Abstract
The Comparative Toxicogenomics Database (CTD) is a free resource that describes chemical-gene-disease networks to help understand the effects of environmental exposures on human health. The database contains more than 13,500 chemical-disease and 14,200 gene-disease interactions. In CTD, chemicals and genes are associated with a disease via two types of relationships: as a biomarker or molecular mechanism for the disease (M-type) or as a real or putative therapy for the disease (T-type). We leveraged these curated datasets to compute similarity indices that can be used to produce lists of comparable diseases (“DiseaseComps”) based upon shared toxicogenomic profiles. This new metric now classifies diseases with common molecular characteristics, instead of the traditional approach of using histology or tissue of origin to define the disorder. In the dawning era of “personalized medicine”, this feature provides a new way to view and describe diseases and will help develop testable hypotheses about chemical-gene-disease networks.
Availability
The database is available for free at http://ctd.mdibl.org/
Keywords: disease, gene, chemical, database, curation
Background
The Comparative Toxicogenomics Database (CTD) is a public resource that promotes understanding about the effects of environmental chemicals on human health [1]. CTD biocurators manually curate interactions from the scientific literature in a structured format using controlled vocabularies for chemicals, genes, diseases, molecular interactions, and organisms [2]. These datasets can be used to explore relationships and to generate novel, testable hypotheses about chemical-gene-disease pathways.
Analyses of the human disease network strive to categorize diseases with respect to common genes and molecular pathways, enabling hypotheses about shared or predisposing co-disorders and putative genetic susceptibilities [3]. Disease names, especially cancers, were often originally based upon histology (e.g., carcinoma) or tissue of origin (e.g., liver cancer). With the advent of molecular genotyping, however, many cancers are now being further differentiated by their unique molecular signatures (e.g., HER2-positive vs. HER2-negative breast cancer). Bioinformatics analyses of human disease networks have also discovered sets of interacting proteins and molecular pathways often shared by multiple diseases [4]. This shift to analyzing, classifying, and describing diseases via their molecular perspective can help discover new therapeutic approaches not previously considered. For example, shared molecular connections between diabetes and dementia are now fueling research into the possible use of insulin to treat Alzheimer disease [5].
Personalized medicine (which seeks to improve a drug's therapeutic potential as well as minimize its side-effects) is dependent upon understanding the unique molecular profile of the patient and their disease [6]. Genes alone, however, are not solely responsible for all complex diseases, since the environment also plays an important role [7]. Thus, CTD, which integrates datasets for all three components of environmental chemicals, genes, and diseases, can be uniquely leveraged to further advance hypotheses on human disorders. Discovering analogous diseases (based upon their shared toxicogenomic portrait) could promote alternative methods for classifying diseases beyond the standard histological techniques, and more towards a molecular basis.
Here we report a new bioinformatics approach to discovering analogous diseases based upon shared chemical and/or gene relationship profiles in CTD, which we call DiseaseComps (for comparable diseases). This metric parallels our previous implementation of GeneComps and ChemComps, which organized analogous genes and chemicals, respectively, based upon common toxicogenomic interactions [8].
Methodology:
CTD biocurators manually curate the literature to capture chemical-disease and gene-disease relationships [2]. A chemical or gene can have an M-type relationship (wherein the molecule acts as either a biomarker or plays a role in the molecular mechanism of the disease) or a T-type relationship (wherein the molecule is described as either a real or putative therapeutic for the disease). Here we used the data available in CTD in September 2011, which included 13,530 chemical-disease relationships (for 2,652 chemicals and 1,180 diseases) and 14,173 gene-disease relationships (for 5,470 genes and 4,149 diseases). Similarity indices were computed using a modification of the Jaccard index, whose value ranges between 0 and 1 [8]. DiseaseComps are delineated by the type of relationship (M or T-type). For example, chemicals with a T-type relationship to a disease can be used to find comparable diseases wherein the chemicals have the same T-type relationship. Six types of DiseaseComps are generated: (1) via all chemical relationships (M- and T-type combined), (2) via only chemical M-type relationships, (3) via only chemical T-type relationships, (4) via all gene relationships (M- and T-type combined), (5) via only gene M-type relationships, and (6) via only gene T-type relationships.
Utility:
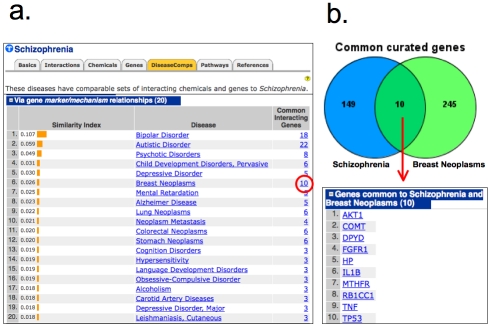
CTD computes values that reflect the degree of similarity between the molecular interaction profiles of each curated disease and generates a list of DiseaseComps, delineated by the six possible types of relationships. DiseaseComps provide a simple approach to view diseases that share common molecular interactions, allowing disorders to be classified in a novel manner without regard to histology or tissue of origin. Every curated disease in CTD now includes a DiseaseComps data tab that lists the top 20 comparable diseases based upon their ranked similarity index. For example, the disease schizophrenia has 160 genes curated with an M-type relationship in CTD. DiseaseComps identifies the top comparable diseases for schizophrenia that share the greatest number of those 160 Mtype genes to produce a ranked list that includes bipolar disorder, autism, and psychoses, as well as non-intuitive diseases such as breast, lung, and colorectal cancers (Figure 1a), suggesting that schizophrenia shares many of the same molecular networks common to some cancers. The shared genes can be viewed by clicking on the hyperlinked numeral in the “Common Interacting Gene” column (Figure ab). This new CTD metric provides researchers with additional predictive information that can help construct novel, testable hypotheses about the mechanisms (and potentially treatments or targets) underlying schizophrenia based upon its shared molecular profile with other diseases.
Figure 1.
(a) DiseaseComps (orange tab) for schizophrenia via gene marker/mechanism (M-type) relationships include familiar diseases such as bipolar disorder, autism, and psychoses (ranked #1-3), but also discovers non-intuitive diseases like breast cancer (ranked #6) that share ten M-type genes with schizophrenia (red circle). (b) The common curated genes can be viewed by clicking on the hyperlinked numeral (red circle) to produce a Venn diagram and a list of the ten genes shared between schizophrenia and breast cancer.
Future development:
DiseaseComps find similar diseases based upon shared chemical or gene relationships. The algorithm can be reversed to now find similar genes (or chemicals) based upon shared diseases, a feature we hope to soon add to our already established GeneComps and ChemComps data-tabs at CTD [8]. CTD is also expanding its content via the targeted curation of over 50,000 additional toxicology publications selected for four disease areas (cardiovascular, renal, hepatic, and neurological disorders). This will provide significantly more data as input for the generation of DiseaseComps calculations.
Acknowledgments
This project is supported by R01ES014065 from the National Institute of Environmental Health Sciences (NIEHS) and the National Library of Medicine (NLM), R01ES014065-04S1 from NIEHS, and P20RR016463 from the National Center for Research Resources (NCRR). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Citation:Davis et al, Bioinformation 7(4): 154-156 (2011)
References
- 1.AP Davis, et al. Nucleic Acids Res. 2011;39:D1067. doi: 10.1093/nar/gkq813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.AP Davis, et al. Database (Oxford) 2011 [Google Scholar]
- 3.M Vidal, et al. Cell. 2011;144:986. [Google Scholar]
- 4.KI Goh, et al. Proc Natl Acad Sci USA. 2007;104:8685. [Google Scholar]
- 5.S Craft, et al. Arch Neurol. 2011 [Google Scholar]
- 6.K Offit. Hum Genet. 2011;130:3. doi: 10.1007/s00439-011-1021-x. [DOI] [PubMed] [Google Scholar]
- 7.JM Gohlke, et al. BMC Syst Biol. 2009;3:46. doi: 10.1186/1752-0509-3-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.AP Davis, et al. Bioinformation. 2009;4:173. [Google Scholar]