Abstract
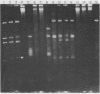
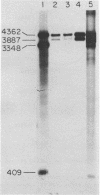
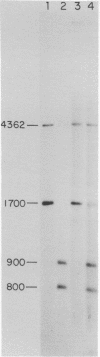
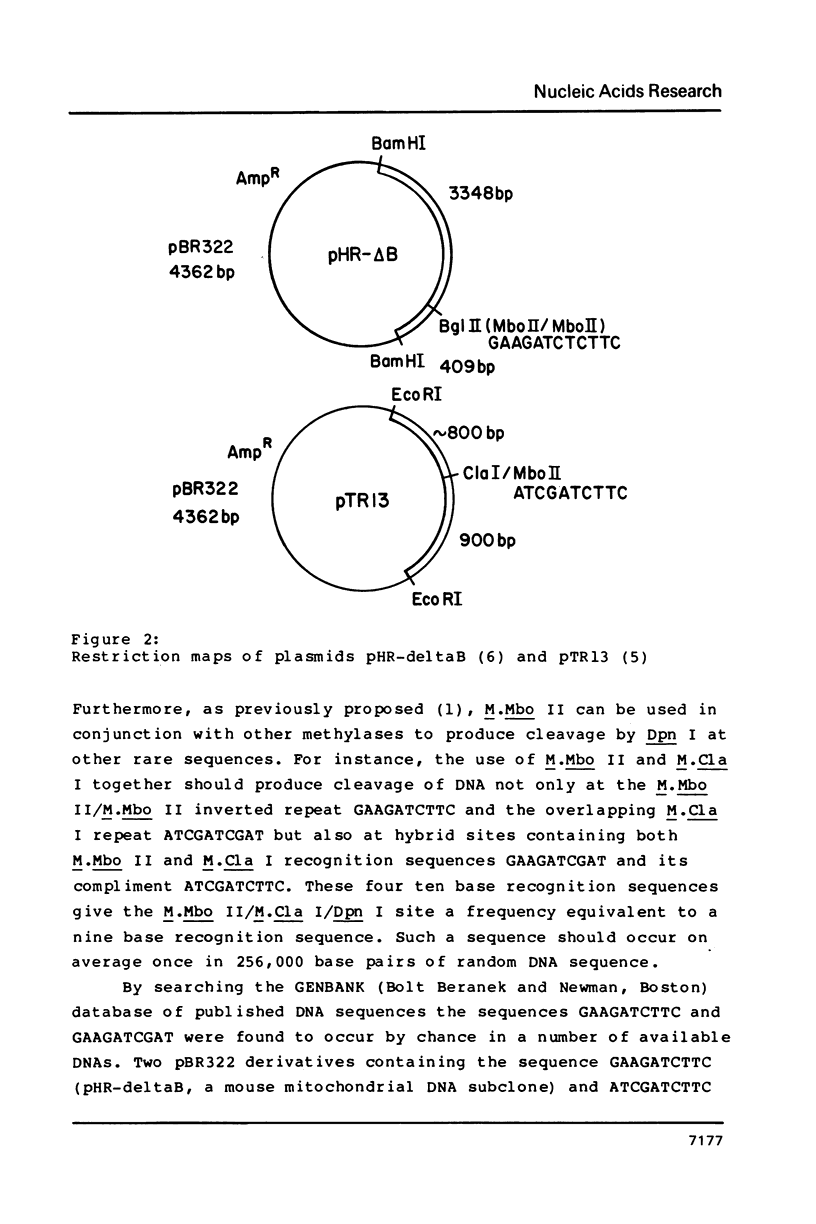
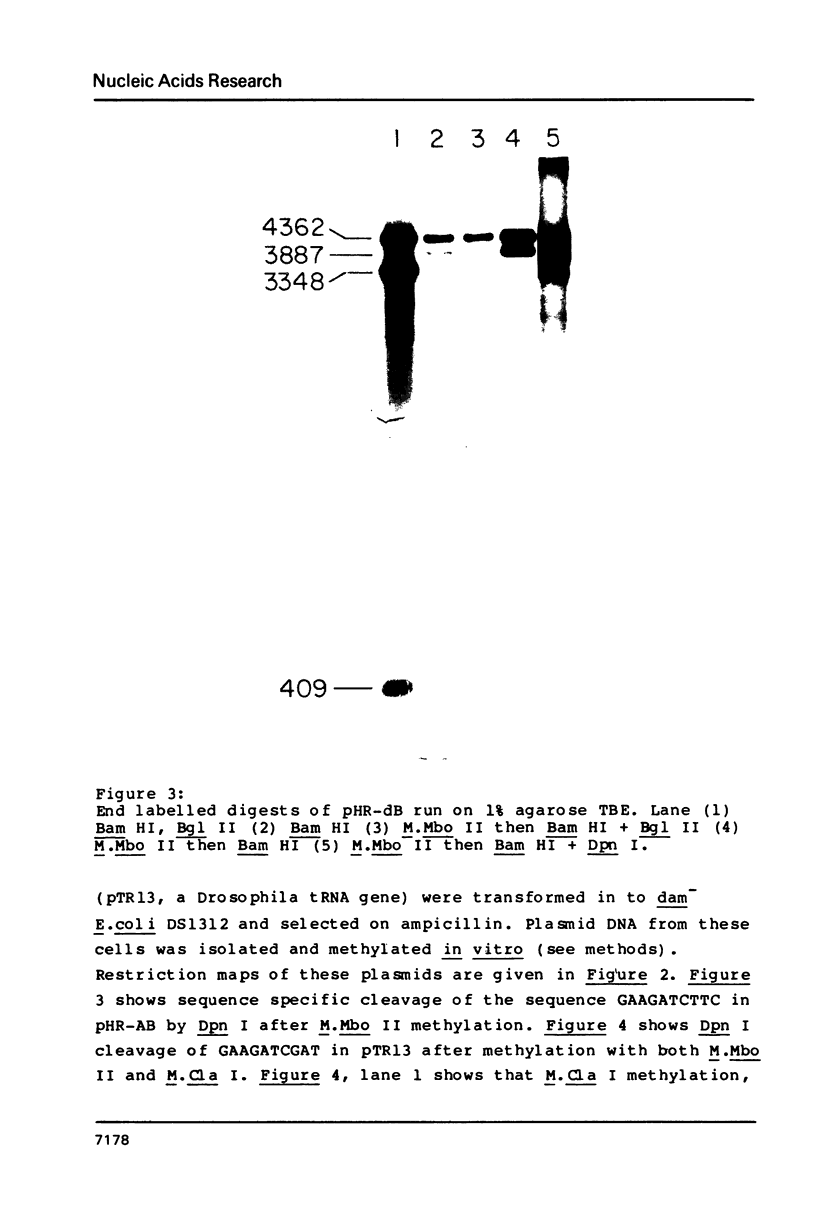
The restriction modification methylase M. Mbo II has been purified using a sensitive oligonucleotide linker assay. The enzyme methylates the Mbo II recognition sequence* GAAGA at adenine to produce GAAGmA. M. Mbo II can be used in conjunction with the methylation dependent restriction endonuclease Dpn I (GmATC) to produce cleavage at the 10 base sequence GAAGATCTTC. When M. Mbo II is used in combination with M. Cla I (ATCGATCGAT), cleavage by Dpn I occurs at the four ten base sequences GAAGATCTTC, GAAGATCGAT, ATCGATCTTC and ATCGATCGAT, which is equivalent to a nine base recognition site. The use of combinations of adenine methylases and Dpn I to generate highly selective DNA cleavages at a variety of sequences up to fourteen base pairs is discussed.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bibb M. J., Van Etten R. A., Wright C. T., Walberg M. W., Clayton D. A. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981 Oct;26(2 Pt 2):167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks J. E., Roberts R. J. Modification profiles of bacterial genomes. Nucleic Acids Res. 1982 Feb 11;10(3):913–934. doi: 10.1093/nar/10.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N. L., Hutchison C. A., 3rd, Smith M. The specific non-symmetrical sequence recognized by restriction endonuclease MboII. J Mol Biol. 1980 Jun 15;140(1):143–148. doi: 10.1016/0022-2836(80)90360-5. [DOI] [PubMed] [Google Scholar]
- Bächi B., Reiser J., Pirrotta V. Methylation and cleavage sequences of the EcoP1 restriction-modification enzyme. J Mol Biol. 1979 Feb 25;128(2):143–163. doi: 10.1016/0022-2836(79)90123-2. [DOI] [PubMed] [Google Scholar]
- Gelinas R. E., Myers P. A., Roberts R. J. Two sequence-specific endonucleases from Moraxella bovis. J Mol Biol. 1977 Jul;114(1):169–179. doi: 10.1016/0022-2836(77)90290-x. [DOI] [PubMed] [Google Scholar]
- Hosbach H. A., Silberklang M., McCarthy B. J. Evolution of a D. melanogaster glutamate tRNA gene cluster. Cell. 1980 Aug;21(1):169–178. doi: 10.1016/0092-8674(80)90124-5. [DOI] [PubMed] [Google Scholar]
- Joyce C. M., Kelley W. S., Grindley N. D. Nucleotide sequence of the Escherichia coli polA gene and primary structure of DNA polymerase I. J Biol Chem. 1982 Feb 25;257(4):1958–1964. [PubMed] [Google Scholar]
- McClelland M., Kessler L. G., Bittner M. Site-specific cleavage of DNA at 8- and 10-base-pair sequences. Proc Natl Acad Sci U S A. 1984 Feb;81(4):983–987. doi: 10.1073/pnas.81.4.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Nelson M. The effect of site specific methylation on restriction endonuclease digestion. Nucleic Acids Res. 1985;13 (Suppl):r201–r207. doi: 10.1093/nar/13.suppl.r201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M. Purification and characterization of two new modification methylases: MClaI from Caryophanon latum L and MTaqI from Thermus aquaticus YTI. Nucleic Acids Res. 1981 Dec 21;9(24):6795–6804. doi: 10.1093/nar/9.24.6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M., Christ C., Schildkraut I. Alteration of apparent restriction endonuclease recognition specificities by DNA methylases. Nucleic Acids Res. 1984 Jul 11;12(13):5165–5173. doi: 10.1093/nar/12.13.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J. Restriction and modification enzymes and their recognition sequences. Nucleic Acids Res. 1985;13 (Suppl):r165–r200. doi: 10.1093/nar/13.suppl.r165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S., Nakazawa K., Shinomiya T. A DNA methylase from Thermus thermophilus HB8. J Biochem. 1980 Sep;88(3):737–747. doi: 10.1093/oxfordjournals.jbchem.a133026. [DOI] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- van Ormondt H., Lautenberger J. A., Linn S., de Waard A. Methylated oligonucleotides derived from bacteriophage fd RF-DNA modified in vitro by E. coli B modification methylase. FEBS Lett. 1973 Jul 1;33(2):177–180. doi: 10.1016/0014-5793(73)80186-3. [DOI] [PubMed] [Google Scholar]