Abstract
The PPARγ2 gene single nucleotide polymorphism (SNP) Pro12Ala has shown variable association with metabolic syndrome traits in healthy subjects. The RISCK Study investigated the effect of interaction between genotype and the ratio of polyunsaturated:saturated (P:S) fatty acid intake on plasma lipids in 367 white subjects (ages 30-70 years) at increased cardiometabolic risk. Interaction was determined after habitual diet at recruitment, at baseline after a 4-week high-SFA (HS) diet, and after a 24-week reference (HS), high-MUFA (HM), or low-fat (LF) diet. At recruitment, there were no significant associations between genotype and plasma lipids; however, P:S × genotype interaction influenced plasma total cholesterol (TC) (P = 0.02), LDL-cholesterol (LDL-C) (P = 0.002), and triglyceride (TG) (P = 0.02) concentrations. At P:S ratio ≤ 0.33, mean TC and LDL-C concentrations in Ala12 allele carriers were significantly higher than in noncarriers (respectively, P = 0.003; P = 0.0001). Significant trends in reduction of plasma TC (P = 0.02) and TG (P = 0.002) concentrations occurred with increasing P:S (respectively, ≤0.33 to >0.65; 0.34 to >0.65) in Ala12 allele carriers. There were no significant differences between carriers and noncarriers after the 4-week HS diet or 24-week interventions. Plasma TC and TG concentrations in PPARG Ala12 allele carriers decrease as P:S increases, but they are not dependent on a reduction in SFA intake.
Keywords: peroxisome proliferator-activated receptor-γ, PPARγ, single nucleotide polymorphism, polyunsaturated fatty acid, saturated fatty acid, gene-nutrient interaction
The transcription factor peroxisome proliferator-activated receptor-γ (PPARγ) is one of three PPARs and a member of the nuclear hormone receptor superfamily (1). The major natural ligands are polyunsaturated fatty acids and prostanoids (2), suggesting a role in transducing nutritional to metabolic signals (3). An increase in PPARγ mRNA in adipose tissue of mice exposed to a high-fat diet (4) suggested that dietary modulation might influence adipogenesis induced by PPARγ in response to raised plasma concentration of fatty acid ligands.
Variants of the PPARγ2 gene PPARG could alter transcriptional activity of the activator through DNA- and/or ligand-binding affinity. The Pro12Ala single nucleotide polymorphism (SNP) (rs1801282) is present at a frequency of approximately 7.6% in Caucasians (5). Previous studies have investigated genotype associations with risk of obesity and diabetes, with equivocal results (6–8), suggesting that environmental influences such as dietary intake may be involved. Fatty acid affinities for PPARγ depend largely on their chain length and degree of saturation (2). Thus, the metabolic impact of this polymorphism is potentially dependent on gene interaction with different types of dietary fat. Luan et al. (9) found that body mass index (BMI) in Ala12 allele carriers (but not Pro12 homozygotes) was influenced by the ratio of habitual dietary polyunsaturated:saturated fatty acid (P:S) intake, and Memisoglu et al. (10) found MUFA (but not total fat intake) was inversely associated with BMI only in Ala12 allele carriers. In vitro, the PPARγ2 Ala-variant exhibits reduced binding to DNA and modest impairment of transcriptional activation following treatment with pharmacological ligand thiazolidinediones (TZD) (11, 12). This raised the possibility that differential responses by Ala12 allele carriers and noncarriers to unsaturated fatty acid ligands might influence expression of PPARγ target genes, including several involved in cholesterol and triglyceride metabolism (13–15).
We hypothesized that the P:S intake ratio might interact with PPARG Pro12Ala genotype to influence concentration of plasma lipids. The RISCK Study investigated 367 white men and women aged 30-70 years who are at increased risk of metabolic syndrome (16) and who underwent a 4-week run-in on a diet high in SFA (HS diet), followed by randomization to 24-week reference (HS), high-MUFA (HM), and low-fat (LF) diets. We utilized habitual intake at recruitment to investigate the effect of P:S ratio, as PUFA intake was constant in the subsequent interventions. A recent study has found interaction between PPARG Pro12Ala genotype with the intake of saturated fat as a determinant of LDL-C peak particle diameter (17). We used data from the interventions to investigate the specific effect of reduction in SFA with respect to PUFA intake in determination of plasma lipid concentrations.
METHODS
Subjects
Ethical approval for the RISCK Study (ISRCTN29111298) was granted from the National Research Ethics Service. Written informed consent from participants was obtained, including for subsequent genetic analyses. Men and women (age range, 30-70 years), who were recruited from the general population, attended a clinic in a fasting state at the participating centers [University of Reading, Imperial College London, University of Surrey, the Medical Research Council-Human Nutrition Research Unit (MRC-HNR) and King's College London]. Eligibility for entry to the study was assessed by a point system and implementation of exclusion criteria described previously (16). A total of 549 subjects completed the study. Self-reported ethnicity was recorded as White, South Asian, Black African, or Other.
Study design
The RISCK Study is a parallel 2 × 2 factorial design compared with a control intervention (16). At screening, unweighed 4-day food diaries (3 week days and 1 weekend day) were collected to record the habitual diet. Nutrient intakes were estimated by using the food-composition database software DINO as described previously (18). The intervention diets were planned to provide similar intakes of dietary energy but to vary in the amount and type of fats and carbohydrates. All participants followed a 4-week run-in period during which they consumed a high-saturated fat “reference diet” before being randomized to the reference diet or one of four isoenergetic dietary interventions designed to lower saturated fat. In this study, the dietary intervention groups differing in carbohydrate quality were combined to focus the analyses on the manipulation of dietary fat. The resulting three dietary groups were: high saturated fat “reference diet” (HS) designed to reflect a higher saturated fat intake than the habitual Western diet (∼18% of energy SFA, 12% MUFA, 38% total fat, 45% CHO), high-MUFA diet (HM) in which SFA was reduced and replaced with MUFA (∼10% of energy SFA, 20% MUFA, 38% total fat, 45% CHO) and low-fat diet (LF), in which SFA was reduced through replacement of total fat with carbohydrate (∼10% of energy SFA, 11% MUFA, 28% total fat, 55% CHO). The dietary intervention is described in detail elsewhere (18). Measurements made after the run-in diet are referred to as “baseline.” All participants followed their randomly prescribed diets for 24 weeks, after which a blood sample was collected and anthropometry was measured. Weight (in light clothing) and height (without shoes) were measured. An indwelling venous cannula was inserted into the forearm.
Biochemical analysis
Blood samples for analysis were drawn after a minimum 8 h overnight fast, and serum was stored at −45°C until analyzed. Fasting lipids, including total cholesterol (TC), HDL-cholesterol (HDL-C), and triglycerides (TG), were measured as described previously (16). LDL-cholesterol (LDL-C) was derived from the Friedwald equation.
DNA extraction and genotyping
Buffy coats removed from blood samples were stored in EDTA at −20°C. Genomic DNA was extracted from 200 μl buffy coat using an Illustra blood genomic prep mini spin kit (GE Healthcare, Amersham, UK) according to manufacturer's instructions. The PPARG Pro12Ala SNP (rs1801282) was genotyped by KBiosciences (Hoddesdon, UK). Genotype accuracy as assessed by inclusion of duplicates in the array was 98%, and negative controls (water blanks) were included on each plate. Genotyping success rate was 89%.
Statistical analysis
PPARG Pro12Ala genotype distributions were tested for deviation from the Hardy-Weinberg equilibrium by a χ2 test with 1 df (P > 0.05). Statistical analyses were carried out using the SPSS version 17.0 for Windows (SPSS Inc, Chicago, IL). Where needed, variables were log-transformed to obtain better approximations of the normal distribution prior to analysis. SNP genotype association with plasma lipid (TC, LDL-C, or TG) concentration was tested using analysis of covariance (ANCOVA), with BMI, age, gender, and diet as covariates. Outliers, defined as points >2.5 times the interquartile range from the median on the transformed scale at recruitment or after HS diet, were excluded. ANCOVA was also used to test interaction between genotypes and P:S quartiles. In this model, the dependent variable was the analyzed plasma lipid and fixed factors were the genotypes and P:S quartiles, with BMI, age, and gender as covariates. All data presented in text and tables are expressed as means or geometric means ± SD or 95% CI. Statistical significance was taken at P < 0.05.
RESULTS
PPARG Pro12Ala allele and genotype frequencies
All available DNA samples were genotyped initially (n = 466), and data was obtained for 415 subjects. The Ala12 allele frequency in white RISCK subjects was 0.10, greater than 0.076 in HapMap-CEU (European) subjects recorded on the NCBI SNP database (5). The SNP was absent in black RISCK subjects, as recorded in HapMap-YRI (sub-Saharan African). There are no comparative data available for South Asians. The genotype distributions did not deviate from Hardy-Weinberg expectations. The numbers of each genotype in white subjects were as follows: Pro/Pro: 258 (80%); Pro/Ala: 61 (19%); Ala/Ala: 3 (1%); total n = 322.
Characteristics of subjects
A total of 549 subjects completed the RISCK Study. According to the criteria of the International Diabetes Federation (19), 47.5% had metabolic syndrome. On the basis of self-reported ethnicity, individuals of white, South Asian, black African, and other ancestry were distinguished. In view of the small sample size of the South Asian and other ancestries and absence of the Ala12 allele in blacks, we chose to focus our genetic investigation on the white subjects only. The characteristics at recruitment of the white participants (n = 367) who completed the study and for whom DNA was available are presented in Table 1.
TABLE 1.
Characteristics of white RISCK Study subjects at recruitment screening
| Phenotype | Male(n = 155) | Female(n = 212) |
| Age (y) | 54 ± 10 | 53 ± 10 |
| Waist circumference (cm) | 103.1 ± 10.7 | 95.4 ± 12.6 |
| BMI (kg/m ) | 28.6 ± 4.0 | 29.1 ± 5.3 |
| TG (mmol/l)a | 1.4 ± 0.8 | 1.2 ± 0.7 |
| TC (mmol/l) | 5.6 ± 0.8 | 5.7 ± 1.0 |
| LDL-C (mmol/l) | 3.6 ± 0.8 | 3.5 ± 0.9 |
| HDL-C (mmol/l)a | 1.2 ± 0.3 | 1.5 ± 0.4 |
Data measured at recruitment is presented for all white subjects who completed the study and for whom DNA samples were available (n = 367). Values are mean ± SD unless indicated otherwise.
Log transformed mean ± SD.
Interaction between PPARG Pro12Ala genotype and habitual dietary P:S ratio
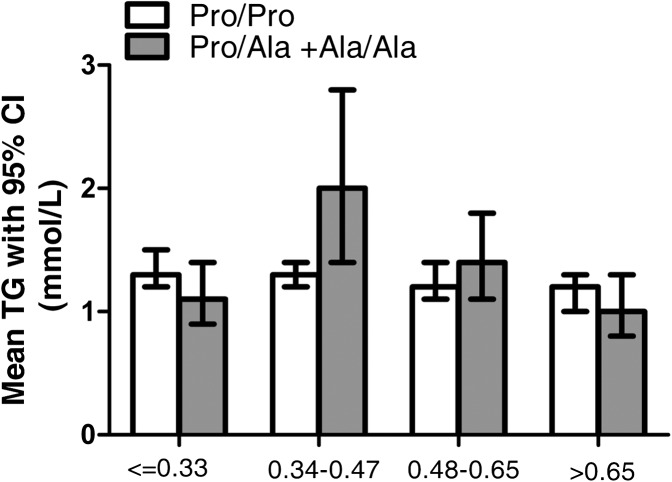
After adjustment for BMI, gender, and age there were no significant associations between PPARG Pro12Ala genotype and plasma concentrations of TC, LDL-C, or TG in white subjects at recruitment screening P = 0.05. Table 2 shows plasma TC, LDL-C, and TG concentrations with respect to genotype in quartiles of habitual P:S intake. There was a significant interaction between dietary P:S ratio and genotype as a determinant of plasma concentrations of TC (P = 0.02), LDL-C (P = 0.002), and TG (P = 0.02) after adjustment for BMI, age, and gender. Interaction between P:S ratio × genotype × gender was not significant, so we did not test the effect of P:S ratio × genotype interaction in males and females separately. When the P:S ratio was low (≤0.33), mean plasma TC concentration in Ala12 carriers was significantly higher than in noncarriers (P = 0.003). As P:S increased, the concentration of TC fell by 10%. The trend in reduction as the ratio increased from ≤0.33 to >0.65 was significant (P = 0.02). An even more significant difference was seen in LDL-C concentration between carriers and noncarriers in the lowest P:S quartile (P = 0.0001). As P:S increased, the concentration fell by 19.5% in Ala12 carriers, but the trend was not significant (P > 0.05). There were no significant differences in plasma TG concentrations between Ala12 carriers and noncarriers in any P:S quartile. However, there was a significant trend in the reduction of plasma TG in Ala12 carriers as the P:S ratio increased from 0.34 to >0.65, in which concentration fell by 50% (P = 0.002). Plasma TG concentrations stratified by genotype and P:S quartile are shown in Fig. 1.
TABLE 2.
Plasma lipid concentrations with respect to Pro12Ala genotype and quartiles of habitual dietary P:S intake ratio
| TC |
LDL-C |
TG |
|||||||||||||
| P:S quartile | Pro/Pro |
Pro/Ala + Ala/Ala |
P | Pro/Pro |
Pro/Ala + Ala/Ala |
P | Pro/Pro |
Pro/Ala + Ala/Ala |
P | ||||||
| Mean (95% CI) | n | Mean (95% CI) | n | Mean (95% CI) | n | Mean (95% CI) | n | Mean (95% CI) | n | Mean (95% CI) | n | ||||
| ≤0.33 | 5.4 (5.2,5.6) | 64 | 6.1 (5.7,6.6) | 16 | 0.003 | 3.3 (3.1,3.5) | 64 | 4.1 (3.7,4.6) | 16 | 0.0001 | 1.3 (1.2,1.5) | 64 | 1.1 (0.9,1.4) | 16 | 0.09 |
| 0.34-0.47 | 5.7 (5.5,5.9) | 76 | 5.8 (5.1,6.5) | 8 | 0.92 | 3.6 (3.4,3.8) | 76 | 3.3 (2.6,3.9) | 9 | 0.16 | 1.3 (1.2,1.4) | 76 | 2.0 (1.4,2.8) | 8 | 0.09 |
| 0.48-0.65 | 5.6 (5.3,5.8) | 59 | 5.5 (5.1,5.9) | 19 | 0.71 | 3.5 (3.3,3.7) | 59 | 3.3 (2.9,3.8) | 19 | 0.51 | 1.2 (1.1,1.4) | 59 | 1.4 (1.1,1.8) | 19 | 0.19 |
| >0.65 | 5.8 (5.5,6.0) | 49 | 5.5 (5.0,6.0) | 17 | 0.33 | 3.7 (3.4,3.9) | 49 | 3.6 (3.1,4.0) | 17 | 0.67 | 1.2 (1.0,1.3) | 49 | 1.0 (0.8,1.3) | 17 | 0.37 |
Data is presented for subjects for whom genotypic and phenotypic data was available (n = 367). Mean (95% CI) or geometric mean (95% CI) values for TG (mmol/l), stratified by genotype, are shown after habitual diet. Association was tested by univariate ANOVA based on a dominant model. P-values, adjusted for BMI, age, and gender, are shown in bold when nominally significant (< 0.05).
Fig. 1.
Mean TG concentrations with respect to quartiles of habitual dietary P:S ratio and PPARG Pro12Ala genotype. The numbers of genotyped subjects with measurements in each quartile of P:S ratio were as shown in Table 2. Geometric mean concentrations of TG are shown. Bars represent 95% CI. Dietary P:S ratio × genotype interaction determined by univariate ANCOVA significantly influenced plasma TG concentration (P = 0.02, after adjustment for BMI, gender, and age). There was a significant trend in reduction of plasma TG concentration between P:S ratio 0.34 and >0.65 (P = 0.002) in Ala12 allele carriers.
Change in plasma lipid concentrations after dietary intervention
After the 4-week run-in on HS diet, subjects were randomly assigned to continue the HS reference diet or to begin the HM or LF diet. The HM group had lower plasma phospholipid % SF than the LF group (P ≤ 0.03) and higher % MUFA (P = 0.0001). The dietary interventions did not affect other fatty acid classes [(n-3) PUFA, (n-6) PUFA and trans FA] (18). TC and LDL-C concentrations were significantly lower with the HM and LF diets than with the HS diet (P < 0.001 and P < 0.001). Apo B concentrations differed between treatment groups (P < 0.001) and were lower with the HM and LF diets than with the HS diet. HDL-C concentrations were lower with the LF diet than with the HS or HM diet (P < 0.001 and P = 0.002, respectively). There were no significant changes in concentration of plasma TG following interventions (16).
PPARG Pro12Ala genotype associations with change in plasma lipid concentrations after dietary intervention
In the HS diet consumed during the 4-week run-in to baseline, SFA composed 18% of energy intake. The P:S ratio in whites at baseline was 0.35 [i.e., in the second quartile of habitual P:S intake (0.34-0.47)]. At baseline, carriers of the Ala12 allele (n = 64) had higher plasma concentrations compared with noncarriers (n = 258) of TC [mean (95% CI) 5.6 (5.5, 5.7) versus 5.8 (5.6, 6.1) mmol/l]; LDL-C [3.5 (3.4, 3.6) versus 3.7 (3.5, 3.9) mmol/l)] and apoB [0.95 (0.29, 0.02) versus 1.03 (0.33, 0.04) g/l)], but differences were not significant after adjustment for BMI, gender, and age.
To investigate the effect of decrease in SFA without alteration in MUFA intake, we compared change in plasma lipid concentrations after continuation on the HS with switching to LF diet (18% versus 10% SFA), with respect to PPARG Pro12Ala genotype. Both diets contained the same proportion of PUFA. There was no significant difference in the change in plasma TC, LDL-C, or TG concentrations with respect to genotype (n =193), respectively, P = 0.72, P = 0.60, and P = 0.69 after adjustment for change in BMI, age, and gender (supplementary Table I). To examine the effect of increased intake of MUFA without alteration in SFA, we compared change in plasma lipid concentrations after the HM and LF diets (20% versus 11% MUFA), which also contained the same proportion of PUFA. There was no significant difference in the change in plasma TC, LDL-C, or TG concentrations with respect to genotype (n =268), respectively, P = 0.74, P = 0.94, and P = 0.43 after adjustments (supplementary Table I).
DISCUSSION
Numerous studies have investigated associations between PPARG Pro12Ala genotype and risk of obesity and diabetes, with equivocal outcomes (6–8). These inconsistencies suggest that environmental modifiers of the effects of genetic variation in PPARγ2 may be involved. We investigated associations between genotype and plasma lipid concentrations in white subjects at risk of the metabolic syndrome following diets differing in proportions of saturated and unsaturated fatty acids. There were no significant associations between genotype and plasma lipids after habitual intake or dietary interventions. However, habitual dietary P:S ratio × genotype interaction influenced plasma TC, LDL-C, and TG concentrations. At low P:S ratio (≤0.33), mean TC and LDL-C concentrations in Ala12 allele carriers were significantly higher than in noncarriers. The trends for reduction in plasma TC and TG concentrations with increasing P:S intake were significant in Ala12 allele carriers. Paired comparisons of outcomes after dietary intervention suggest that lower SFA intake was not responsible for the effect.
Memisoglu et al. (10) showed that the responsiveness of Ala12 carriers to dietary manipulation only emerged when MUFA rather than total fat intake was analyzed. Luan et al. (9) had previously shown greater sensitivity of Ala12 allele carriers to dietary PUFA in determination of BMI. Genotype was not significantly associated with BMI without reference to diet, but interaction between the P:S ratio and genotype in its determination was highly significant. As the ratio of P:S increased, BMI decreased in Ala12 carriers but not in Pro12 homozygotes. Both findings (9, 10) are compatible with unsaturated fats acting as specific ligands for PPARγ (2) and lower transcriptional activity of the PPARγ-Ala variant reducing PPARγ-mediated adipogenesis (11).
Our study is the first to report significant interaction between the P:S ratio and Pro12Ala genotype influencing plasma TC, LDL-C, and TG concentrations. At low (<0.33) ratio of P:S in habitual intake, TC and LDL-C concentrations in carriers of the less transcriptionally active PPARγ-Ala variant were significantly higher than in those homozygous for the normal PPARγ-Pro form. As the P:S ratio increased, the concentration of plasma TC and LDL-C fell in Ala12 carriers, with a significant trend seen in the former. When PUFA replaces SFA in the diet, the major portion of cholesterol lowering is seen in the LDL fraction (20). Increased plasma LDL-C has been observed following TZD treatment (21); however, a mechanistic link to PPARγ target gene activation that might infer association of the less active PPARγ-Ala form with lower LDL-C concentration has not been established.
Lipoprotein lipase activity is a rate-limiting determinant of TG hydrolysis in plasma. Plasma TG concentration in Ala12 allele carriers fell consistently beyond the second P:S quartile, showing a significant trend. It is well known that n-3 fatty acids decrease the concentration of serum TG (22). PPARγ may mediate this effect, as PUFAs are PPARγ ligands (2) and LPL is a PPARγ target gene (15). Lindi et al. (23) found a significantly greater decrease in serum TG concentration in Ala12 allele carriers than in Pro12 homozygotes in response to n-3 fatty acid supplementation when the intake of SFA was below 10% (i.e., at high P:S intake). This is consistent with our finding of a fall in plasma TG concentration in Ala12 allele carriers as P:S intake increased, but it is inconsistent with reduced lipase activity associated with a less active PPARγ-Ala form.
To establish whether effects of interaction between with the P:S ratio of habitual intake and genotype that we had observed were related to increased PUFA (as distinct from decreased SFA), we first compared change in plasma lipid concentrations after HS and LF diets, in which SFA was reduced and MUFA remained constant. Carriage of the Ala12 allele was not significantly associated with change in either plasma LDL-C or TG concentration, so the decrease in SFA had no significant effect. This was not unexpected, in view of the finding that SFAs fail to interact efficiently with PPARγ in vitro (2). We obtained the same results when we compared changes in lipids after HM and LF diets, in which MUFA was raised and SFA remained constant. An increase in MUFAs might have been expected to have some effect, but they are weaker PPARγ activators than are PUFAs (2). Therefore, we cannot confirm that the interaction between the P:S ratio of habitual intake and Pro12Ala genotype in determining plasma TC, LDL-C, and TG concentrations depends specifically on an increase in consumption of PUFA, but it seems not to depend on a decrease in SFA.
Limitations of our study include a relatively small number of genotyped subjects with plasma lipid measurements (n = 367) and the small observed changes in plasma lipid concentrations. To demonstrate a significant difference in LDL-C concentration in Ala12 allele carriers compared with noncarriers across all P:S quartiles, a total sample size of 1,600 would be required for α = 0.05 and a power of 0.95. For TG concentration, the equivalent sample size needed would be 1,800. The significance of the effect of dietary P:S × gene interactions on plasma TC, LDL-C, and TG concentrations should be treated with caution, as they were of modest significance in mainly overweight subjects. Replication in other studies with maximal correspondence in ethnic origin, age, and gender would be required to minimize the risk of false positive or negative gene-diet associations. If substantiated in a larger cohort, a recommendation to Ala12 carriers to maintain a high dietary intake of PUFA:SFA and to reduce plasma concentrations of atherogenic cholesterol and TG would be justified. Identification of individuals who are genetically more likely to respond to particular dietary changes may be important for successful intervention in the prevention of cardiovascular disease.
Supplementary Material
Acknowledgments
The authors acknowledge the contributions of the additional RISCK Study Group members: University of Reading: Hannah Farrant (local coordinator); Claire Lawrence, Edel Magee, and Kit Tsoi (research assistants); Darren Cole (database manager); Anna Gent, Celia Greenberg, and Caroline Stokes (coding and analyses of dietary data); and Mario Siervo and Rosemary Hall (clinicians); Imperial College London: Louise Goff (local coordinator); Claire Howard, Namrata Dhopatkar and Bushra Siddiqui (research assistants); and Anne Dornhurst (clinician); King's College London: Fiona Lewis (local coordinator), Samantha Bowen, L. Chen, and Robert Gray (research assistants); Roy Sherwood (sample analyses of clinical biochemistry); and Anthony Leeds, A. Shah, G. Saran, J. Niehuser-Saran, and J. A. Cockburn (clinicians); University of Reading: Rachel Gitau (local coordinator); Katie Newens and Sean Lovegrove (research assistants); University of Reading and University of Surrey: John Wright (clinician); and University of Surrey: Margaret Griffin (local coordinator) and Nicola Harman (lead for lipid subclasses).
Footnotes
Abbreviations:
- BMI
- body mass index
- CHO
- carbohydrate
- CI
- confidence interval
- HDL-C
- high density lipoprotein cholesterol
- HM
- high MUFA (diet)
- HS
- high SFA (diet)
- LDL-C
- low density lipoprotein cholesterol
- LF
- low fat (diet)
- NCBI
- National Center for Biotechnology Information
- PPAR
- peroxisome proliferator-activated receptor
- P:S
- polyunsaturated:saturated fatty acid
- RISCK (study)
- Reading, Imperial, Surrey, Cambridge, King's
- SNP
- single nucleotide polymorphism
- TC
- total cholesterol
- TG
- triglyceride
- TZD
- thiazolidinedione
This work was supported by the UK Food Standards Agency Project NO2031 (G.S.F., B.A.G., J.A.L., S.A.J., and T.A.B.S.). Foods were supplied by Unilever Food and Health Research Institute (Unilever R&D, Vlaardingen, The Netherlands); Cereal Partners UK (Welwyn Garden City, Hertfordshire, United Kingdom); Grampian (Banff, United Kingdom); Weetabix Ltd. (Kettering, United Kingdom); and Sainsbury's Supermarkets Ltd. (London, United Kingdom). None of these providers had any role in the design and implementation of the study or analysis and interpretation of the data. A.A. was supported by a studentship from the Saudi Arabian Ministry of Higher Education. The authors and their research groups have a number of links with the food industry. In a personal capacity, G.S.F. is a consultant to Coca-Cola, Premier Foods, and Unilever. T.A.B.S. has been a consultant to Seven Seas, is a member of the Scientific Advisory Committee for the Global Dairy Platform and External Scientific Review Committee of the Malaysian Palm Oil Board, and chairs Cadbury's Global Nutrition Advisory Panel. T.A.B.S., B.A.G., J.A.L., S.A.J., and G.S.F. have received ad hoc honoraria for lectures or writing articles. In a nonpersonal capacity, B.A.G. was formerly a member of an expert group known as the Fat Panel, which was supported by Dairy Crest, Kerry Gold, and Unilever. S.A.J. serves on scientific advisory boards for Coca-Cola, Heinz, PepsiCo, Nestlé, and Kellogg's and sits on UK government advisory boards that include food industry members. All research groups received products from a range of food companies gratis for research purposes, including Archer Daniel Mills, Croda, Matthews Foods, Nestlé, PepsiCo, Jordan, GSK, and Unilever. A.A. and S.D.O. reported no conflicts of interest.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one tables.
REFERENCES
- 1.Desvergne B., Wahli W. 1999. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr. Rev. 20: 649–688. [DOI] [PubMed] [Google Scholar]
- 2.Xu H. E., Lambert M. H., Montana V. G., Parks D. J., Blanchard S. G., Brown P. J., Sternbach D. D., Lehmann J. M., Wisely G. B., Willson T. M., et al. 1999. Molecular recognition of fatty acids by peroxisome proliferator-activated receptors. Mol. Cell. 3: 397–403. [DOI] [PubMed] [Google Scholar]
- 3.Semple R. K., Chatterjee V. K., O'Rahilly S. 2006. PPARγ and human metabolic disease. J. Clin. Invest. 116: 581–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vidal-Puig A., Jimenez-Linan M., Lowell B. B., Hamann A., Hu E., Spiegelman B., Flier J. S., Moller D. E. 1996. Regulation of PPAR gamma gene expression by nutrition and obesity in rodents. J. Clin. Invest. 97: 2553–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.NCBI dbSNP [database]. Accessed December 15, 2010, at: http://www.ncbi.nlm.nih.gov/snp.
- 6.Altshuler D., Hirschhorn J. N., Klannemark M., Lindgren C. M., Vohl M. C., Nemesh J., Lane C. R., Schaffner S. F., Bolk S., Brewer C., et al. 2000. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat. Genet. 26: 76–80. [DOI] [PubMed] [Google Scholar]
- 7.Tönjes A., Scholz M., Loeffler M., Stumvoll M. 2006. Association of Pro12Ala polymorphism in peroxisome proliferator-activated receptor gamma with prediabetic phenotypes: meta-analysis of 57 studies on nondiabetic individuals. Diabetes Care. 29: 2489–2497. [DOI] [PubMed] [Google Scholar]
- 8.Masud S., Ye S., Group S. A. S. 2003. Effect of the peroxisome proliferator activated receptor-gamma gene Pro12Ala variant on body mass index: a meta-analysis. J. Med. Genet. 40: 773–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luan J., Browne P. O., Harding A. H., Halsall D. J., O'Rahilly S., Chatterjee V. K., Wareham N. J. 2001. Evidence for gene-nutrient interaction at the PPARgamma locus. Diabetes. 50: 686–689. [DOI] [PubMed] [Google Scholar]
- 10.Memisoglu A., Hu F. B., Hankinson S. E., Manson J. E., De Vivo I., Willett W. C., Hunter D. J. 2003. Interaction between a peroxisome proliferator-activated receptor gamma gene polymorphism and dietary fat intake in relation to body mass. Hum. Mol. Genet. 12: 2923–2929. [DOI] [PubMed] [Google Scholar]
- 11.Deeb S. S., Fajas L., Nemoto M., Pihlajamäki J., Mykkänen L., Kuusisto J., Laakso M., Fujimoto W., Auwerx J. 1998. A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat. Genet. 20: 284–287. [DOI] [PubMed] [Google Scholar]
- 12.Masugi J., Tamori Y., Mori H., Koike T., Kasuga M. 2000. Inhibitory effect of a proline-to-alanine substitution at codon 12 of peroxisome proliferator-activated receptor-gamma 2 on thiazolidinedione-induced adipogenesis. Biochem. Biophys. Res. Commun. 268: 178–182. [DOI] [PubMed] [Google Scholar]
- 13.Kast-Woelbern H. R., Dana S. L., Cesario R. M., Sun L., de Grandpre L. Y., Brooks M. E., Osburn D. L., Reifel-Miller A., Klausing K., Leibowitz M. D. 2004. Rosiglitazone induction of Insig-1 in white adipose tissue reveals a novel interplay of peroxisome proliferator-activated receptor gamma and sterol regulatory element-binding protein in the regulation of adipogenesis. J. Biol. Chem. 279: 23908–23915. [DOI] [PubMed] [Google Scholar]
- 14.Ogata M., Tsujita M., Hossain M. A., Akita N., Gonzalez F. J., Staels B., Suzuki S., Fukutomi T., Kimura G., Yokoyama S. 2009. On the mechanism for PPAR agonists to enhance ABCA1 gene expression. Atherosclerosis. 205: 413–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schoonjans K., Peinado-Onsurbe J., Lefebvre A. M., Heyman R. A., Briggs M., Deeb S., Staels B., Auwerx J. 1996. PPARα and PPARγ activators direct a distinct tissue-specific transcriptional response via a PPRE in the lipoprotein lipase gene. EMBO J. 15: 5336–5348. [PMC free article] [PubMed] [Google Scholar]
- 16.Jebb S. A., Lovegrove J. A., Griffin B. A., Frost G. S., Moore C. S., Chatfield M. D., Bluck L. J., Williams C. M., Sanders T. A. RISCK Study Group. 2010. Effect of changing the amount and type of fat and carbohydrate on insulin sensitivity and cardiovascular risk: the RISCK (Reading, Imperial, Surrey, Cambridge, and Kings) trial. Am. J. Clin. Nutr. 92: 748–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bouchard-Mercier A., Godin G., Lamarche B., Pérusse L., Vohl M. C. 2011. Effects of peroxisome proliferator-activated receptors, dietary fat intakes and gene-diet interactions on peak particle diameters of low-density lipoproteins. J. Nutrigenet. Nutrigenomics. 4: 36–48. [DOI] [PubMed] [Google Scholar]
- 18.Moore C., Gitau R., Goff L., Lewis F. J., Griffin M. D., Chatfield M. D., Jebb S. A., Frost G. S., Sanders T. A., Griffin B. A., et al. RISCK Study Group. 2009. Successful manipulation of the quality and quantity of fat and carbohydrate consumed by free-living individuals using a food exchange model. J. Nutr. 139: 1534–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alberti K. G., Zimmet P., Shaw J. 2006. Metabolic syndrome - a new world-wide definition. A consensus statement from the International Diabetes Federation. Diabet. Med. 23: 469–480. [DOI] [PubMed] [Google Scholar]
- 20.Vega G. L., Groszek E., Wolf R., Grundy S. M. 1982. Influence of polyunsaturated fats on composition of plasma lipoproteins and apolipoproteins. J. Lipid Res. 23: 811–822. [PubMed] [Google Scholar]
- 21.Ovalle F., Bell D. S. 2002. Lipoprotein effects of different thiazolidinediones in clinical practice. Endocr. Pract. 8: 406–410. [DOI] [PubMed] [Google Scholar]
- 22.Harris W. S., Lu G., Rambjor G. S., Walen A. I., Ontko J. A., Chang Q., Windsor S. L. 1997. Influence of n-3 fatty acid supplementation on the endogenous activities of plasma lipases. Am. J. Clin. Nutr. 66: 254–260. [DOI] [PubMed] [Google Scholar]
- 23.Lindi V., Schwab U., Louheranta A., Laakso M., Vessby B., Hermansen K., Storlien L., Riccardi G., Rivellese A. KANWU Study Group. 2003. Impact of the Pro12Ala polymorphism of the PPAR-gamma2 gene on serum triacylglycerol response to n-3 fatty acid supplementation. Mol. Genet. Metab. 79: 52–60. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.