). For vehicle treated C81S/C86S S100A4 we observed a mass of 11 564.3 Da, which is consistent with the expected mass of 11 565.2 Da (Figure 2C). Following treatment with NSC 95397, we detected an abundant protein species of 11 563.5 Da that represented the unmodified protein and a second lower abundance species with a mass of 11 795.1 Da, which corresponds to covalent modification of each S100A4 subunit with 1 mol of NSC 95397 (Figure 2D). This protein species likely represents modification of Cys3, which occurs in the absence of Cys81 and Cys86, as Cys76 is buried at the dimer interface and is resistant to chemical modification.21
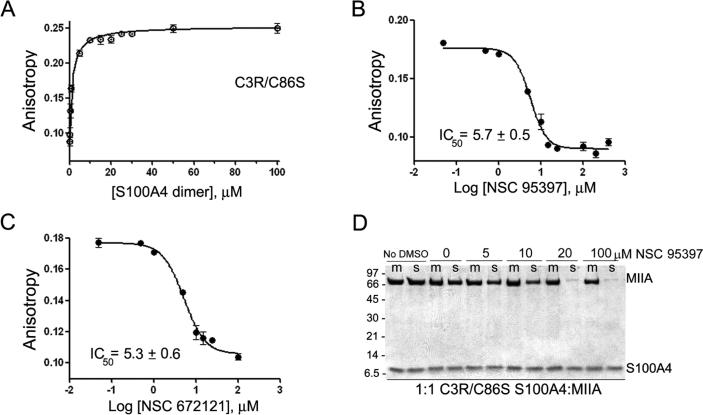
To evaluate the contribution of Cys81 to the S100A4/myosin-IIA interaction and determine if modification of this residue is required for NSC 95397 to disrupt S100A4 function, we substituted Cys86 with serine and replaced Cys3 with an Arg, which is found at position 3 in the mouse and rat S100A4 proteins. In the anisotropy assay, C3R/C86S S100A4 bound the FITC-MIIA1908−1923 peptide with a Kd of 1.4 ± 0.1 μM (Figure 3A), which is comparable to the Kd observed for WT S100A4 (2.3 ± 0.3 μM, Figure 1A). In competitive binding assays, titration of the C3R/C86S S100A4-FITC-MIIA1908−1923 complex with NSC 95397 demonstrated that it competed for myosin-IIA binding with an IC50 of 5.7 ± 0.5 μM (Figure 3B). NSC 672121 also disrupted the C3R/C86S S100A4-FITCMIIA1908−1923 complex with an IC50 of 5.3 ± 0.6 μM, which is similar to the IC50 for the WT S100A4 (1.8 ± 0.6 μM) (Figure 3C). To determine if modification of only Cys81 inhibits S100A4 function with respect to myosin-IIA depolymerization, we examined the ability of NSC 95397 to block disassembly of myosin-IIA filaments by C3R/C86S S100A4. The C3R/C86S S100A4 promoted myosin-IIA filament disassembly in a comparable manner to WT S100A4 since at a 1:1 molar ratio of C3R/C86S S100A4 dimer per myosin-IIA rod 93% of the myosin-IIA rods were in the supernatant (Figure 3D). In the presence of 10 μM NSC 95397, only 57% of the myosin-IIA rods were found in the supernatant, which is comparable to the extent of inhibition observed for the WT S100A4 following treatment with NSC 95397 (Figure 1D). These observations suggest that modification of just Cys81 is suffcient to block S100A4-mediated depolymerization of myosin-IIA filaments.
Figure 3.
Napthoquinones disrupt the binding of C3R/C86S S100A4 binding to myosin-IIA. Assays were performed with 2 μM S100A4 dimer, 100 nM FITC-MIIA1908−1923, and 0−600 μM compound in 20 mM Tris pH 7.5, 150 mM KCl, 1 mM DTT, 0.02% NaN3, and 0.5 mM CaCl2, 2% DMSO. (A) Fluorescence anisotropy measurements of C3R/C86S S100A4 binding to FITC-MIIA1908−1923. Values represent the mean ± standard deviation from two independent experiments. A Kd of 1.4 ± 0.1 μM was determined from the fit to a single site saturation binding curve. Competition assays with the napthoquinones NSC 95397 (B) and NSC 672121 (C). Values represent the mean ± standard deviation from 2 to 5 independent experiments. (D) Myosin-IIA disassembly assays performed at a ratio of 1:1 C3R/C86S S100A4 dimer:myosin-IIA rod. NSC 95397 inhibits the ability of C3R/C86S S100A4 to promote myosin-IIA filament disassembly (m = reaction mixture, s = supernatant). Representative gel from two independent experiments.
Role of Cys81 in the S100A4/Myosin-IIA Interaction
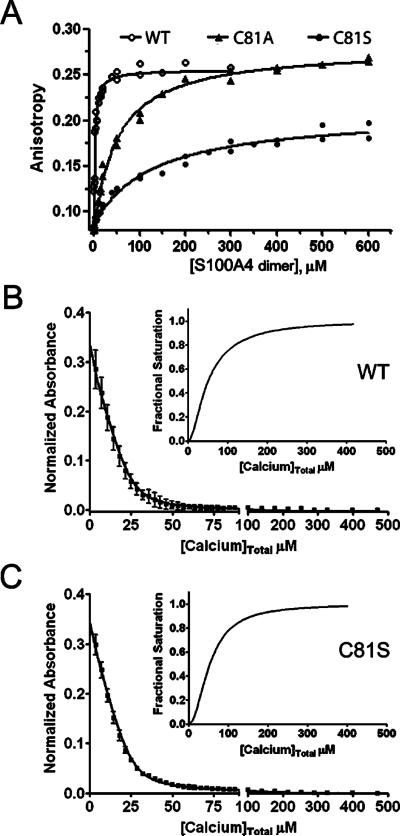
To examine the role of Cys81 in mediating the interaction of S100A4 and myosin-IIA, we created S100A4 proteins incorporating single amino acid substitutions at this position. For C81A and C81S S100A4 the dissociation constants for myosin-IIA binding were 46.6 ± 3.2 and 128.4 ± 15.7 μM, respectively (Figure 4A). These data show that substitution of Cys81 with either alanine or serine reduced the affnity of S100A4 for the FITC-MIIA1908−1923 peptide by 20−56-fold. The reduced myosin-IIA binding affnities exhibited by C81S and C81A S100A4 does not result from significant structural alterations in these proteins as circular dichroism spectroscopy showed that the secondary structures of the C81S and C81A S100A4 proteins were comparable to the wild-type protein (data not shown).
Figure 4.
Biochemical characterization of S100A4 C81 mutants. (A) Fluorescence anisotropy measurements of wild-type S100A4 or C81 mutants. Dissociation constants of 46.6 ± 3.2 and 128.4 ± 15.7 μM were obtained for C81A and C81S, respectively. Values represent the mean ± standard deviation from two independent experiments. Ca2+-binding to wild-type S100A4 (B) and C81S S100A4 (C). Dissociation constants for Ca2+-binding were determined using a competition assay with the chromophoric Ca2+ chelator 5,5′Br2-BAPTA. The data represent two independent experiments. The insets show the saturation curve representation for the best fit in CaLigator. Wild-type and C81S S100A4 bind Ca2+ at EF2 with comparable affnities of 5.8 ± 0.6 and 4.2 ± 0.2 μM, respectively.
Calcium-binding to the canonical EF-hand (EF2) mediates conformational rearrangements required for target binding.27−29 On the basis of the X-ray structure of the Ca2+−S100A4 complex, Cys81 is distal from the canonical EF-hand and is not involved in Ca2+ ion ligation;27−29 however, to confirm that the reduced myosin-IIA binding affnity exhibited by C81S S100A4 is not a consequence of alterations in Ca2+-binding, we measured Ca2+ affnities in the absence of target using a competition assay with the chromophoric Ca2+ chelator 5,5′Br2-BAPTA.23 For the WT and C81S S100A4, the Kd's for the high-affnity EF-hands (EF2) were 5.8 ± 0.6 and 4.2 ± 0.2 μM, respectively (Figure 4B,C). Since 5,5′Br2-BAPTA has a relatively high affnity for Ca2+ (2.3 μM), only limiting values of ≥500 μM for WT and C81S S100A4, respectively, could be estimated for the low affnity pseudo EF-hands (EF1) as they do not compete for Ca2+-binding in this assay. These values are consistent with previous Ca2+-binding measurements for S100A4, which reported dissociation constants in the low micromolar range for EF2 in the absence of target21,27,30 and demonstrate that the WT and C81S S100A4 exhibited similar Ca2+-binding affnities.
Cys81 Substitutions and Filament Disassembly
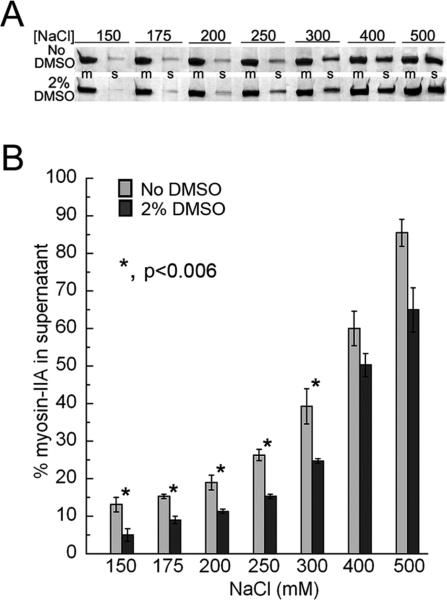
Since promotion of disassembly assays performed in the presence of small molecules contained low levels of DMSO (final concentration 2%), we examined whether DMSO itself has any effect on myosin-IIA assembly. At salt concentrations equivalent to or below 300 mM NaCl, 2% DMSO modestly enhanced myosin-IIA filament assembly by approximately 6−15% (Figure 5A,B). At physiological salt (150 mM), we observed a 6% increase in myosin-IIA filaments.
Figure 5.
DMSO increases the stability of myosin-IIA filaments. Myosin-IIA assembly was monitored as a function of salt concentration (millimolar) using a standard sedimentation assay. (A) Coomassie-stained SDS-PAGE showing representative reaction mixtures (m) and supernatants (s) from assembly assays performed in the absence or presence of 2% DMSO. (B) Myosin-IIA assembly assay. The amount of myosin-IIA recovered in the supernatant was determined by densitometry of Coomassie-stained gels. Values represent the mean ± standard deviation for three independent experiments.
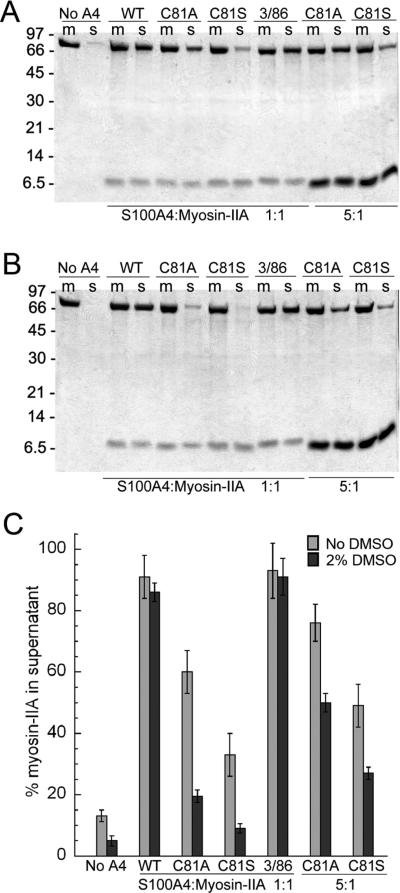
Next, we examined whether substitution of Cys81 with serine or alanine affected the ability of S100A4 to promote myosin-IIA filament disassembly in both the absence and presence of 2% DMSO. DMSO had no effect on the ability of WT or C3R/C86S S100A4 to depolymerize myosin-IIA filaments (Figure 6A−C), and at a 1:1 stoichiometry (S100A4 dimer:myosin-IIA rod) ~90% of the myosin-IIA rods were found in the supernatant. Both C81A and C81S S100A4 exhibited impaired myosin-IIA depolymerization activity, which was more apparent in the presence of DMSO. In the absence of DMSO, 60% and 33% of the myosin-IIA rods were disassembled by C81A and C81S S100A4, respectively. The presence of two percent DMSO attenuated the ability of C81A and C81S S100A4 to depolymerize myosin-IIA filaments with only 20% and 9% of the myosin-IIA rods present in the supernatant. In the absence of DMSO at ratios of 5:1 approximately 76% and 49% of the myosin-IIA had shifted to the supernatant fraction, consistent with the reduced affnities that these proteins exhibited for myosin-IIA.
Figure 6.
Mutation of C81 disrupts S100A4-mediated disassembly of myosin-IIA filaments. Coomassie-stained SDS-PAGE showing representative reaction mixtures (m) and supernatants (s) from disassembly assays performed with wild-type and substituted S100A4 proteins (C81A, C81S, C3R/C86S) in the absence (A) or presence of 2% DMSO (B). Substitution of C81 with either alanine or serine inhibited S100A4-mediated depolymerization of myosin-IIA filaments, which was more pronounced in the presence of DMSO. (C) Quantification of disassembly assays performed at 1:1 and 5:1 S100A4 dimers:myosin-IIA rod. Values represent the mean ± standard deviation for 2−5 independent experiments.
DISCUSSION
Using an anisotropy assay that monitors the binding of S100A4 to myosin-IIA, we identified NSC 95397 as a small molecule inhibitor that covalently modifies Cys81 and Cys86 of S100A4. NSC 95397 has been reported to inhibit the activities of protein tyrosine phosphatases (e.g., Cdc25 dual specificity phosphatase, mitogen-activated protein kinase phosphatase-1 and -3, and CD45 tyrosine phosphatase),31−33 the cysteine proteases papain and cathepsin B,34 and tissue transglutaminase.35 Inhibition of these enzymes is thought to occur primarily as a consequence of covalent modification of an active site cysteine.34−36 To our knowledge, this is the first report that NSC 95397 can modify a protein that performs in a noncatalytic function and can modulate activity through the regulation of protein−protein interactions.
NSC 95397 as well as other quinones are reported to exert their biochemical and biological effects via two potential mechanisms. Quinones can undergo redox cycling to generate reactive oxygen species (ROS) such as H2O2, which can irreversibly oxidize reactive cysteine residues. In addition, they can function as Michael acceptors that can covalently bind thiolates and other electrophiles. In vitro studies demonstrate that NSC 95397 produces significant amounts of H2O2 in the presence of strong reducing agents such as DTT.37 Although our biochemical analysis of S100A4 modification and activity was performed in the presence of millimolar concentrations of DTT, we observed only the formation of a S100A4 adduct rather than oxidation of cysteine residues to sulfinic or sulfonic acid, suggesting that H2O2 production was not required for NSC 95397 inhibition of S100A4 activity.
Given that NSC 95397 is a relatively promiscuous inhibitor that has pleiotropic effects on cellular function,32,38,39 it will likely not have utility as an S100A4-selective inhibitor. Nonetheless, the identification of NSC 95397 as a S100A4 inhibitor led to the discovery that cysteine residues are critical for the S100A4/myosin-IIA interaction. In the context of the wild-type S100A4, NSC 95397 forms covalent adducts with both Cys81 and Cys86, which are located on helix 4 of the S100A4 target binding cleft. Our mutagenesis studies demonstrate that NSC 95397-mediated modification of Cys81 alone is suffcient to disrupt S100A4 function with respect to myosin-IIA binding and depolymerization. The importance of Cys81 in mediating the S100A4/myosin-IIA interaction is also supported by studies with a fluorescent S100A4 biosensor, in which Cys81 was modified with a merocyanine dye.21 The merocyanine-labeled S100A4 exhibited comparable binding affnities to myosin-IIA filaments as the WT S100A4 but was defective in depolymerizing these filaments. Substitution of Cys81 with either alanine or serine substantially reduced S100A4 activity. The reduced activity of the C81S mutant is particularly informative as it suggests that the mechanistic contribution of Cys81 is not solely its hydrogen-bonding capacity. Given the relative pKa's of cysteine and serine (approximately 8 and 14, respectively), it is plausible to suggest that the thiolate anion may play a direct role in target protein recognition.
Prior to these studies, the only functional analysis of residues involved in modulating the S100A4/myosin-IIA interaction focused on the C-terminal tail. Deletion of the three basic residues at the extreme C-terminal end of S100A4 or substitution of the last two lysine residues with alanine reduced the affnity for myosin-IIA by ~18-fold as measured by surface plasmon resonance.18 In addition to these functional studies on the C-terminal tail, titration of Ca2+−S100A4 with the MIIA1908−1923 peptide demonstrated perturbations in the amide 1H and 15N backbone resonances of residues in the hinge (Glu41, Gly47, Lys48, Thr50, and Asp51), helix 3 (Ser60 and Leu62), and helix 4 (Phe78, Leu79, Met85, and Cys86).27 The observed changes in chemical shift for these residues likely results from the direct interaction of the myosin-IIA peptide with the Ca2+−S100A4 complex; however, it is also possible that these changes in chemical shift reflect conformational rearrangements of residues near the myosin-IIA binding site which are induced by myosin-IIA peptide binding. To establish the direct involvement of these residues in myosin-IIA binding will require further detailed mutagenesis and biochemical studies.
The identification of Cys81 as a crucial residue for the S100A4/myosin-IIA interaction raises the possibility that modification of this residue may provide an additional regulatory mechanism for mediating the binding of S100A4 to myosin-IIA. Notably Cys81 and Cys86 are invariant within the S100A4 family, and Cys86 is conserved in 10 of the S100 proteins. Two of the most common reversible covalent cysteine modifications are glutathionylation and nitrosylation. At present, five S100 proteins (S100A1, S100A4, S100A6, S100A8, and S100A9) have been reported to be modified by either glutathionylation or nitrosylation;40−43 however, the biochemical consequence of most of these modifications has not been examined. The one exception is S100A1, where glutathionylation of Cys85 (comparable to Cys86 in S100A4) is reported to increase the Ca2+-binding affnities for both EF1 and EF2.40 Unmodified S100A1 exhibits a relatively weak affnity for calcium even in the presence of target;44 thus, glutathionylation may allow for Ca2+-binding under physiologically relevant intracellular calcium concentrations.45 The exposure of human cancer cells to ionizing radiation induces cysteinylation and glutathionylation of both S100A6 and S100A4.41 The single cysteine on the S100A6 N-terminus is the proposed site of modification; moreover, post-translational modification is associated with translocation of S100A6 from the nucleus to the cytoplasm, which may reflect changes in the interaction of S100A6 with its binding partners. For S100A4, which contains four cysteines, the sites of modification were not identified; however, multiple S100A4 charge variants have been detected in other normal and cancer lines, consistent with post-translational modification.46 Altogether these observations suggest that modification of S100A4 via cysteine or other residues may provide a mechanism for regulating S100A4 function through the modulation of its Ca2+-binding or protein−protein interaction activities.
ACKNOWLEDGMENTS
We thank Dr. Sarah Garrett for assistance with calcium-binding assays.
Funding
This work was supported in part with grants from the National Institutes of Health grant CA129598 (A.R.B.) and in part with funds from the National Cancer Institute, National Institutes of Health, under Contract HHSN26120080001E.
ABBREVIATIONS
- EF1
N-terminal pseudo-EF-hand
- EF2
C-terminal canonical EF-hand
- WT
wild-type
- MIIA
nonmuscle myosin-IIA
- FITC
fluorescein isothiocyanate
- LOPAC
library of pharmacologically active compounds
- DMSO
dimethyl sulfoxide
- BAPTA
1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid
- ROS
reactive oxygen species
- DTT
dithiothreitol
REFERENCES
- 1.Zimmer DB, Cornwall EH, Landar A, Song W. The S100 protein family: history, function, and expression. Brain Res. Bull. 1995;37:417–429. doi: 10.1016/0361-9230(95)00040-2. [DOI] [PubMed] [Google Scholar]
- 2.Marenholz I, Heizmann CW, Fritz G. S100 proteins in mouse and man: from evolution to function and pathology (including an update of the nomenclature). Biochem. Biophys. Res. Commun. 2004;322:1111–1122. doi: 10.1016/j.bbrc.2004.07.096. [DOI] [PubMed] [Google Scholar]
- 3.Rety S, Sopkova J, Renouard M, Osterloh D, Gerke V, Tabaries S, Russo-Marie F, Lewit-Bentley A. The crystal structure of a complex of p11 with the annexin II N-terminal peptide. Nat. Struct. Biol. 1999;6:89–95. doi: 10.1038/4965. [DOI] [PubMed] [Google Scholar]
- 4.Rustandi RR, Baldisseri DM, Weber DJ. Structure of the negative regulatory domain of p53 bound to S100B(betabeta). Nat. Struct. Biol. 2000;7:570–574. doi: 10.1038/76797. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharya S, Large E, Heizmann CW, Hemmings B, Chazin WJ. Structure of the Ca2+/S100B/NDR kinase peptide complex: insights into S100 target specificity and activation of the kinase. Biochemistry. 2003;42:14416–14426. doi: 10.1021/bi035089a. [DOI] [PubMed] [Google Scholar]
- 6.Donato R. Intracellular and extracellular roles of S100 proteins. Microsc. Res. Tech. 2003;60:540–551. doi: 10.1002/jemt.10296. [DOI] [PubMed] [Google Scholar]
- 7.Santamaria-Kisiel L, Rintala-Dempsey AC, Shaw GS. Calcium-dependent and -independent interactions of the S100 protein family. Biochem. J. 2006;396:201–214. doi: 10.1042/BJ20060195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heizmann CW, Ackermann GE, Galichet A. Pathologies involving the S100 proteins and RAGE. Subcell Biochem. 2007;45:93–138. doi: 10.1007/978-1-4020-6191-2_5. [DOI] [PubMed] [Google Scholar]
- 9.Schneider M, Hansen JL, Sheikh SP. S100A4: a common mediator of epithelial-mesenchymal transition, fibrosis and regeneration in diseases? J. Mol. Med. 2008;86:507–522. doi: 10.1007/s00109-007-0301-3. [DOI] [PubMed] [Google Scholar]
- 10.Grigorian M, Ambartsumian N, Lukanidin E. Metastasis-inducing S100A4 protein: implication in non-malignant human pathologies. Curr. Mol. Med. 2008;8:492–496. doi: 10.2174/156652408785747942. [DOI] [PubMed] [Google Scholar]
- 11.Helfman DM, Kim EJ, Lukanidin E, Grigorian M. The metastasis associated protein S100A4: role in tumour progression and metastasis. Br. J. Cancer. 2005;92:1955–1958. doi: 10.1038/sj.bjc.6602613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garrett SC, Varney KM, Weber DJ, Bresnick AR. S100A4, a mediator of metastasis. J. Biol. Chem. 2006;281:677–680. doi: 10.1074/jbc.R500017200. [DOI] [PubMed] [Google Scholar]
- 13.Takenaga K, Nakamura Y, Sakiyama S, Hasegawa Y, Sato K, Endo H. Binding of pEL98 protein, an S100-related calcium-binding protein, to nonmuscle tropomyosin. J. Cell Biol. 1994;124:757–768. doi: 10.1083/jcb.124.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kriajevska MV, Cardenas MN, Grigorian MS, Ambartsumian NS, Georgiev GP, Lukanidin EM. Non-muscle myosin heavy chain as a possible target for protein encoded by metastasis-related mts-1 gene. J. Biol. Chem. 1994;269:19679–19682. [PubMed] [Google Scholar]
- 15.Grigorian M, Andresen S, Tulchinsky E, Kriajevska M, Carlberg C, Kruse C, Cohn M, Ambartsumian N, Christensen A, Selivanova G, Lukanidin E. Tumor suppressor p53 protein is a new target for the metastasis-associated Mts1/S100A4 protein: functional consequences of their interaction. J. Biol. Chem. 2001;276:22699–22708. doi: 10.1074/jbc.M010231200. [DOI] [PubMed] [Google Scholar]
- 16.Kriajevska M, Fischer-Larsen M, Moertz E, Vorm O, Tulchinsky E, Grigorian M, Ambartsumian N, Lukanidin E. Liprin beta 1, a member of the family of LAR transmembrane tyrosine phosphatase-interacting proteins, is a new target for the metastasis-associated protein S100A4 (Mts1). J. Biol. Chem. 2002;277:5229–5235. doi: 10.1074/jbc.M110976200. [DOI] [PubMed] [Google Scholar]
- 17.Li ZH, Bresnick AR. The S100A4 metastasis factor regulates cellular motility via a direct interaction with myosin-IIA. Cancer Res. 2006;66:5173–5180. doi: 10.1158/0008-5472.CAN-05-3087. [DOI] [PubMed] [Google Scholar]
- 18.Ismail TM, Fernig DG, Rudland PS, Terry CJ, Wang G, Barraclough R. The basic C-terminal amino acids of calcium-binding protein S100A4 promote metastasis. Carcinogenesis. 2008;29:2259–2266. doi: 10.1093/carcin/bgn217. [DOI] [PubMed] [Google Scholar]
- 19.Li ZH, Dulyaninova NG, House RP, Almo SC, Bresnick AR. S100A4 regulates macrophage chemotaxis. Mol. Biol. Cell. 2010;21:2598–2610. doi: 10.1091/mbc.E09-07-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malashkevich VN, Dulyaninova NG, Ramagopal UA, Liriano MA, Varney KM, Knight D, Brenowitz M, Weber DJ, Almo SC, Bresnick AR. Phenothiazines inhibit S100A4 function by inducing protein oligomerization. Proc. Natl. Acad. Sci. U. S. A. 2010;107:8605–8610. doi: 10.1073/pnas.0913660107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garrett SC, Hodgson L, Rybin A, Toutchkine A, Hahn KM, Lawrence DS, Bresnick AR. A biosensor of S100A4 metastasis factor activation: inhibitor screening and cellular activation dynamics. Biochemistry. 2008;47:986–996. doi: 10.1021/bi7021624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z-H, Spektor A, Varlamova O, Bresnick AR. Mts1 regulates the assembly of nonmuscle myosin-IIA. Biochemistry. 2003;42:14258–14266. doi: 10.1021/bi0354379. [DOI] [PubMed] [Google Scholar]
- 23.Andre I, Linse S. Measurement of Ca2+-binding constants of proteins and presentation of the CaLigator software. Anal. Biochem. 2002;305:195–205. doi: 10.1006/abio.2002.5661. [DOI] [PubMed] [Google Scholar]
- 24.Lazo JS, Nemoto K, Pestell KE, Cooley K, Southwick EC, Mitchell DA, Furey W, Gussio R, Zaharevitz DW, Joo B, Wipf P. Identification of a potent and selective pharmacophore for Cdc25 dual specificity phosphatase inhibitors. Mol. Pharmacol. 2002;61:720–728. doi: 10.1124/mol.61.4.720. [DOI] [PubMed] [Google Scholar]
- 25.Nishikawa Y, Carr BI, Wang M, Kar S, Finn F, Dowd P, Zheng ZB, Kerns J, Naganathan S. Growth inhibition of hepatoma cells induced by vitamin K and its analogs. J. Biol. Chem. 1995;270:28304–28310. doi: 10.1074/jbc.270.47.28304. [DOI] [PubMed] [Google Scholar]
- 26.Kar S, Lefterov IM, Wang M, Lazo JS, Scott CN, Wilcox CS, Carr BI. Binding and inhibition of Cdc25 phosphatases by vitamin K analogues. Biochemistry. 2003;42:10490–10497. doi: 10.1021/bi027418p. [DOI] [PubMed] [Google Scholar]
- 27.Malashkevich VN, Varney KM, Garrett SC, Wilder PT, Knight D, Charpentier TH, Ramagopal UA, Almo SC, Weber DJ, Bresnick AR. Structure of Ca2+-bound S100A4 and its interaction with peptides derived from nonmuscle myosin-IIA. Biochemistry. 2008;47:5111–5126. doi: 10.1021/bi702537s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gingras AR, Basran J, Prescott A, Kriajevska M, Bagshaw CR, Barsukov IL. Crystal structure of the Ca(2+)-form and Ca(2+)-binding kinetics of metastasis-associated protein, S100A4. FEBS Lett. 2008;582:1651–1656. doi: 10.1016/j.febslet.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 29.Pathuri P, Vogeley L, Luecke H. Crystal structure of metastasis-associated protein S100A4 in the active calcium-bound form. J. Mol. Biol. 2008;383:62–77. doi: 10.1016/j.jmb.2008.04.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Badyal SK, Basran J, Bhanji N, Kim JH, Chavda AP, Jung HS, Craig R, Elliott PR, Irvine AF, Barsukov IL, Kriajevska M, Bagshaw CR. Mechanism of the Ca(2)+-dependent interaction between S100A4 and tail fragments of nonmuscle myosin heavy chain IIA. J. Mol. Biol. 2011;405:1004–1026. doi: 10.1016/j.jmb.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brisson M, Nguyen T, Wipf P, Joo B, Day BW, Skoko JS, Schreiber EM, Foster C, Bansal P, Lazo JS. Redox regulation of Cdc25B by cell-active quinolinediones. Mol. Pharmacol. 2005;68:1810–1820. doi: 10.1124/mol.105.016360. [DOI] [PubMed] [Google Scholar]
- 32.Vogt A, McDonald PR, Tamewitz A, Sikorski RP, Wipf P, Skoko JJ, 3rd, Lazo JS. A cell-active inhibitor of mitogen-activated protein kinase phosphatases restores paclitaxel-induced apoptosis in dexamethasone-protected cancer cells. Mol. Cancer Ther. 2008;7:330–340. doi: 10.1158/1535-7163.MCT-07-2165. [DOI] [PubMed] [Google Scholar]
- 33.Panchal RG, Ulrich RL, Bradfute SB, Lane D, Ruthel G, Kenny TA, Iversen PL, Anderson AO, Gussio R, Raschke WC, Bavari S. Reduced expression of CD45 protein-tyrosine phosphatase provides protection against anthrax pathogenesis. J. Biol. Chem. 2009;284:12874–12885. doi: 10.1074/jbc.M809633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valente C, Moreira R, Guedes RC, Iley J, Jaffar M, Douglas KT. The 1,4-naphthoquinone scaffold in the design of cysteine protease inhibitors. Bioorg. Med. Chem. 2007;15:5340–5350. doi: 10.1016/j.bmc.2007.04.068. [DOI] [PubMed] [Google Scholar]
- 35.Lai TS, Liu Y, Tucker T, Daniel KR, Sane DC, Toone E, Burke JR, Strittmatter WJ, Greenberg CS. Identification of chemical inhibitors to human tissue transglutaminase by screening existing drug libraries. Chem. Biol. 2008;15:969–978. doi: 10.1016/j.chembiol.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Contour-Galcera MO, Sidhu A, Prevost G, Bigg D, Ducommun B. What's new on CDC25 phosphatase inhibitors. Pharmacol. Ther. 2007;115:1–12. doi: 10.1016/j.pharmthera.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 37.Soares KM, Blackmon N, Shun TY, Shinde SN, Takyi HK, Wipf P, Lazo JS, Johnston PA. Profiling the NIH Small Molecule Repository for compounds that generate H2O2 by redox cycling in reducing environments. Assay Drug Dev. Technol. 2010;8:152–174. doi: 10.1089/adt.2009.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han Y, Shen H, Carr BI, Wipf P, Lazo JS, Pan SS. NAD(P)H:quinone oxidoreductase-1-dependent and -independent cytotoxicity of potent quinone Cdc25 phosphatase inhibitors. J. Pharmacol. Exp. Ther. 2004;309:64–70. doi: 10.1124/jpet.103.059477. [DOI] [PubMed] [Google Scholar]
- 39.Peyregne VP, Kar S, Ham SW, Wang M, Wang Z, Carr BI. Novel hydroxyl naphthoquinones with potent Cdc25 antagonizing and growth inhibitory properties. Mol. Cancer Ther. 2005;4:595–602. doi: 10.1158/1535-7163.MCT-04-0274. [DOI] [PubMed] [Google Scholar]
- 40.Goch G, Vdovenko S, Kozlowska H, Bierzynski A. Affinity of S100A1 protein for calcium increases dramatically upon glutathionylation. FEBS J. 2005;272:2557–2565. doi: 10.1111/j.1742-4658.2005.04680.x. [DOI] [PubMed] [Google Scholar]
- 41.Orre LM, Pernemalm M, Lengqvist J, Lewensohn R, Lehtio J. Up-regulation, modification, and translocation of S100A6 induced by exposure to ionizing radiation revealed by proteomics profiling. Mol. Cell. Proteomics. 2007;6:2122–2131. doi: 10.1074/mcp.M700202-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Lim SY, Raftery M, Cai H, Hsu K, Yan WX, Hseih HL, Watts RN, Richardson D, Thomas S, Perry M, Geczy CL. S-nitrosylated S100A8: novel anti-inflammatory properties. J. Immunol. 2008;181:5627–5636. doi: 10.4049/jimmunol.181.8.5627. [DOI] [PubMed] [Google Scholar]
- 43.Lim SY, Raftery MJ, Goyette J, Geczy CL. S-glutathionylation regulates inflammatory activities of S100A9. J. Biol. Chem. 2010;285:14377–14388. doi: 10.1074/jbc.M109.075242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wright NT, Cannon BR, Wilder PT, Morgan MT, Varney KM, Zimmer DB, Weber DJ. Solution structure of S100A1 bound to the CapZ peptide (TRTK12). J. Mol. Biol. 2009;386:1265–1277. doi: 10.1016/j.jmb.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 46.Haugen MH, Flatmark K, Mikalsen SO, Malandsmo GM. The metastasis-associated protein S100A4 exists in several charged variants suggesting the presence of posttranslational modifications. BMC Cancer. 2008;8:172. doi: 10.1186/1471-2407-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]