Abstract
Scavenger receptor BI (SR-BI) is an HDL receptor. It binds HDL and mediates the uptake of cholesteryl ester from HDL. Early studies have pointed out that the extracellular domain of SR-BI is critical for SR-BI-mediated cholesteryl ester uptake. However, the extracellular loop of SR-BI is large: it contains 403 amino acids. The HDL binding site and the modulation of SR-BI-mediated cholesteryl ester uptake remain to be identified. In this study, using C323G mutant SR-BI, we showed that C323G mutant SR-BI lost its HDL binding and cholesteryl ester uptake activity, indicating that the highly conserved C323 is required for SR-BI-mediated HDL binding and cholesteryl ester uptake. Using a blocking antibody against C323 region, we demonstrated that C323 is directly involved in HDL binding and likely an HDL binding site. Using C323G mutant transgenic mouse model, we further demonstrated that C323 of SR-BI is required for regulating plasma cholesterol levels in vivo. Using redox reagents, we showed that physiological relevant levels of H2O2 upregulated the SR-BI-mediated cholesteryl ester uptake activity by 65%, whereas GSH or DTT significantly downregulated SR-BI-mediated cholesteryl ester uptake activity by 45%. C323 of SR-BI is critical for SR-BI-mediated HDL binding and cholesteryl ester uptake, and changes in redox status may be a regulatory factor modulating SR-BI-mediated cholesterol transport.
Keywords: scavenger receptor class B type I, cholesterol efflux, high density lipoprotein
Scavenger receptor class B type I (SR-BI or Scarb1) is a physiological receptor for HDL (1, 2). It regulates plasma cholesterol levels through a process called reverse cholesterol transport, in which SR-BI uptakes cholesteryl ester from HDL into hepatocytes and subsequently secretes cholesterol from bile. SR-BI-mediated cholesteryl ester uptake is also required for steroidogenesis, in which SR-BI uptakes cholesteryl ester from HDL into endocrine tissues for steroid synthesis (3–7). The importance of SR-BI-mediated cholesterol transport has been established in mouse models. Mice deficient in SR-BI have a 2-fold increase in total cholesterol concentration and a 3-fold increase in free cholesterol concentration in circulation, and they develop cardiovascular diseases (8–13). Mice overexpressing SR-BI have lower plasma cholesterol levels (14–17) and are protected against the development of atherosclerosis (11, 18). A recent report showed that a family with functional mutation in SR-BI has elevated plasma HDL and changes in cholesterol metabolism in macrophages and platelets, indicating a role of SR-BI in humans (19). In addition to modulating HDL metabolism, recent studies revealed that SR-BI is a multifunctional protein. It activates eNOS in endothelial cells in the presence of HDL (20–23), induces apoptosis in the absence of HDL/eNOS (24), protects against nitric oxide (NO)-induced oxidative damage (25), and prevents endotoxic and septic animal death (25–27).
SR-BI facilitates intracellular uptake of HDL cholesteryl esters in a two-step process involving binding of HDL through its extracellular domain and transferring cholesteryl ester from HDL into cells. The importance of extracellular domain of SR-BI in cholesteryl ester uptake has been explored by the use of chimeric receptors (28), by insertion of epitope tags into various regions of the domain of SR-BI (29), by blocking antibody against the extracellular domain, and by mutations (30, 31). However, SR-BI has a large extracellular domain that contains 403 amino acid residues, and the HDL binding site remains to be determined. Identifying the HDL binding site and understanding the regulation of SR-BI-mediated cholesterol transport may provide a target for promoting plasma cholesterol clearance and for the intervention of inflammatory disease, such as atherosclerosis and sepsis.
We previously identified C323 of SR-BI as a highly conserved site in SR-BI. Single site mutation at C323 completely abolishes SR-BI-induced apoptosis (24) and its protection against nitric oxide-induced oxidative cell damage (25), indicating that the C323 is critical for its functions. Interestingly, our study also showed that the mutation at C323 does not affect the ability of SR-BI to suppress LPS-induced inflammatory response in macrophages (27). These studies indicate that SR-BI is a multifunctional molecule, and C323 may play different roles in modulating the functions of SR-BI. In this study, we demonstrated that C323 is involved in HDL binding and required for SR-BI-mediated cholesteryl ester uptake.
MATERIALS AND METHODS
Materials
Anti-SR-BI serum was custom made by Sigma-Genosys using a 15-amino-acid peptide derived from the C-terminal of human SR-BI. Blocking antibody against C323 region was generated by Sigma-Genosys using a 15-amino-acid peptide PNEGFCPCLESGIQN conjugated to KLH. The blocking antibody and nonimmune serum were affinity purified with a protein A column. Enzymatic deglycosylation kit was from Calbiochem. Cell surface protein isolation kit was from Pierce Biotechnology. Serum total and free cholesterol quantification kits were from Wako.
Generation of transgenic mice
SR-BI and SR-BIC323G transgenic mice were generated as previously described (27). To exclude the effect of endogenous SR-BI, the transgenic mice were crossed to SR-BI−/− mice (in C57BL/6×129 background) to generate transgenic and nontransgenic littermates in SR-BI−/− background. The breeders were fed with 0.2% Probucol to correct female infertility. Transgenic mice were genotyped by PCR using tail DNA. The animals were fed with standard laboratory chow diet and used at ages 12 to 15 weeks. Animal care and experiments were approved by the Institutional Animal Care and Use Committee of the University of Kentucky.
CHO cell lines expressing mutant SR-BI
Chinese hamster ovary (CHO) cell lines stably expressing vector, wild-type, and C323G SR-BI were generated as previously described (20). CHO cells stably expressing C251G, C280G, C321G, G327A, C334G, and C384G were generated using the same method. The cells were cultured in Ham's F-12 medium containing 5% FBS, 2 mmol/l L-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin, and 0.5 mg/ml G418.
Isolation of caveolae
Caveolae, cytosol, and internal membranes were isolated using Opti-Prep method as described previously (7, 8). This method generates a highly purified fraction of caveolae that is enriched in caveolin-1 and free of bulk plasma membrane markers.
Preparation of [125I]apolipoprotein- and [3H]cholesteryl oleoyl ether-labeled HDL
Human HDL was isolated from normolipidemic subjects by sequential ultracentrifugation as described previously (32). The HDL was iodinated with 125I (GE Healthcare), and HDL-associated cholesteryl ester was traced with nonhydrolyzable [3H] cholesteryl oleoyl ether (COE; GE Healthcare) as described previously (33, 34).
HDL binding and selective uptake assays
The binding and uptake assay were performed as previously described (33, 34). Briefly, the cells were cultured in a 12-well dish to 95% confluency in Ham's F-12 medium containing 5% FBS. The cells were rinsed three times with PBS and incubated in Ham's F12 containing 5% human lipoprotein-deficient serum (binding buffer) at 37°C for 2 h. Then, [3H]COE-[125I]HDL was added at 10 µg/ml for the indicated times. After washing with cold PBS, the cells were lysed in 0.5 N NaOH. The 125I in the cell lysate (representing cell-associated HDL) were counted with a γ-counter. [3H]COE was extracted from the cell lysate by Dole reagent and counted with a scintillation counter. The degradation of HDL was less than 10% when tested by TCA precipitation method. The [125I]HDL associated with cells was subtracted from the COE measured as extractable 3H radioactivity (expressed as HDL equivalents) to give the selective uptake of cholesterol. The data are from three independent experiments with triplicate measurements in each experiment.
Cholesterol efflux assay
Cells were incubated for 48 h with 0.2 µCi/ml [3H]cholesterol. Cholesterol efflux was determined after incubating cells with 40 µg/ml HDL as the acceptor for the indicated times at 37°C. Results are expressed as a ratio of the radioactivity released in the culture media to the total radioactivity in media and cells. The data are from three independent experiments with triplicate measurements in each experiment.
Western blot
Cells were lysed in lysis buffer consisting of 25 mmol/l 2-(N-morpholino) ethane sulfonic acid (MES), 137 mmol/l NaCl, 1% Triton X-100, 60 mmol/l octyl glucoside, 0.1% SDS, and 1% proteinase inhibitor cocktail (Sigma), pH 6.4, and analyzed by Western blot as previously described (20, 35).
Cell surface protein isolation
Cell surface proteins were isolated with the cell surface protein isolation kit following the manufacturer's instructions. Briefly, cells were cultured to 90% confluency and labeled with biotin. The cell lysate was applied to an avidin affinity column. The nonbiotin-labeled proteins passed through the column and were collected as nonbiotin-labeled fraction; biotin-labeled proteins (cell surface proteins) were eluted with a buffer containing DTT and collected as biotin-labeled fraction. Cell lysate (10 µg of protein, one fraction), nonlabeled (two fractions), and biotin-labeled (two fractions) fractions were analyzed by Western blot.
Deglycosylation
Cell lysates were deglycosylated by using the enzymatic deglycosylation kit following the manufacturer's instructions. After deglycosylation, samples were analyzed by Western blot.
Lipoprotein profiling by FPLC
Serum (50 µl) was resolved by gel filtration chromatography with FPLC equipped with a Superose 6 column (Pharmacia LKB Biotechnology, Inc.). The column was eluted at a flow rate of 0.5 ml/min, and 0.5 ml/fraction was collected. One hundred microliters of sample was mixed with 100 µl of 2× assay reagent to determine the cholesterol content of fractions (Wako Chemicals).
Statistical analysis
Significance in experiments comparing two groups was determined by 2-tailed Student t-test. Significance in experiments comparing more than two groups was evaluated by one way ANOVA, followed by post hoc analysis using Tukey's test. Values are reported as the mean ± SD. A value of P < 0.05 was considered significant.
RESULTS
SR-BIC323G was located in caveolae on plasma membrane
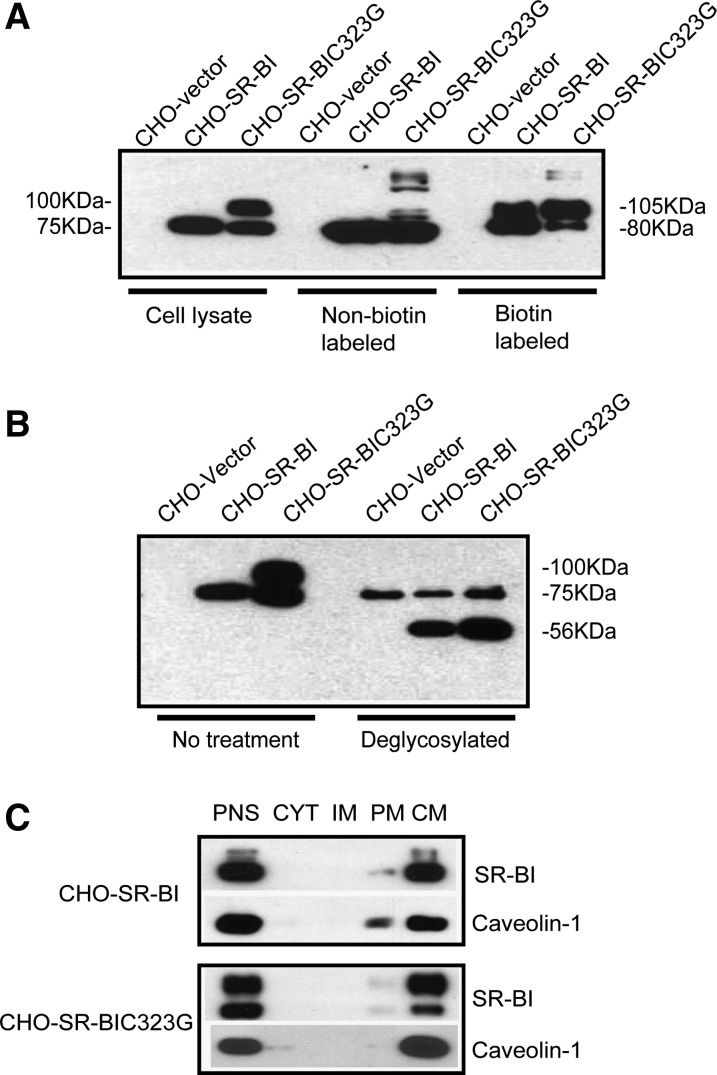
The CHO cell system has been widely used to study the functions of SR-BI, including SR-BI-mediated HDL binding and cholesteryl ester uptake. Thus, we employed CHO-SR-BIC323G cells to determine the importance of C323 in HDL binding and cholesteryl ester uptake. We first verified membrane targeting of C323G mutant SR-BI with biotin labeling assay. About 40% of SR-BIC323G appeared in cell surface fraction, and 60% of SR-BIC323G appeared in nonlabeled fraction, which is similar to wild-type SR-BI (Fig. 1A).
Fig. 1.
Subcellular location of C323G mutant SR-BI. A: Detection of cell surface SR-BI expression by biotin labeling assay. CHO cells expressing vector, wild-type, or C323G mutant SR-BI were labeled with biotin. Biotinylated proteins were isolated with an avidin affinity column and analyzed by Western blot. B: Deglycosylation of wild-type and C323G mutant SR-BI. The CHO cells were lysed, treated with a mixture of enzymes to remove saccharide from protein, and then analyzed by Western blot. C: Subcellular targeting of SR-BI. Subcellular fractions were isolated from the CHO cells and analyzed by Western blot. Postnuclear supernatant (PNS), cytosol (CYT), intracellular membrane (IM), plasma membrane (PM), and caveolae membrane (CM) fractions.
We observed a higher molecular weight (100 kDa) band in the lysate of CHO-SR-BIC323G cells, and this higher molecular weight (105 kDa) band appeared in the biotin-labeled fractions from wild-type and C323G mutant SR-BI. We speculated that this higher molecular weight SR-BI band is highly glycosylated SR-BI. To test it, we treated the cell lysate with deglycosylation enzymes, including N-glycosidase, F exoglycosidases, and endo-α-N-acetylgalactosaminidase, which remove all N-linked and O-inked oligosaccharides from protein. As shown in Fig. 1B, both CHO-SR-BIC323G and CHO-SR-BI displayed a 56 kDa band upon deglycosylation, which corresponds to the size of nonglycosylated SR-BI, indicating that the 100 kDa band is highly glycosylated SR-BI. Of note, the 75 kDa band that appeared in the deglycosylated sample was considered a nonspecific band because the CHO vector also displayed such a band (Fig. 1B).
SR-BI has been shown to locate in caveolae. We verified its caveola targeting with Opti-prep analysis using caveolin-1 as a marker of caveola. Opti-prep analysis showed that C323G mutant SR-BI is colocalized with caveolin-1, indicating that SR-BIC323G is located in caveolae on plasma membrane (Fig. 1C).
C323 was required for SR-BI-mediated HDL binding, cholesteryl ester uptake, and cholesterol efflux
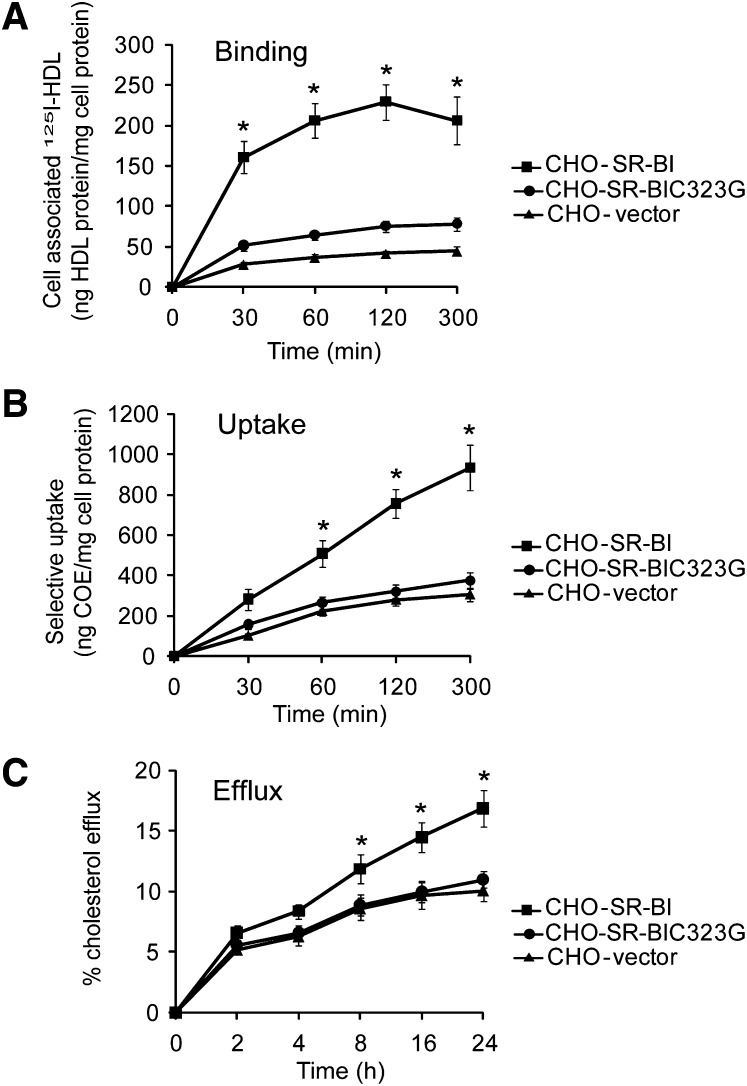
We conducted HDL binding and cholesteryl ester uptake assays using CHO-SR-BIC323G cells. In contrast to wild-type SR-BI, which binds HDL and significantly promotes cholesteryl ester uptake, the C323G mutant lost most SR-BI-mediated HDL binding and cholesteryl ester uptake, similar to the levels of CHO vector cells, indicating C323 is required for HDL binding and SR-BI-mediated cholesteryl ester uptake (Fig. 2A, B).
Fig. 2.
C323 is required for SR-BI-mediated cholesterol transport. A and B: SR-BI mediated HDL binding and cholesteryl ester uptake. [3H]COE/[125I] double-labeled HDL3 (10 µg/ml) were incubated with CHO-vector, CHO-SR-BI, or CHO-C323G cells at 37°C for the indicated times, and then the HDL binding (A) and selective cholesteryl ester uptake (B) were determined. C: SR-BI-mediated cholesterol efflux. The CHO cells were prelabeled with [3H]cholesterol for 48 h. Cholesterol efflux was determined after incubating cells with 40 µg/ml HDL as the acceptor for the indicated times at 37°C. Results are expressed as a ratio of the radioactivity released in the culture media to the total radioactivity in media and cells. *P < 0.05 versus CHO-SR-BI cells.
In addition to mediating cholesteryl ester uptake from HDL, SR-BI has the ability to stimulate the release of free cholesterol from cells (36). To determine whether C323G alters SR-BI-mediated cholesterol efflux, CHO-SR-BIC323G cells were assayed for their ability to promote efflux of cholesterol to HDL. Compared with CHO-SR-BI cells, CHO-SR-BIC323G cells displayed significantly reduced levels of cholesterol efflux to HDL, similar to the levels of CHO vector cells (Fig. 2C). The data indicate that C323 is also required for SR-BI-mediated cholesterol efflux.
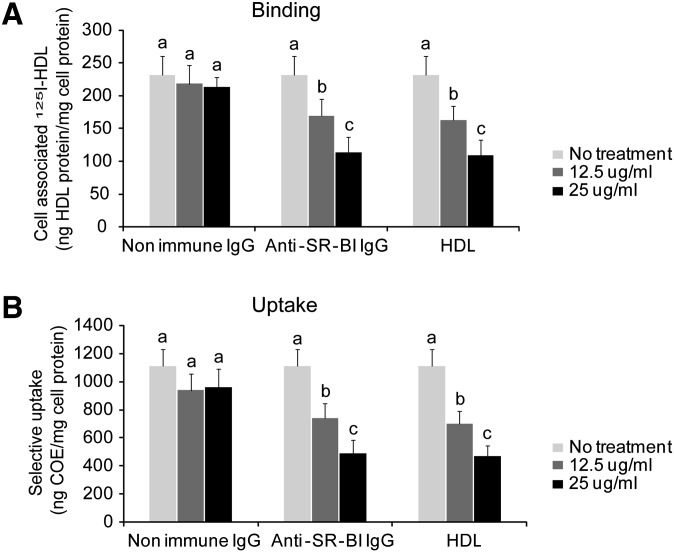
To determine whether C323 is directly involved in HDL binding, we generated a blocking antibody using a 15-amino-acid peptide flanking C323. The blocking antibody abolished SR-BI-mediated HDL binding and cholesteryl ester uptake in a dose-dependent manner (Fig. 3A, B). The blocking antibody worked as efficiently as unlabeled HDL did. These data suggest that C323 is likely an HDL binding site.
Fig. 3.
C323 of SR-BI is required for HDL binding and cholesteryl ester uptake. A blocking antibody against the C323 region and nonimmune IgGs or nonlabeled HDL were added to CHO-SR-BI cells 30 min prior to adding 10 µg/ml of [3H]COE/[125I] double-labeled HDL. The HDL binding (A) and selective cholesteryl ester uptake (B) were determined. a,b,cP < 0.05, data labeled with different letters are considered statistically different.
C323 of SR-BI was required for regulating plasma cholesterol levels in mice
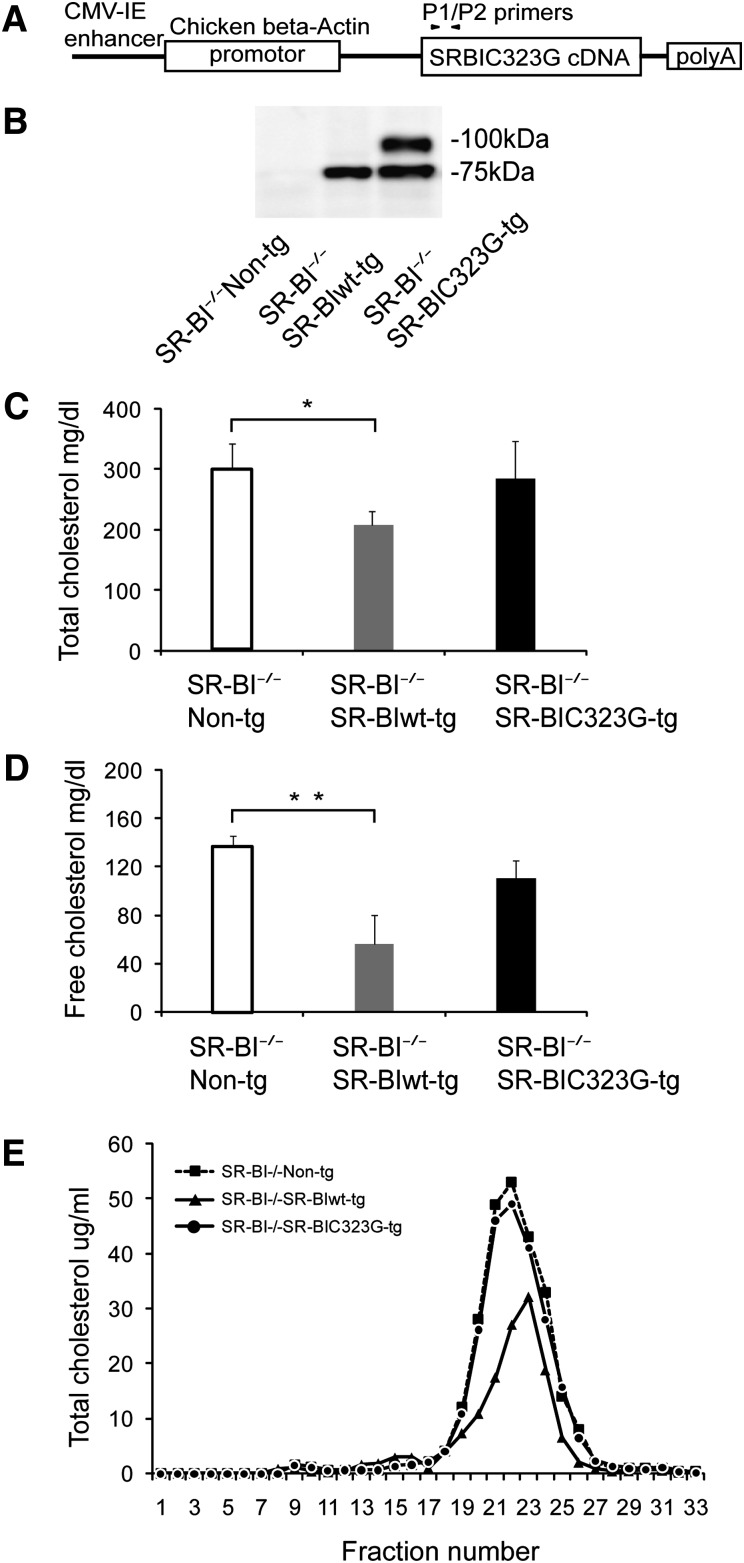
To determine the role of C323 in vivo, we generated transgenic mice expressing wild-type and C323G mutant SR-BI (Fig. 4A). To exclude the effect of endogenous SR-BI, we used transgenic mice in SR-BI−/− background. When assessed by Western blot, the wild-type SR-BI expressed as a single band at 75 kDa, and C323G mutant SR-BI expressed as two bands at 75 kDa and 100 kDa in the liver (Fig. 4B). As we expected, transgenic expression of wild-type SR-BI in SR-BI−/−SR-BIwt-tg mice significantly reduced the total and free cholesterol levels compared with SR-BI−/−non-tg littermates (Fig. 4C). However, transgenic expression of C323G mutant SR-BI in SR-BI−/−SR-BIC323G-tg mice had no significant effect on the total or free cholesterol levels compared with SR-BI−/−non-tg littermates (Fig. 4C, D). These data indicated that C323 of SR-BI is required for regulating plasma cholesterol levels in mice. Mice deficient in SR-BI have abnormally large HDL particles. We compared the size of HDL particles between SR-BI−/−SR-BIC323G-tg mice and SR-BI−/−SR-BI-wt-tg mice with FPLC. As shown in Fig. 4E, expression of wild-type SR-BI reduced the size of HDL particles but expression of C323G mutant had no such effect.
Fig. 4.
C323G of SR-BI is required for regulating plasma cholesterol levels in mice. A: Schematic model of SR-BIC323G transgenic construct. B: Western blot analysis of SR-BI expression in the liver of SR-BI−/−non-tg, SR-BI−/−SR-BIwt-tg, and SR-BI−/−SR-BIC323G-tg mice. C and D: Total and free plasma cholesterol levels in SR-BI−/−non-tg, SR-BI−/−SR-BIwt-tg, and SR-BI−/−SR-BIC323G-tg mice. N = 6 mice per group. *P < 0.05 and **P < 0.01 versus SR-BI−/−non-tg littermates. E: Lipoprotein profiling of SR-BI−/−non-tg, SR-BI−/−SR-BIwt-tg, and SR-BI−/−SR-BIC323G-tg mice. SR-BI−/−non-tg littermates were used as control. N = 2 mice per group.
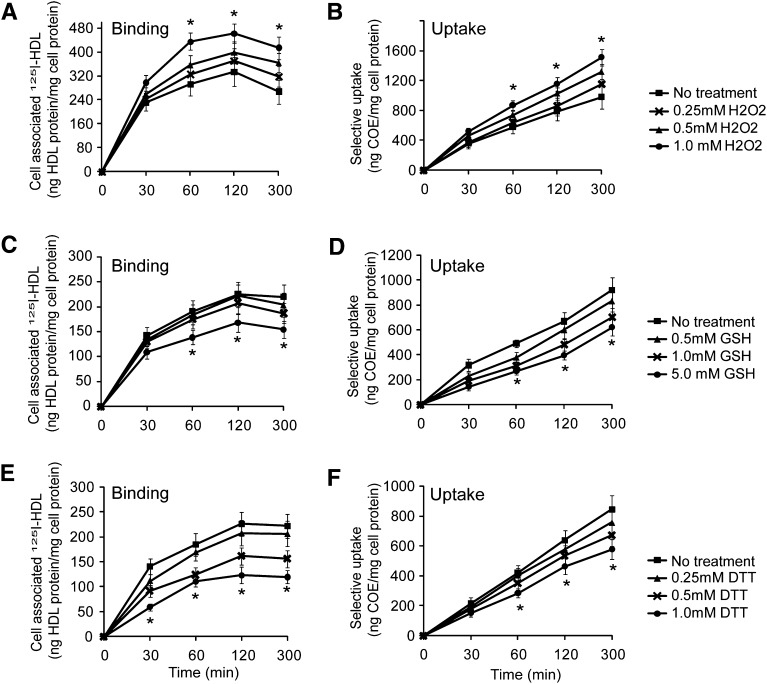
Changes in redox status regulated SR-BI-mediated HDL binding and cholesteryl ester uptake
The redox regulatory nature of cysteine suggests that a change in redox status may modulate SR-BI-mediated HDL binding and cholesteryl ester uptake. To test this hypothesis, we conducted HDL binding and cholesteryl ester uptake assays in the presence of redox reagents. The H2O2 treatment markedly enhanced SR-BI-mediated HDL binding and cholesteryl ester uptake by 65%, and GSH treatment markedly downregulated SR-BI-mediated HDL binding and cholesteryl ester uptake by 45% (Fig. 5AndashD). Similar downregulation of SR-BI-mediated HDL binding and cholesteryl ester uptake was observed with DTT treatment (Fig. 5E, F). These data indicated that an oxidative status promotes SR-BI-mediated HDL binding and cholesteryl ester uptake, whereas a reductive status suppresses SR-BI-mediated HDL binding and cholesteryl ester uptake. Note that the extracellular compartment is an oxidative environment and that H2O2 is one of the major oxidants in the extracellular compartment, which presents as high as submillimole per liter levels (37). Our data suggest that physiological relevant levels of H2O2 can promote SR-BI-mediated HDL binding and cholesteryl ester uptake.
Fig. 5.
Changes in redox status modulate SR-BI-mediated HDL binding and selective uptake activity. [3H]COE/[125I] double-labeled HDL (10 µg/ml) were incubated with CHO-SR-BI cells in the presence of H2O2, GSH, or DTT, and the effects of H2O2 (A, B), GSH (C, D), and DTT (E, F) on HDL binding and CE selective uptake were determined. *P < 0.05 versus CHO-SR-BI cells without redox reagent treatment.
DISCUSSION
The importance of extracellular domain of SR-BI in cholesteryl ester uptake has been explored by the use of chimeric receptors (28), by insertion of epitope tags into various regions of the domain of SR-BI (29), by blocking antibody against the extracellular domain, and by mutations (30, 31). However, the HDL binding site and the factors that regulate SR-BI-mediated cholesteryl ester uptake remain to be identified. In this study, using C323G mutant SR-BI, we demonstrated that the highly conserved C323 is required for SR-BI-mediated HDL binding and cholesteryl ester uptake. Using a blocking antibody against C323 region, we showed that C323 is involved in HDL binding and that it is likely an HDL binding site. Using transgenic mouse models, we further demonstrated that C323 of SR-BI is required for regulating plasma cholesterol levels in vivo.
A potential problem with the point mutation approach is that a single site mutation may disrupt the conformation of a protein, resulting in loss of function. The following evidence suggests that it may not be the case for SR-BIC323G mutant: i) our early study showed that SR-BIC323G mutant still had full activity of suppressing LPS-induced inflammatory response in macrophages (27), and ii) the blocking antibody effectively suppressed HDL binding to CHO-SR-BI cells, suggesting that C323 of SR-BI is likely an HDL binding site directly involved in SR-BI-mediated cholesteryl ester uptake.
SR-BIC323G mutant is highly glycosylated, which may affect its HDL binding and cholesterol ester uptake activity. To determine whether the extra glycosylation affects the activity of SR-BI, we generated CHO cells stably expressing cysteine mutants, including C251G, C280G, C321G, C334G, and C384G (supplementary Fig. I-A). Among these mutants, CHO-SR-BIC321G, CHO-SR-BIC334G, and CHO-SR-BIC384G cells displayed the highly glycosylated SR-BI as CHO-SR-BIC323G cells did (supplementary Fig. I-A, B). However, both CHO-SR-BIC321G and CHO-SR-BIC384G cells had almost full HDL binding and cholesterol ester uptake (supplementary Fig. I-D, E), indicating that extra glycosylation of SR-BI does not affect HDL binding and cholesterol ester uptake.
Different from a reducing environment in the intracellular compartment, the extracellular compartment is highly oxidative (37). Cysteinyl residues in the extracellular compartment can undergo a variety of oxidative modifications. For example, the cysteinyl residues of many proteins participate in reversible oxidation reactions leading to the formation of disulfides (-S-S-), thiolates (-S-), thiyl radicals (-S·) et al., as well as irreversible oxidation reactions leading to formation of sulfinic acid (-SO2H) and sulfonic acid (-SO3H). In addition, protein thiols can react to form mixed disulfides between proteins, with extracellular GSH (S-glutathiolation), or with other small thiols, or to form S-nitrosthiol derivatives (RSNO). Given the critical role of cysteinyl residues in SR-BI-mediated HDL binding and cholesteryl uptake and the redox regulatory nature of cysteine, we speculated that changes in redox status modulate SR-BI-mediated HDL binding and cholesteryl ester uptake. To test this, we conducted HDL binding and cholesteryl ester uptake assays in the presence of redox reagents. We found that H2O2 upregulated the SR-BI-mediated cholesteryl ester uptake by 65%, whereas GSH or DTT significantly downregulated SR-BI-mediated cholesteryl ester uptake by 45%. Of note, the extracellular compartment is highly oxidative (37). The extracellular compartment contains high levels (up to submilimole per liter) of H2O2 (38). Our data indicate that physiological relevant levels of H2O2 markedly enhance SR-BI-mediated HDL binding and cholesteryl ester uptake. To understand how redox reagents regulate SR-BI-mediated cholesterol uptake, we treated the CHO-SR-BI cells with redox regents, isolated subcellular fractions with Opti-Prep method, and assessed the effect of redox reagents on SR-BI subcellular distribution. As shown in supplementary Fig. II, treatment of CHO-SR-BI cells with 1 µM DTT moderately increased SR-BI caveolae distribution, and treatment of CHO-SR-BI cells with 1 µM H2O2 slightly decreased SR-BI caveolae distribution (based on CM/PNS ratio). It seems that the changes in SR-BI caveolae distribution may not account for the changes induced by redox reagents.
Note that H2O2, DTT, and GSH are mild redox reagents that modify cysteinyl residue in a revisable manner. So, technically, we cannot treat the cells or HDL with the redox regents first, remove the reagents, and then perform HDL binding/cholesterol uptake assay. Therefore, the current approach could not distinguish whether the effect is actually on SR-BI or HDL. Nevertheless, we could get an idea that changes in redox status may regulate SR-BI-mediated HDL binding and cholesteryl ester uptake. Further study is warranted to determine redox regulation of SR-BI-mediated cholesterol transport and how an increase in oxidative status, such as in case of hypercholesterolemia, alters SR-BI-mediated cholesterol transport in vivo.
In summary, this study demonstrated that C323 is required for SR-BI-mediated HDL binding/cholesteryl ester uptake and for regulating plasma cholesterol levels in vivo and that change in redox status might be a regulatory factor that modulates SR-BI-mediated cholesterol transport.
Supplementary Material
Footnotes
Abbreviations:
- CHO
- Chinese hamster ovary
- COE
- cholesteryl oleoyl ether
- SR-BI
- scavenger receptor BI
This work was supported by grants to X-A.L. from the American Heart Association (0530241N), the National Institutes of Health (R01 GM-085231), and the Children's Miracle Network. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two figures.
REFERENCES
- 1.Acton S., Rigotti A., Landschulz K. T., Xu S., Hobbs H. H., Krieger M. 1996. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science. 271: 518–520. [DOI] [PubMed] [Google Scholar]
- 2.Krieger M. 2001. Scavenger receptor class B type I is a multiligand HDL receptor that influences diverse physiologic systems. J. Clin. Invest. 108: 793–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reaven E., Cortez Y., Leers-Sucheta S., Nomoto A., Azhar S. 2004. Dimerization of the scavenger receptor class B type I: formation, function, and localization in diverse cells and tissues. J. Lipid Res. 45: 513–528. [DOI] [PubMed] [Google Scholar]
- 4.Azhar S., Tsai L., Reaven E. 1990. Uptake and utilization of lipoprotein cholesteryl esters by rat granulosa cells. Biochim. Biophys. Acta. 1047: 148–160. [DOI] [PubMed] [Google Scholar]
- 5.Out R., Kruijt J. K., Rensen P. C., Hildebrand R. B., de Vos P., Van Eck M., Van Berkel T. J. 2004. Scavenger receptor BI plays a role in facilitating chylomicron metabolism. J. Biol. Chem. 279: 18401–18406. [DOI] [PubMed] [Google Scholar]
- 6.Brodeur M. R., Luangrath V., Bourret G., Falstrault L., Brissette L. 2005. Physiological importance of SR-BI in the in vivo metabolism of human HDL and LDL in male and female mice. J. Lipid Res. 46: 687–696. [DOI] [PubMed] [Google Scholar]
- 7.Brundert M., Ewert A., Heeren J., Behrendt B., Ramakrishnan R., Greten H., Merkel M., Rinninger F. 2005. Scavenger receptor class B type I mediates the selective uptake of high-density lipoprotein-associated cholesteryl ester by the liver in mice. Arterioscler. Thromb. Vasc. Biol. 25: 143–148. [DOI] [PubMed] [Google Scholar]
- 8.Rigotti A., Trigatti B. L., Penman M., Rayburn H., Herz J., Krieger M. 1997. A targeted mutation in the murine gene encoding the high density lipoprotein (HDL) receptor scavenger receptor class B type I reveals its key role in HDL metabolism. Proc. Natl. Acad. Sci. USA. 94: 12610–12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arai T., Wang N., Bezouevski M., Welch C., Tall A. R. 1999. Decreased atherosclerosis in heterozygous low density lipoprotein receptor-deficient mice expressing the scavenger receptor BI transgene. J. Biol. Chem. 274: 2366–2371. [DOI] [PubMed] [Google Scholar]
- 10.Braun A., Trigatti B. L., Post M. J., Sato K., Simons M., Edelberg J. M., Rosenberg R. D., Schrenzel M., Krieger M. 2002. Loss of SR-BI expression leads to the early onset of occlusive atherosclerotic coronary artery disease, spontaneous myocardial infarctions, severe cardiac dysfunction, and premature death in apolipoprotein E-deficient mice. Circ. Res. 90: 270–276. [DOI] [PubMed] [Google Scholar]
- 11.Kozarsky K. F., Donahee M. H., Glick J. M., Krieger M., Rader D. J. 2000. Gene transfer and hepatic overexpression of the HDL receptor SR-BI reduces atherosclerosis in the cholesterol-fed LDL receptor-deficient mouse. Arterioscler. Thromb. Vasc. Biol. 20: 721–727. [DOI] [PubMed] [Google Scholar]
- 12.Yu H., Zhang W., Yancey P. G., Koury M. J., Zhang Y., Fazio S., Linton M. F. 2006. Macrophage apolipoprotein E reduces atherosclerosis and prevents premature death in apolipoprotein E and scavenger receptor-class BI double-knockout mice. Arterioscler. Thromb. Vasc. Biol. 26: 150–156. [DOI] [PubMed] [Google Scholar]
- 13.Van Eck M., Twisk J., Hoekstra M., Van Rij B. T., Van Der Lans C. A., Bos I. S., Kruijt J. K., Kuipers F., Van Berkel T. J. 2003. Differential effects of scavenger receptor BI deficiency on lipid metabolism in cells of the arterial wall and the liver. J. Biol. Chem. 278: 23699–23705. [DOI] [PubMed] [Google Scholar]
- 14.Fu T., Kozarsky K. F., Borensztajn J. 2003. Overexpression of SR-BI by adenoviral vector reverses the fibrateinduced hypercholesterolemia of apolipoprotein E-deficient mice. J. Biol. Chem. 278: 52559–52563. [DOI] [PubMed] [Google Scholar]
- 15.Wang N., Arai T., Ji Y., Rinninger F., Tall A. R. 1998. Liver-specific overexpression of scavenger receptor BI Decreases levels of very low density lipoprotein ApoB, low density lipoprotein ApoB, and high density lipoprotein in transgenic mice. J. Biol. Chem. 273: 32920–32926. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y., Da Silva J. R., Reilly M., Billheimer J. T., Rothblat G. H., Rader D. J. 2005. Hepatic expression of scavenger receptor class B type I (SR-BI) is a positive regulator of macrophage reverse cholesterol transport in vivo. J. Clin. Invest. 115: 2870–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozarsky K. F., Donahee M. H., Rigotti A., Iqbal S. N., Edelman E. R., Krieger M. 1997. Overexpression of the HDL receptor SR-BI alters plasma HDL and bile cholesterol levels. Nature. 387: 414–417. [DOI] [PubMed] [Google Scholar]
- 18.Ueda Y., Gong E., Royer L., Cooper P. N., Francone O. L., Rubin E. M. 2000. Relationship between expression levels and atherogenesis in scavenger receptor class B, type I transgenics. J. Biol. Chem. 275: 20368–20373. [DOI] [PubMed] [Google Scholar]
- 19.Vergeer M., Korporaal S. J., Franssen R., Meurs I., Out R., Hovingh G. K., Hoekstra M., Sierts J. A., Dallinga-Thie G. M., Motazacker M. M., et al. 2011. Genetic variant of the scavenger receptor BI in humans. N. Engl. J. Med. 364: 136–145. [DOI] [PubMed] [Google Scholar]
- 20.Li X. A., Titlow W. B., Jackson B. A., Giltiay N., Nikolova-Karakashian M., Uittenbogaard A., Smart E. J. 2002. High density lipoprotein binding to scavenger receptor, Class B, type I activates endothelial nitric-oxide synthase in a ceramide-dependent manner. J. Biol. Chem. 277: 11058–11063. [DOI] [PubMed] [Google Scholar]
- 21.Yuhanna I. S., Zhu Y., Cox B. E., Hahner L. D., Osborne-Lawrence S., Lu P., Marcel Y. L., Anderson R. G., Mendelsohn M. E., Hobbs H. H., et al. 2001. High-density lipoprotein binding to scavenger receptor-BI activates endothelial nitric oxide synthase. Nat. Med. 7: 853–857. [DOI] [PubMed] [Google Scholar]
- 22.Mineo C., Yuhanna I. S., Quon M. J., Shaul P. W. 2003. High density lipoprotein-induced endothelial nitric-oxide synthase activation is mediated by Akt and MAP kinases. J. Biol. Chem. 278: 9142–9149. [DOI] [PubMed] [Google Scholar]
- 23.Gong M., Wilson M., Kelly T., Su W., Dressman J., Kincer J., Matveev S. V., Guo L., Guerin T., Li X. A., et al. 2003. HDL-associated estradiol stimulates endothelial NO synthase and vasodilation in an SR-BI-dependent manner. J. Clin. Invest. 111: 1579–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X. A., Guo L., Dressman J. L., Asmis R., Smart E. J. 2005. A Novel ligand-independent apoptotic pathway induced by scavenger receptor class B, type I and suppressed by endothelial nitric-oxide synthase and high density lipoprotein. J. Biol. Chem. 280: 19087–19096. [DOI] [PubMed] [Google Scholar]
- 25.Li X. A., Guo L., Asmis R., Nikolova-Karakashian M., Smart E. J. 2006. Scavenger receptor BI prevents nitric oxide-induced cytotoxicity and endotoxin-induced death. Circ. Res. 98: e60–e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai L., Ji A., de Beer F. C., Tannock L. R., van der Westhuyzen D. R. 2008. SR-BI protects against endotoxemia in mice through its roles in glucocorticoid production and hepatic clearance. J. Clin. Invest. 118: 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo L., Song Z., Li M., Wu Q., Wang D., Feng H., Bernard P., Daugherty A., Huang B., Li X. A. 2009. Scavenger receptor BI protects against septic death through its role in modulating inflammatory response. J. Biol. Chem. 284: 19826–19834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Connelly M. A., de la Llera-Moya M., Monzo P., Yancey P. G., Drazul D., Stoudt G., Fournier N., Klein S. M., Rothblat G. H., Williams D. L. 2001. Analysis of chimeric receptors shows that multiple distinct functional activities of scavenger receptor, class B, type I (SR-BI), are localized to the extracellular receptor domain. Biochemistry. 40: 5249–5259. [DOI] [PubMed] [Google Scholar]
- 29.Connelly M. A., De La Llera-Moya M., Peng Y., Drazul-Schrader D., Rothblat G. H., Williams D. L. 2003. Separation of lipid transport functions by mutations in the extracellular domain of scavenger receptor class B, type I. J. Biol. Chem. 278: 25773–25782. [DOI] [PubMed] [Google Scholar]
- 30.Parathath S., Sahoo D., Darlington Y. F., Peng Y., Collins H. L., Rothblat G. H., Williams D. L., Connelly M. A. 2004. Glycine 420 near the C-terminal transmembrane domain of SR-BI is critical for proper delivery and metabolism of high density lipoprotein cholesteryl ester. J. Biol. Chem. 279: 24976–24985. [DOI] [PubMed] [Google Scholar]
- 31.Papale G. A., Nicholson K., Hanson P. J., Pavlovic M., Drover V. A., Sahoo D. 2010. Extracellular hydrophobic regions in scavenger receptor BI play a key role in mediating HDL-cholesterol transport. Arch. Biochem. Biophys. 496: 132–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bilheimer D. W., Eisenberg S., Levy R. I. 1972. The metabolism of very low density lipoprotein proteins. I. Preliminary in vitro and in vivo observations. Biochim. Biophys. Acta. 260: 212–221. [DOI] [PubMed] [Google Scholar]
- 33.Graf G. A., Connell P. M., van der Westhuyzen D. R., Smart E. J. 1999. The class B, type I scavenger receptor promotes the selective uptake of high density lipoprotein cholesterol ethers into caveolae. J. Biol. Chem. 274: 12043–12048. [DOI] [PubMed] [Google Scholar]
- 34.Temel R. E., Walzem R. L., Banka C. L., Williams D. L. 2002. Apolipoprotein A-I is necessary for the in vivo formation of high density lipoprotein competent for scavenger receptor BI-mediated cholesteryl ester-selective uptake. J. Biol. Chem. 277: 26565–26572. [DOI] [PubMed] [Google Scholar]
- 35.Li X., Hatanaka K., Guo L., Tsushima M., Kitamura Y., Yamamoto A. 1993. A sensitive peroxidase staining immunoblotting method for measuring total protein S in human plasma. Thromb. Haemost. 69: 331–334. [PubMed] [Google Scholar]
- 36.Ji Y., Jian B., Wang N., Sun Y., Moya M. L., Phillips M. C., Rothblat G. H., Swaney J. B., Tall A. R. 1997. Scavenger receptor BI promotes high density lipoprotein-mediated cellular cholesterol efflux. J. Biol. Chem. 272: 20982–20985. [DOI] [PubMed] [Google Scholar]
- 37.Ottaviano F. G., Handy D. E., Loscalzo J. 2008. Redox regulation in the extracellular environment. Circ. J. 72: 1–16. [DOI] [PubMed] [Google Scholar]
- 38.Lacy F., Kailasam M. T., O'Connor D. T., Schmid-Schonbein G. W., Parmer R. J. 2000. Plasma hydrogen peroxide production in human essential hypertension: role of heredity, gender, and ethnicity. Hypertension. 36: 878–884. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.