Abstract
The endoplasmic reticulum (ER) uses an elaborate surveillance system called the ER quality control (ERQC) system. The ERQC facilitates folding and modification of secretory and membrane proteins and eliminates terminally misfolded polypeptides through ER-associated degradation (ERAD) or autophagic degradation. This mechanism of ER protein surveillance is closely linked to redox and calcium homeostasis in the ER, whose balance is presumed to be regulated by a specific cellular compartment. The potential to modulate proteostasis and metabolism with chemical compounds or targeted siRNAs may offer an ideal option for the treatment of disease.
The ER-associated degradation (ERAD) machinery eliminates terminally misfolded polypeptides. Proteins that are difficult to fold properly (e.g., CFTR) are frequently degraded by ERAD, even under normal conditions.
The endoplasmic reticulum (ER) serves as a protein-folding factory where elaborate quality and quantity control systems monitor an efficient and accurate production of secretory and membrane proteins, and constantly maintain proper physiological homeostasis in the ER including redox state and calcium balance. In this article, we present an overview the recent progress on the ER quality control system, mainly focusing on the mammalian system.
TRANSLATION OF ER-TARGETED PROTEINS
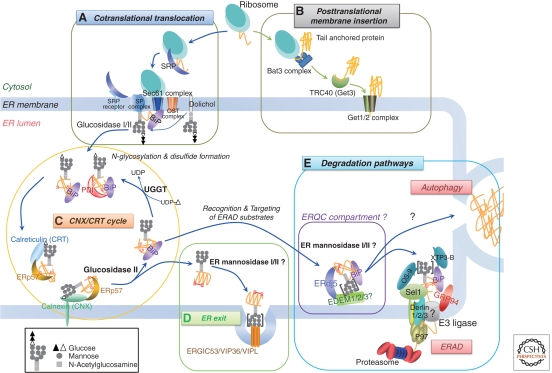
Most mammalian secretory and membrane proteins are cotranslationally imported into the ER (Fig. 1A). Signal sequence on the newly synthesized polypeptides are caught by the signal-recognition particle (SRP), whose binding slows protein synthesis in a process known as elongation arrest, and directs polypeptides to the translocon, composed of the Sec61αβγ complex in the ER membrane (Johnson and Van Waes 1999; Saraogi and Shan 2011). After arriving at the translocon, translation resumes in a process called cotranslational translocation (Hegde and Kang 2008; Zimmermann et al. 2010). Numerous ER-resident chaperones and enzymes aid in structural and conformational maturation necessary for proper protein folding, including signal-peptide cleavage, N-linked glycosylation, disulfide bond formation, and glycophosphatidylinositol (GPI)-anchor addition (Ellgaard and Helenius 2003).
Figure 1.
Schematic model of the ER quality control system. Selected components are depicted in this model. (A) Cotranslational translation: The ER signal sequence of a newly synthesized polypeptide is bound by a signal-recognition particle (SRP). The SRP–ribosome complex is guided to the Sec complex by an ER membrane-localized SRP receptor. After release of the SRP and SRP receptor, the polypeptide begins translocation. Subsequently, the signal sequence is processed by the signal peptidase (SP) complex (Paetzel et al. 2002). Transfer of oligosaccharides is catalyzed by the oligosaccharyl transferase (OST) complex, and the two outermost glucose residues are sequentially removed by glucosidases I and II. (B) Posttranslational membrane insertion: a tail-anchored (TA) protein is posttranslationally inserted into the ER membrane. The carboxy-terminal single trans-membrane domain of the TA protein is recognized by the Bat3 complex and transferred to the cytoplasmic chaperone TRC40 for targeting to the ER-membrane localized Get1/2 receptor. (C) CNX/CRT cycle: a monoglucosylated N-glycan of the polypeptide recruits the ER lectin chaperones, calnexin (CNX) and/or calreticulin (CRT), which promote proper folding by preventing aggregation and premature export from the ER. ERp57, a CNX/CRT-bound oxidoreductase, catalyzes disulfide formation. Trimming of the innermost glucose residue by glucosidase II then releases the polypeptide from CNX/CRT. UDP-glucose/glycoprotein glucosyl transferase (UGGT) monitors the folding state of released glycoprotein and, if the correct conformation has not been achieved, UGGT reglucosylates it to be reengaged by CNX/CRT. (D) ER exit: The natively folded protein is released from the CNX/CRT cycle and transported to its destination. In the early secretory pathway, lectins (ERGIC53, VIP36, and VIPL) support the sorting or trafficking of glycosylated proteins from the ER to the Golgi (Kamiya et al. 2008; Dancourt and Barlowe 2010). “[ ]” indicates that there are several possibilities for N-glycan formation. (E) Degradation pathways: Terminally misfolded proteins are degraded primarily through ER-associated degradation (ERAD) or autophagic degradation. Before degradation, N-glycans on ERAD substrates are extensively trimmed for efficient degradation, possibly in a specific compartment within the ER known as the ER quality control (ERQC) compartment, where ERAD machineries such as ER mannosidase I and EDEM family proteins are enriched. Subsequently, ERAD substrates are retrotranslocated into the cytosol, possibly through an E3 ligase complex, and finally degraded by the ubiquitin proteasome pathway. A recent report suggests that misfolded TA proteins are also degraded by the ERAD pathway (Claessen et al. 2010). Autophagy also degrades some ERAD substrates, but its recognition mechanism is not well understood.
Recently, the synthesis of posttranslationally inserted proteins, known as tail-anchored (TA) proteins, was elucidated (Fig. 1B) (Rabu et al. 2009; Brodsky 2010; Borgese and Fasana 2011). Tail-anchored (TA) proteins are translated in the cytosol, and the carboxy-terminal single trans-membrane domain (TMD) is recognized by the cytoplasmic chaperones TRC40 (Get3) together with the three-protein complex Bat3 (Sgt2 in yeast), TRC35 (Get4), and Ubl4A (Get5). The proteins guide and facilitate the insertion of the TA protein into the ER membrane with the aid of the ER-membrane localized Get1/2 receptor in a Sec61-independent manner (Mateja et al. 2009; Mariappan et al. 2010; Wang et al. 2010a).
PROTEIN FOLDING AND POSTTRANSLATIONAL MODIFICATION
Glycosylation
The covalent addition of N-linked glycans to proteins is one of the major biosynthetic functions of the ER and occurs in 90% of all glycoproteins (Helenius 1994). Polypeptides entering the ER lumen are covalently modified by attachment of the preformed oligosaccharide Glc3Man9GlcNAc2 (Glc: glucose, Man: mannose, GlcNAc: N-acetylglucosamine) to asparagine side chains in Asn-Xxx-Ser/Thr sequons, catalyzed by oligosaccharyltransferase (OST), a multisubunit enzyme associated with the translocon complex (Fig. 1A) (Shibatani et al. 2005; Ruiz-Canada et al. 2009). The transfer of N-glycans occurs cotranslocationally in a single enzymatic step, and immediately the two outermost glucose residues of the N-glycans are sequentially removed by glucosidases I and II, thereby generating monoglucosylated oligosaccharides (GlcMan9GlcNAc2) (Grinna and Robbins 1979). These N-glycans are recognized by ER lectin-like chaperones calnexin (CNX) and/or calreticulin (CRT), which promote proper folding by preventing aggregation and premature export from the ER (Fig. 1C) (Williams 2006; Rutkevich and Williams 2010). Trimming of the innermost glucose residue by glucosidase II releases those polypeptides from CNX/CRT. UDP-glucose/glycoprotein glucosyl transferase (UGGT) senses the folding state of released glycoproteins and, if the correct conformation has not been achieved, UGGT reglucosylates the N-glycan again to be reengaged by CNX/CRT (Solda et al. 2007; D'Alessio et al. 2010). In this way, the structure of the N-glycan codes the mandatory information for folding state of the glycoproteins (Hebert et al. 2005; Aebi et al. 2010). Correctly folded proteins are released from this cycle and transported to their destinations (Fig. 1D) (Molinari 2007; Lederkremer 2009).
Folding by Chaperones and Co-Chaperones
In addition to the CNX/CRT complex, the other major chaperone system in the ER is the BiP/GRP78 or Hsp70 system (Hendershot 2004; Dudek et al. 2009; Otero et al. 2010). BiP (binding immunoglobulin protein) is one of the most abundant ER chaperones and serves multiple roles in the ER ranging from productive folding to ERAD. BiP is composed of two domains; an ATPase domain and a substrate-binding domain (SBD) that contains a hydrophobic region suitable for binding to unfolded substrates. After the hydrolysis of ATP, ADP-bound BiP acquires a high affinity for substrates in which the hydrophobic region is closed. By binding to substrates with high affinity, BiP prevents unfolded proteins from forming aggregates. As such, BiP recognizes and helps to assemble unfolded or misfolded regions of the polypeptide (Hendershot 2004).
To date, seven BiP cochaperones, known as DnaJ/Hsp40 family members (ERdj1–7), have been identified in the ER. They contain a J-domain with a His-Pro-Asp (HPD) motif required for the binding with Hsp70 or BiP. These cochaperones play a decisive and critical role not only in stimulating ATP hydrolysis of BiP, but also in regulating its various activities. The ADP form of BiP is converted to the ATP form by nucleotide exchange factors (NEFs) including GRP170 and Sil1/BAP, which direct BiP to the open and accessible form for the substrates (Chung et al. 2002; Kampinga and Craig 2010). These hydrolytic cycles of BiP regulated by the DnaJ family cochaperones and NEFs ensure the solubility of nascent and misfolded proteins in the ER by preventing their aggregation.
Of the Hsp40 family proteins, ERdj1, ERdj2, ERdj4, and ERdj7 have trans-membrane domains, whereas the others are ER luminal proteins. ERdj3–6 have been reported to be up-regulated by ER stresses, whereas ERdj1 and 2 are not. ERdj1 and 2 (homologs of yeast Sec63) presumably recruit BiP to the translocon gate to facilitate the translocation of newly synthesized polypeptides into the ER, and also close the translocon channel to maintain the ER environment. ERdj3 was identified in canine pancreatic microsomes as a component of a multiprotein complex that directly binds to immunoglobulin G during folding in the ER (Meunier et al. 2002; Shen and Hendershot 2005; Jin et al. 2008). ERdj4 and ERdj5 are reported to enhance the ERAD of misfolded proteins. ERdj5 was shown to interact with EDEM (ER degradation-enhancing α-mannosidase-like protein), and overexpressed ERdj4 and ERdj5 interact with p97, a component of the ERAD machinery (Dong et al. 2008; Ushioda et al. 2008; Ushioda and Nagata 2011) (see also later section). ERdj6, designated p58IPK, was initially reported to negatively regulate PKR and PERK phosphorylation in the cytosol (Gale et al. 1998; Yan et al. 2002). However, it was later determined that ERdj6 is also localized to the ER by an ER-targeting signal and that ERdj6 binds to BiP through its J-domain or the tetratricopeptide (TPR) repeat domain and functions as a cochaperone (Rutkowski et al. 2007; Petrova et al. 2008). ERdj6 is most probably involved in the productive folding of newly synthesized proteins in the ER lumen. Proteomics analysis with canine pancreatic microsomes revealed another Hsp40 family protein ERdj7, which possesses a trans-membrane and luminal domain (Zahedi et al. 2009). As such, ER resident-Hsp40 family proteins cooperatively regulate a wide spectrum of BiP functions.
Recently, a novel ER membrane cochaperone called DNAJB12 was identified. It contains a cytosolic J-domain that interacts with cytosolic Hsp70 and plays a role in the degradation of membrane proteins, including CFTR and TCRα, which suggests that it functions in ERAD on the cytosolic side (Grove et al. 2010; Yamamoto et al. 2010).
Disulfide Bond Formation
Another important maturation step in the ER is the formation of disulfide bonds, which are crucial for protein function and stability (Appenzeller-Herzog 2011). The oxidative environment of the ER is suitable for the oxidation of free sulfhydryl (SH) groups on cysteines to form disulfide (S–S) bonds. Oxidoreductases, called PDI family proteins, catalyze these reactions by acting as an oxidase and isomerase, thereby promoting the formation of native disulfides. PDI family members are defined by the presence of at least one thioredoxin (Trx)-like domain containing Cys-X-X-Cys (CXXC) motifs in the active site. To date, approximately 20 PDI family members have been identified, including soluble and trans-membrane-containing proteins, most of which are ubiquitously expressed (Ellgaard and Ruddock 2005). PDI is a canonical member of the PDI family and also functions as a molecular chaperone (Hatahet et al. 2009). PDI is composed of two active Trx-like domains (called the a and a′ domains) that are linked by two inactive Trx-like domains (called the b and b′ domains), and its primary substrate binding site is located in the b′ domain. ERp57, the other member, stably binds to CNX or CRT and acts as an oxidoreductase, especially for glycoproteins (Oliver et al. 1999). ERp57 also acts as a thiol oxidoreductase of heavy chain (HC) oxidation in MHC class I biogenesis, and as a structural component of the peptide loading complex (PLC), which consists of the HC-β2m heterodimer, CRT (or CNX), and the additional components tapasin, TAP and Bap31 (Chapman and Williams 2010). PDI may also come into play and reoxidize HC (Park et al. 2006). Other ER proteins, such as ERp44, which is localized to the ER-Golgi intermediate compartment (ERGIC), engages in the folding/oligomerization or retention of some proteins in the ER (Anelli and Sitia 2008; Cortini and Sitia 2010). ERdj5 participates in ERAD as a reductase (Ushioda et al. 2008; Hagiwara et al. 2011) (see later section). The ER also contains a number of selenoproteins. One of these is Sep15, which binds to UGGT and presumably works as a reductase, as suggested by the reducing potential of selenocysteine (Korotkov et al. 2001).
Why are there so many oxidoreductases in the ER? The reason is not entirely clear, but some oxidoreductases appear to have a specific function, whereas others have redundant functions (Feige and Hendershot 2010; Rutkevich et al. 2010). Specific functions or redundancies are often inferred from their direct or indirect binding partners (e.g., ERp57-CNX/CRT, BiP-P5, ERdj5-BiP-EDEM, ERp44-ERGIC53, etc.), which define substrate specificities and specific cellular compartments in which the oxidoreductases localize (Jessop et al. 2009). Knockout mice models also elucidate specific and redundant functions of ER oxidoreductases. For example, ERp57 knockout mice showed embryonic lethality, suggesting that ERp57 has a specific role in early development (Garbi et al. 2006; Coe et al. 2010). Knockout mice of ERdj5, Prdx4, and Ero1α/β (see later section) showed less severe phenotypes, which suggests that their roles can be compensated for by other factors (Iuchi et al. 2009; Hosoda et al. 2010; Zito et al. 2010a). In addition, some ER proteins appear to possess functions outside the ER, such as in mitochondria or nucleus (P5 and ERp57) (Coppari et al. 2002; Kimura et al. 2008). Clarifying their detailed roles and regulatory mechanisms will be an exciting topic for future work (Appenzeller-Herzog and Ellgaard 2008).
Other Specific Chaperones
A number of substrate-specific chaperones have been reported, among which collagen-specific or related chaperones have been well illustrated. HSP47/SERPINH1 specifically and transiently binds to various types of collagen in the ER and is believed to facilitate the triple-helical structure of collagen (Nagata 2003). The P4H complex, consisting of α2β2 tetramers, in which the β-subunits are identical to PDI, and the P3H complex, containing cartilage-associated protein (CRTAP), prolyl 3-hydroxylase 1 (P3H1), and cyclophilin B, are also known to be essential assembling factors and collagen chaperones (Ishikawa et al. 2009; Gorres and Raines 2010; Morello and Rauch 2010). Mutation or knockout of these factors results in embryonic lethality or osteogenesis imperfecta, which clearly shows their importance for productive procollagen folding and maturation (Nagai et al. 2000; Morello et al. 2006; Cabral et al. 2007; Choi et al. 2009). Microsomal triglyceride transfer protein (MTP) plays a pivotal role in lipoprotein assembly, and receptor-associated protein (RAP) participates in the maturation of several membrane receptor proteins, such as low-density lipoprotein receptor-related protein (LRP) and lipoprotein receptor 11 (SorLA/LR11) (Orlando 2004; Blasiole et al. 2007).
ERAD
Terminally misfolded or unassembled proteins that are unable to acquire their native structure must be degraded to prevent fruitless folding attempts and the accumulation of misfolded polypeptides in the ER (Fig. 1E). This degradation process is known as ER-associated degradation (ERAD), which occurs in three primary steps: (1) recognition and targeting (substrate recognition within the ER and targeting to the retrotranslocon), (2) retrotranslocation (substrate delivery from the ER to the cytosol), and (3) degradation (ubiquitin–proteasome dependent degradation) (Hegde and Ploegh 2010).
Recognition and Targeting
The mechanism by which ERAD substrates are recognized and distinguished from properly folded proteins or those that are in the process of being correctly folded remains largely unknown. A large portion (around 75%) of proteins such as CFTR, apolipoprotein A, or the erythropoietin receptor, which are difficult to fold properly, are degraded even under normal conditions (Kopito 1999; Sanders and Myers 2004). On the other hand, influenza HA protein folds with near 100% efficiency (Braakman et al. 1991). A recently proposed model suggests that the efficiency of productive folding and trafficking cannot be defined by a single feature, but rather by the combination of multiple factors, including protein stability, folding rate, enzyme distribution, and metabolism (redox state, calcium flux, etc.), collectively called ERAF (ER-associated folding) (Sekijima et al. 2005). Therefore, protein quality control, now conceptualized as proteostasis (protein homeostasis), is maintained by the teleological relationship between conformational maturation (ERAF) and recognition for disposal in the ER lumen (ERAD), and thereby determines the probability that newly synthesized proteins will acquire their native structure (Kowalski et al. 1998; Kjeldsen et al. 2002; Brodsky 2007; Wiseman et al. 2007; Hutt and Balch 2010).
Because different quality control (QC) machineries detect structurally different defects in different environments, three subdivisions of the ERAD pathway have been proposed in budding yeast dependent on the site of the defect, ERAD-C (cytosol), ERAD-M (membrane), and ERAD-L (lumen) (Taxis et al. 2003; Vashist and Ng 2004; Carvalho et al. 2006), although this classification might be oversimplified (Goeckeler and Brodsky 2010).
ERAD machineries often organize higher molecular weight complexes around E3 ligase (Fig. 2) (Kawaguchi and Ng 2007). Yeast has two major membrane-associated E3 ligases containing RING domain, Doa10p (degradation of Mat-α2-10 protein) and Hrd1p (HMG-CoA reductase degradation 1 protein) (Carvalho et al. 2006; Denic et al. 2006). Doa10p works on substrates with misfolded regions on the cytosolic side (ERAD-C), whereas the Hrd1p complex, comprised of the ubiquitin ligases Hrd1p, Hrd3p, Usa1p, and Der1p, acts on substrates with defects in the luminal region (ERAD-L). ERAD-M requires a subset of these components required for ERAD-L (Bordallo et al. 1998; Bays et al. 2001a; Swanson et al. 2001). Following polyubiquitination, these pathways merge at an ATPase complex consisting of the ATPase Cdc48p and two cofactors, Ufd1p and Npl4p (Bays et al. 2001b; Braun et al. 2002; Jarosch 2002; Rabinovich et al. 2002; Zito et al. 2010b). In contrast to yeast, mammalian ERAD E3 ligases are more diverse (HRD1, RMA1, Parkin, CHIP, gp78, TEB4, TRC8, etc.) and additional ERAD E3 ligases continue to be identified, which indicates that various complexes are needed to monitor different classes of substrates (Hirsch et al. 2009; Tsai and Weissman 2010; Neutzner et al. 2011).
Figure 2.
Schematic models of mammalian E3 ligase complexes. Selected components are depicted in each model (Tsai and Weissman 2010). The majority of motif annotations are taken from the Pfam and SMART databases (see Tables 1 and 2; Letunic et al. 2009; Finn et al. 2010). (A) HRD1 ligase complex: This complex is a well-illustrated E3 complex that primarily targets proteins for ERAD-L. OS-9 and XTP3-B recognize aberrant nonglycosylated or glycosylated proteins in the ER lumen. Both associate with the HRD1 complex through SEL1L in a mutually exclusive manner. BiP and GRP94 presumably cooperate to regulate the assembly/disassembly of the HRD1 complex and sequester misfolded proteins to prevent other interactions until retrotranslocation. Derlin family proteins (Derlin 1, 2, or 3) might participate in substrate retrotranslocation from the ER lumen into the cytosol. UBXD8 and UBXD2 bind to p97/VCP through their UBX domain and accelerate the degradation of ERAD substrates. E2 ubiquitin-conjugating enzymes (Ube2j1, Ube2k) mediate substrate ubiquitination (Burr et al. 2011), whereas the p97/VCP hexamer promotes substrate extraction into the cytosol. Ubiquilin-1 is suggested to act as a ubiquitin–proteasome shuttle protein. Other ubiquitin-chain modifiers may also come into play, such as E4 ubiquitin-conjugating enzyme (Ufd2), Ufd2 inhibitor (Ufd3), and deubiquitinases (Ataxin-3, YOD1, VCIP135) (Rumpf and Jentsch 2006). The deglycosylating enzyme PNGase releases N-linked glycan chains from the glycopeptide (Tanabe et al. 2006). The proteasome then captures the polyubiquitin chains on the substrate through specific subunits (Rpn10/13, Rpt5) and degrades it. (B) RMA1 ligase complex: RMA1/RNF5 associates with Derlin-1 and E2 Ube2j1. BAP31, known to be an ER sorting factor of diverse membrane proteins, interacts with RMA1 as well as components of the Sec61 pore, which suggests that BAP31 might recruit the ERAD complex to the translocon channel to clear newly synthesized misfolded membrane proteins from the channel. In addition, DNAJB12, which contains the cytosolic J-domain, may also participate in the degradation of membrane proteins together with HSP70. All of these factors have been reported to play a role in the degradation of cystic fibrosis trans-membrane conductance regulator (CFTR) and its mutant (CFTRΔ508) during the early steps of translation (Younger et al. 2006; Morito et al. 2008). (C) Cytosolic E3 ligases: CHIP (carboxyl terminus of Hsp70-interacting protein) possesses a U-box domain, which has a structure similar to the RING-finger domain, and a tetratricopeptide repeat domain (TPR) that interacts with Hsp70 and Hsp90. In contrast to RMA1, CHIP posttranslationally monitors the folding of CFTR or CFTRΔ508. Parkin, which is responsible for autosomal recessive juvenile Parkinsonism, targets several proteins, such as O-glycosylated α-synuclein, the Pael receptor (Pael-R), Synphilin-1, and Tau. Regarding Pael-R, Parkin and CHIP act together to enhance its ubiquitination and inhibit cell death induced by accumulated unfolded Pael-R. Parkin also works with the E2 proteins Ube2j1 and Ube2g2, which suggests that it is involved in ERAD. The SCF (SKP1-CUL1-F-box protein) is composed of three proteins (Cullin1, Skp1, and RING finger protein Rbx1) and one F-box protein (Fbs1 or Fbs2). Fbs1 and Fbs2 are novel F-box proteins that recognize sugar chains in N-linked glycoproteins and show a chaperone-like activity to prevent their aggregation (Yoshida and Tanaka 2010). (D) gp78 ligase complex: The carboxyl terminus of gp78 is composed of four motifs: RING, CUE, G2BR (Ube2g2 binding region), and VIM (p97/VCP-interacting motif). The gp78 ligase complex usually consists of Ube2g2, Derlin-1, p97/VCP, and UBXD2 or UBXD8. gp78 mediates several ERAD substrates, including T-cell receptor subunits (CD3-δ and TCR-α), apolipoprotein B-100, Insig-1, and HMG-CoA reductase. The latter two substrates suggest that gp78 is involved in cholesterol metabolism (Jo and Debose-Boyd 2010). gp78 also cooperates with RMA1 to degrade CFTRΔ508. SVIP is reported to inhibit the assembly of the gp78 ligase complex (Derlin-1, gp78, and p97), which suggests an inhibitory effect on ERAD (Ballar et al. 2007). (E) TEB4 (Doa10) ligase complex: TEB4 is known to be a mammalian homolog of yeast Doa10. Together with Ube2g2, TEB4 is implicated in the degradation of ER resident type 2 iodothyronine deiodinase (Zavacki et al. 2009). Based on the homology of the yeast Doa10, TEB4 might also interact with Ube2j1 (Ubc6) and UBXD8 (Ubx2).
Table 1.
Selected ERAD-related proteins
| Human | Other name | Localization | Main motifs | Yeast homolog |
|---|---|---|---|---|
| Processing and targeting | ||||
| ER ManI | Membrane | Glyco_hydro_47 | Mns1 | |
| EDEM1-3 | Possibly ER or ER membrane | Glyco_hydro_47 | Htm1(Mnl1) | |
| PDI | ER | Trx | Pdi | |
| BiP | GRP78 | ER | SBD, NBD | Kar2 |
| GRP94 | ER | SBD, NBD | — | |
| ERdj4 | Membrane | DnaJ | — | |
| ERdj5 | JPDI | ER | DnaJ, Trx | — |
| ERFAD | FOXRED2 | ER | FAD/NADPH binding motif | — |
| CyPB | Cyclophilin B | ER | Pro_isomerase | Cpr2 |
| OS-9 | ER | MRH | Yos9 | |
| XTP3-B | ER | MRH | Yos9 | |
| SEL1L | Membrane | SEL1, Fn | Hrd3 | |
| Possible retrotranslocation channel | ||||
| Sec61 complex | Membrane | Sec motifs | Sec61, Ssh1 complex | |
| Derlin1-3 | Membrane | DER1 | Dfm1, Der1 | |
| Other possible component or regulator | ||||
| HERP | Mif1 | Membrane | UBL | Usa1 |
| VIMP | ERASIN, SelS | Membrane | SelS motif | — |
| BAP31 | Membrane | — | Yet3 | |
| JAMP | Membrane | — | — | |
| DNAJB12 | Membrane | DnaJ, TPR | Hlj1 | |
| SPP | HM13 | Membrane | Peptidase_A22B | — |
| TRAP complex | Membrane | TRAP motifs | — | |
| TRAM | Membrane | TRAM motif | — | |
| AUP1 | Membrane | CUE | — | |
| SVIP | Cytosol | VIM | — | |
| E2 ubiquitin-conjugating enzyme | ||||
| UBE2K | UBE2D1 | Cytosol | UBA, UBC | Ubc1 |
| UbcH5 | Ubc5a | Cytosol | UBC | Ubc5 |
| Ube2j1/2 | NCUBE-1/-2 | ER membrane | UBC | Ubc6 |
| Ube2g1/2 | Cytosol | UBC | Ubc7 | |
| Ubc13 | UBE2N | Cytosol | UBC | Ubc13 |
| E3 ubiquitin-protein ligase enzyme | ||||
| HECT (homologous to E6-AP carboxyl terminus) type E3 | ||||
| NEDD4-2 | Cytosol | HECT, C2, WW | Rsp5 | |
| RING (really interesting new gene)-finger type E3 | ||||
| Prakin | PARK2 | Cytosol | RING, UBL, IBR | — |
| RNF5 | RMA1 | Membrane | RING | — |
| RNF45 | AMFR, gp78 | Membrane | RING, CUE, VIM | — |
| HRD1 | Synoviolin | Membrane | RING | Hrd1/Der3 |
| TEB4 | MARCH VI | Membrane | RING | Doa10 |
| RNF139 | TRC8 | Membrane | RING | — |
| RNF77 | RFP2 | Membrane | RING, BBOX | — |
| RNF103 | Kf-1 | Membrane | RING | — |
| RNF19 | Dorfin | Membrane | RING, IBR | — |
| RNF121 | Membrane | RING | — | |
| UBOX type E | ||||
| CHIP | STUB1 | Cytosol | U-box, TPR | — |
| SKP1-CUL1-F-box (SCF) E3 (F-box protein component) | ||||
| SKP1 | Cytosol | Skp1 | skp1 | |
| CUL1 | Cytosol | Cullin | Cdc53 | |
| Fbs1 | FBXO2 | Possibly membrane associated or cytosol | FBA, F-box | — |
| Fbs2 | FBXO6 | Possibly membrane associated or cytosol | F-box, SBD | — |
| Rbx1 | Cytosol | RING | Hrt1 | |
| E4 ubiquitin-conjugating enzyme | ||||
| Ufd2 | Ube4 | Cytosol | U-box, UFD2 core motif | Ufd2 |
| Ufd2 inhibitor | ||||
| Ufd3 | PLAA | Cytosol | PFU, PUL, WD40 | Doa1 |
| Substrate extraction and recruiting | ||||
| p97 | VCP | Possibly membrane associated or cytosol | BS1 box | Cdc48 |
| UFD1 | Cytosol | UFD1 motif | Ufd1 | |
| NPL4 | Cytosol | NPL4 motifs | Npl4 | |
| UBXD family protein | ||||
| Ubxd1 | Ubxn6 | Cytosol | UBX, PUB | — |
| Ubxd2 | Erasin | Membrane associated | UBX, Trx-fold | Ubx7 |
| Ubxd7 | Cytosol | UBX, UBA, UIM, UAS | Ubx5 | |
| Ubxd8 | ETEA | Membrane associated | UBX, UBA | Ubx2 |
| Ubxd10 | SAKS1 | Cytosol | UBX | — |
| Deglycosylating enzyme | ||||
| PNGase | Cytosol | PUB | Png1 | |
| DUB (deubiquitylation) | ||||
| VCIP135 | Cytosol | OTU motif | — | |
| YOD1 | Otu1 | Cytosol | OTU motif | Otu1 |
| Ataxin-3 | Cytosol | UIM, Josephin | — | |
| USP19 | Membrane | UCH, MYND finger | — | |
| Shuttle protein | ||||
| Ubiquilin-1 | Dsk2, UBQLN1, | Possibly membrane associated or cytosol | UBL, UBA, STI1 | Dsk2 |
| HR23A/B | RAD23A/B | Cytosol | UBL, UBA, STI1 | Rad23 |
| Ubiquitin receptor | ||||
| Rpn10 | S5a | Cytosol | VWA, UIM | Rpn10 |
| Rpt5 | S6′ | Cytosol | AAA | Rpt5 |
| Rpn13 | ADRM1 | Cytosol | PH | Rpn13 |
Table 2.
Motif namesa
| Motif name | Pfam annotation or other comment |
|---|---|
| AAA | ATPase family associated with various cellular activities |
| BBOX | B-Box-type zinc finger |
| BS1 box | BS1 (binding site 1) is a p97-interacting domain |
| C2 | Ca2+-dependent membrane-targeting module |
| CUE | CUE domains may be involved in binding ubiquitin-conjugating enzymes (UBCs) or ubiquitin. |
| Cullin | Cullins are a family of hydrophobic proteins that act as scaffolds for ubiquitin ligases (E3). |
| DER1 | Der1-like family |
| DnaJ | DnaJ domain |
| FBA | F-box associated region |
| F-box | The F-box domain has a role in mediating protein–protein interactions. |
| Fn2 | Fibronectin type 2 domain |
| Glyco_hydro_47 | Glycosyl hydrolase family 47 |
| HECT | The name HECT-domain (ubiquitin-transferase) comes from Homologous to the E6-AP Carboxyl Terminus. |
| IBR | The IBR (in between ring fingers) domain is often found to occur between pairs of ring finger. |
| Josephin | The name Josephin comes from Machado–Joseph Disease protein MJD. |
| MRH | Mannose 6-phosphate receptor homology domain |
| MYND finger | The MYND (myeloid, Nervy, and DEAF-1) zinc finger domain might be involved in protein–protein interactions. |
| NBD | Nucleotide binding domain. |
| OTU | The ovarian tumor (OTU) -like cysteine protease |
| Peptidase_A22B | Signal peptide peptidase domain |
| PFU | PFU is the ubiquitin binding domain of Doa1 |
| PH | PH (pleckstrin homology) is involved in intracellular signaling or as constituents of the cytoskeleton. |
| Pro_isomerase | Cyclophilin type peptidyl-prolyl cis-trans isomerase |
| PUB | The PUB (also known as PUG) domain is found in peptide N-glycanase where it functions as a AAA ATPase binding domain. |
| PUL | The PUL (PLAP, Ufd3p and Lub1p) domain is a novel α-helical Ub-associated domain; it directly binds to Cdc48. |
| RING | RING finger is a cysteine-rich domain of 40 to 60 residues that coordinates two zinc ions and plays a key role in the ubiquitination pathway. |
| SBD | Substrate binding domain |
| SEL1 | This short repeat is found in the Sel1 protein; it is related to TPR repeats. |
| Skp1 | SKP1 (together with SKP2) was found to bind several F-box containing proteins (e.g., Cdc4, Skp2, cyclin F) and to be involved in the ubiquitin protein degradation pathway. |
| STI1 | Heat shock chaperonin-binding motif is found in the stress-inducible phosphoprotein STI1. |
| TPR | The tetratrico peptide repeat (TPR) mediates protein–protein interactions and the assembly of multiprotein complexes. |
| Trx | Thioredoxin domain |
| UAS | UAS is a domain of unknown function found in FAF1 proteins (FAS-associated factor 1) and in other proteins. |
| UBA | Ubiquitin associated domain |
| UBC | Ubiquitin-conjugating enzyme (E2) catalytic domain |
| UBL | Ubiquitin-like (UBL) domain |
| U-box | U-box has a similar structure to the RING-finger domain and bears ligase activity. |
| UBX | UBX domain is present in ubiquitin-regulatory proteins and is a general Cdc48-interacting module. |
| UCH | Ubiquitin carboxy-terminal hydrolase |
| UIM | Ubiquitin interacting motif (UIM) containing domains all interact with ubiquitin. |
| VIM | p97/VCP-interacting motif |
| VWA | The von Willebrand factor is a large multimeric glycoprotein found in blood plasma. |
| WD40 | Repeated WD40 motifs act as a site for protein–protein interaction. |
| WW | The WW domain is a protein module with two highly conserved tryptophans that binds proline-rich peptide motifs in vitro. |
The majority of the annotations are taken from the Pfam and SMART databases.
aEven if the human gene lacks a yeast homolog, another functionally relevant gene might exit.
In mammalian ERAD, the degradation of N-glycosylated proteins is well-characterized. As mentioned, the structure of the N-glycan codes the mandatory information on the state of protein folding. CNX/CRT recognizes monoglucosylated oligosaccharides (GlcMan9 GlcNAc2) and the CNX/CRT cycle enhances productive folding and protects immature glycopolypeptides from ERAD. Proteins that will be terminally misfolded are retained longer in this cycle, which may raise the probability of mannose trimming of the polypeptide-bound N-glycans by ER-mannosidase I, a process known as the mannose timer model (Helenius 1994). These mannose trimmed structures are recognized by ERAD factors, most likely EDEM family proteins (Molinari 2007; Hosokawa et al. 2010a).
EDEM family proteins (EDEM1, EDEM2, and EDEM3) have α-mannosidase-like domains with conserved catalytic residues for glycolytic activity (Kanehara et al. 2007). EDEM1 presumably possesses mannosidase activity that trims the C branch of N-glycans on misfolded proteins; however, that activity is apparently not required for ERAD acceleration because mutant EDEM1 that lacks the putative active site for mannosidase is still able to accelerate ERAD (Hosokawa et al. 2001, 2006, 2010b; Molinari et al. 2003; Oda et al. 2003; Olivari et al. 2006). EDEM2 also promotes ERAD, even though it has no enzymatic activity (Mast et al. 2005; Olivari et al. 2005). In contrast, ERAD-acceleration by EDEM3 (Htm1p/Mnl1p in yeast) is dependent on its mannosidase activity (Hirao et al. 2006; Clerc et al. 2009). Therefore, EDEM family proteins may not be functionally redundant, but they all contribute to ERAD acceleration.
Extensive trimming of N-glycans would lead to the increased hydrophobicity of misfolded proteins, and the Man7GlcNAc2 form of N-glycans is recognized by the lectins OS9 and perhaps XTP3-B, which contain one and two MRH (mannose 6-phosphate receptor homology) domains, respectively (Fig. 2A) (Hosokawa et al. 2010a; Satoh et al. 2010). Although OS-9 and XTP3-B do not interact with each other, both associate with the HRD1 E3 ubiquitin ligase complex through SELIL, a multiply glycosylated type I ER membrane protein. OS-9 and XTP3-B recognize aberrant nonglycosylated or glycosylated proteins even when their MRH domains are mutated (Christianson et al. 2008; Hosokawa et al. 2008). OS-9 also associates with BiP/GRP94 (an ER-resident Hsp90 homolog) and SELIL in a mutually exclusive manner, where BiP and GRP94 presumably contribute to regulate the assembly/disassembly of the HRD1 complex and sequester misfolded proteins to prevent other interactions until retrotranslocation (Eletto et al. 2010). Consistent with ERAD in yeast, mammalian HRD1 complexes are required for ERAD- L but are not necessary for ERAD-M (Bernasconi et al. 2010).
Our laboratory recently identified ERdj5 as an EDEM1-binding protein that can accelerate ERAD by reducing the incorrect formation of disulfide bonds in misfolded glycoproteins (Hoseki et al. 2010). Based on its crystal structure, ERdj5 contains a J-domain and six tandem thioredoxin domains, two of which do not contain redox active CXXC motif (Hagiwara et al. 2011). ERdj5 can be structurally divided into two clusters, called the N- and C-clusters. The N-cluster contains the J-domain, and the C-cluster interacts with EDEM1 and can efficiently reduce the disulfide bonds of recruited substrates. ERAD substrates are sequentially transferred from calnexin to the EDEM1-ERdj5 complex, and subsequently to BiP, which tightly binds substrates in a dislocation-competent state until retrotranslocation after stimulation of its ATPase activity by ERdj5. However, we have yet to identify the reductive source of ERdj5. One potential candidate is the recently reported flavoprotein, ERFAD, which interacts with SEL1L and OS9, and might provide reducing equivalents to ERdj5 (Riemer et al. 2009).
For nonglycosylated proteins, the recognition mechanism is apparently somewhat distinct from that of glycosylated proteins. The unfolded regions of nonglycosylated proteins are recognized by ER chaperones, mostly by BiP. The DnaJ family proteins, cochaperones of BiP, play a crucial role in regulating its various activities. Accumulated evidence suggests that ERdj3 and ERdj6 are primarily involved in productive-folding (ERAF), whereas ERdj4 and ERdj5 predominantly affect ERAD (Otero et al. 2010). The process of transition from folding to degradation is far from clear. Presumably, the retention time of a substrate by BiP and its cofactors (e.g., ERdj3/6) may determine its fate. Prolonged retention would recruit other cofactors involved in ERAD, such as ERdj4/5 and p97/VCP (Otero et al. 2010). HERP, a membrane-bound ubiquitin (Ub)-like protein, has been implicated in the efficient delivery of nonglycosylated substrates to the proteasome (Okuda-Shimizu and Hendershot 2007).
Retrotranslocation and Degradation
After recognition and targeting of the substrates, they must be dislocated/retrotranslocated into the cytosol for proteolysis by 26S proteasomes. Although the identities of the components that comprise the retrotranslocation channel remain unclear, Sec61 complex and Derlin-1 are possible candidates (Knop et al. 1996; Wiertz 1996; Pilon et al. 1997; Willer et al. 2008; Schafer and Wolf 2009). Sec61 complex has been reported to interact with several ERAD substrates and ERAD machineries including proteasome, TRAP complex, SPP (signal peptide peptidase), PDI, and BAP31 (Wiertz et al. 1996; Pilon et al. 1997; Loureiro et al. 2006; Nagasawa et al. 2007; Ng et al. 2007; Wang et al. 2008; Lee et al. 2010). Derlin-1 was initially reported to be important for the extraction of MHC class I molecules from the ER membrane in cytomegalovirus-infected cells (Lilley and Ploegh 2004; Ye et al. 2004). Reconstitution assay using a fluorescently labeled substrate also showed that Derlin-1 is involved in substrate dislocation, which is independent of Sec61α (Wahlman 2007). The yeast Hrd1p E3 ligase, which interacts with the ERAD substrate via its trans-membrane region, is also a possible candidate (Carvalho et al. 2010). Thus far, E3 ligase complexes are the predominant candidates because they constitute large protein complexes containing multispanning membrane proteins such as HRD1 and Derlin-1, which can efficiently recognize, target, retrotranslocate, and ubiquitinate ERAD substrates within the organized complexes (Fig. 2) (Bagola et al. 2011).
Ubiquitination takes place in the cytosol, which ensures efficient delivery of the substrates to the proteasome. ERAD substrates are ubiquitinated at serine/threonine residues and less frequently at lysine residues (Shimizu et al. 2010). This process is apparently different from that of other cytosolic ubiquitination mechanisms (Wang et al. 2009; Ishikura et al. 2010). At the cytosolic face of the ERAD complex, the AAA+ ATPase p97/VCP (Cdc48 in yeast) hexamer directs substrate to be drawn into the cytosol (Chapman et al. 2010). p97 binds to several ERAD components, including Derlin-1, VIMP (SelS), UBXD2 (Erasin), and UBXD8, and recruits several ubiquitin-chain modifiers including E3 ligases (gp78, HRD1, etc.), chain elongation factors (Ufd2, E4 ubiquitin ligase), and deubiquitinases (YOD1, Ataxin-3, VCIP135) (Liang et al. 2006; Mueller et al. 2008; Schuberth and Buchberger 2008; Ernst et al. 2009). Ubiquitinated substrates are transferred to the proteasome by shuttle proteins, known as HR23A/B or Ubiquilin-1 (Rad23 and Dsk2 in yeast), which contain ubiquitin-associated (UBA) and ubiquitin-like (UBL) domains that bind to polyubiquitin chains and the proteasome subunits (Rpn10/13, Rpt5), respectively (Deveraux et al. 1994; Lam et al. 2002; Raasi and Wolf 2007; Husnjak et al. 2008; Finley 2009; Lim et al. 2009).
Other Degradation Pathways
Cells possess additional degradation pathways to cope with a variety of situations based on the characteristics of the misfolded proteins (Fu and Sztul 2009; Kroeger et al. 2009; Wong and Cuervo 2010). We have reported previously that the aggregated or insoluble form of type I collagen in the ER is degraded by autophagy-mediated lysosomal degradation, whereas nonaggregated forms are subject to ERAD (Fig. 1E) (Ishida et al. 2009). When ERAF/ERAD or ubiquitin proteasome activities are compromised, autophagy is triggered via signaling pathways that usually involve unfolded protein response (UPR)-dependent elements (Ogata et al. 2006; Fujita et al. 2007; Hosokawa et al. 2007; Kouroku et al. 2007; Yorimitsu and Klionsky 2007b). Additionally, the ER membrane itself may be a source of lipids for autophagosome formation (Hayashi-Nishino et al. 2009; Yla-Anttila et al. 2009). Hence, the ER and the process of autophagy would certainly have a tight physiological relationship. Organelles are also selectively delivered to lysosomes by macroautophagy, known as ERphagy (reticulophagy), mitophagy, or pexophagy (Kim et al. 2007; Yorimitsu and Klionsky 2007a; Ding and Yin 2008; Todde et al. 2009; Manjithaya et al. 2010).
Intriguingly, endogenous EDEM1, a major component of ERAD, is constitutively degraded via an autophagic-like pathway (Cali et al. 2008). In support of this, electron microscopy showed that EDEM1 is primarily localized in double-membrane buds that form outside canonical ER exit sites, known as EDEMosomes (Zuber et al. 2007; Le Fourn et al. 2009; Reggiori et al. 2010). At steady state, short-living ERAD components like EDEM1 and OS-9 appeared to be engulfed in these buds in a COPII-independent manner and degraded without the attachment of nonlipidated LC3. This turnover of ERAD factors, known as ERAD tuning, maintains an extra capacity of ERAD factors at steady state by EDEMosome-linked degradation (Bernasconi and Molinari 2011). It is hypothesized that this makes it possible for the ER to respond quickly to sudden changes without waiting for transcriptional UPR responses. It is still unclear whether EDEM1 is an intrinsic component of the EDEMosome and the mechanism by which these buds are created.
ER HOMEOSTASIS
As discussed above, the ER is the primary site of secretory and membrane protein production. The proteostatic balance is intimately associated with ER redox homeostasis and calcium balance (Fig. 3).
Figure 3.
ER redox and calcium homeostasis on the MAM. On the mitochondria-associated membrane (MAM), ER chaperones (CNX, BiP), oxidoreductases (Ero1α, ERp44), and Ca2+ channels/pumps (IP3R3, SERCAs, and Sig-1R) are enriched, thereby creating an ideal environment for oxidative protein folding, as well as the regulation of Ca2+ flux. (A) ER redox homeostasis: Ero1α serves as the primary oxidase of PDI. Concomitantly, hydrogen peroxide (H2O2) is thought to be produced from oxygen as an electron acceptor. Peroxiredoxin IV (PRDX4) is thought to work as a H2O2 reducer. PRDX4 can also oxidize some PDI family members. Oxidized PDI family members drive oxidative protein folding, as well as oxidize GSH into GSSG, thereby generating an oxidative environment. The FAD, cofactor of Ero1α, may be delivered from mitochondria via unknown transporters. (B) Calcium flux: Mammalian cells contain two main channels responsible for Ca2+ efflux from the ER, inositol 1,4,5-trisphosphate receptors (IP3Rs) and ryanodine receptors (RyRs), and one pump responsible for Ca2+ influx into the ER, sarcoplasmic reticulum Ca2+-ATPase (SERCAs). ERp44 interacts with the luminal loop of IP3R type 1 (IP3R1) and inhibits its activity. ERp57 oxidizes the luminal thiols of SERCA2b in a Ca2+-dependent manner (Li and Camacho 2004). By-product of oxidative protein folding (i.e., ROS) affects the redox states of the channels (RyR and IP3R1) and changes their activities. In this way redox homeostasis and Ca2+ flux are interrelated. Sigma-1 receptor (Sig-1R) works as a Ca2+ sensor and interacts with BiP. On Ca2+ depletion from the ER via IP3R, BiP dissociates from Sig-1R, interacts with IP3R, and protects intrinsically unstable IP3R from degradation. Cytosolic sorting protein PACS-2 recruits CNX to the MAM (Myhill et al. 2008). Both CRT and CNX interact with SERCA2b and inhibit Ca2+ oscillations (John et al. 1998). The MAM is also the place where energy (ATP) and lipids are exchanged between ER and mitochondria. Ca2+ and ATP levels affect the activities of ER chaperones and foldases. Thus, redox homeostasis, Ca2+ flux and the activities of ER foldases integrally affect the oxidative protein folding capacity in the ER.
ER Redox Homeostasis
Conditions in the ER are more oxidizing than those of the cytosol to favor the oxidative protein folding. This oxidative environment was long thought to be maintained by preferential GSSG influx into the ER lumen, but the discovery of an ER-resident flavoprotein, Ero1p (ER oxidoreductin 1) in yeast, changed this concept (Fig. 3A) (Hwang et al. 1992; Frand and Kaiser 1998; Pollard et al. 1998). Ero1, which has two orthologs (Ero1α and Ero1β) in higher eukaryotes, is now thought to be the oxidative engine that serves as the primary oxidase of PDI (Appenzeller-Herzog et al. 2010). Concomitantly, hydrogen peroxide (H2O2) is thought to be produced from oxygen as an electron acceptor (Enyedi et al. 2010). Ero1α is expressed broadly in multiple human tissue-types, whereas Ero1β is well-expressed only in certain tissue-types, such as the pancreas (Dias-Gunasekara et al. 2005). Interestingly, Ero1α activity is regulated by the isomerization/reduction of intramolecular disulfide bonds that are exerted by PDI monitoring the redox state of the ER, whereas Ero1β seems to be less tightly regulated and shows higher oxidase activity in vitro (Inaba et al. 2010; Tavender and Bulleid 2010a; Wang et al. 2011). Surprisingly, a mutant mouse lacking both isoforms of intact Ero1 is viable, even though Ero1 genes in both Saccharomyces cerevisiae and Drosophila are essential (Frand and Kaiser 1998; Tien et al. 2008; Zito et al. 2010a). Recent studies revealed an alternate cascade centered on peroxiredoxin IV (PRDX4), which is thought to work as a H2O2 reducer (Tavender and Bulleid 2010b). PRDX4 can oxidize several different PDI family members, and H2O2 itself may also directly oxidize PDI family members (Karala et al. 2009; Margittai and Banhegyi 2010; Tavender et al. 2010; Zito et al. 2010b). These alternative pathways would serve to alleviate excess ROS production in the ER and suggest the existence of diversified and intricate oxidative cascades (Csala et al. 2010).
ER Calcium Balance
The ER plays a major role in Ca2+ homeostasis and signaling, and contains a total Ca2+ concentration of 1–3 mM and a free Ca2+ concentration of 60–400 µM (Bygrave and Benedetti 1996). A number of Ca2+-binding proteins reside in the ER, including calreticulin, calnexin, BiP, GRP94, calumenin, and the reticulocalbins (Coe and Michalak 2009; Michalak et al. 2009). Their functions are facilitated by high Ca2+ concentrations, whereas Ca2+ depletion with agents such as thapsigargin results in the deterioration of the proteins and triggers the UPR. It has been speculated that the higher Ca2+ concentration in the lumen mimics the effect of extracellular Ca2+ and helps proteins adopt a stable conformation for secretion.
Accumulating evidence shows that the ER calcium flux is linked with the luminal redox condition (Fig. 3B) (Gorlach et al. 2006). The ryanodine receptor is a redox sensitive Ca2+ channel in the membrane of the sarcoplasmic reticulum (Zable et al. 1997; Feng et al. 2000). ERp44, which senses the redox state as well as luminal pH and Ca2+ concentration, interacts with the luminal loop of IP3R type 1 (IP3R1) and directly inhibits its activity (Higo et al. 2005). ERp57, another ER oxidoreductase, has been reported to oxidize the luminal thiols of SERCA2b in a Ca2+-dependent manner, thereby reducing the frequency of SERCA2b-dependent Ca2+ oscillations (Li and Camacho 2004). These interactions maintain ER calcium concentration and play an antiapoptotic role during cellular stress responses (Rizzuto et al. 2009). On the other hand, the ER-resident oxidase Ero1α has a proapoptotic role. Ero1α is induced by the CHOP-dependent stress response and hyperoxidizes the ER environment, which indirectly activates IP3R1 and releases Ca2+ into the cytosol (Li et al. 2009). The released Ca2+ activates calcium/calmodulin-dependent protein kinase II (CaMKII), which triggers apoptosis through both mitochondrial pathways and death receptor (Timmins et al. 2009). Thus, redox and Ca2+ homeostasis are interrelated in both a physical and physiological context.
STIM 1 and 2 (stromal-interacting molecules 1 and 2) were recently discovered to be ER Ca2+ sensors that, under depletion of Ca2+ stores, signal to the outer plasma membrane to activate store-operated Ca2+ channels (e.g., Orai1, Orai2, and Orai3) and inhibit voltage-gated Ca2+ channels (e.g., CaV1.2 channel) (Liou et al. 2005; Park et al. 2010; Wang et al. 2010b). Sigma-1 receptor (Sig-1R), which has a BiP-interacting lumenal domain and is primarily localized to the mitochondria-associated membrane (MAM), also works as a Ca2+ sensor. On Ca2+ depletion from the ER via IP3R type 3 (IP3R3), BiP dissociates from Sig-1R, associates with IP3R3, and protects intrinsically unstable IP3R3 from degradation (Hayashi and Su 2007). Thereby, Ca2+ signaling to the mitochondria is stabilized, which potentially contributes to antiapoptotic regulation.
SPECIALIZED COMPARTMENTS WITHIN THE ER
Although the ER is composed of a continuous and interconnected tubular membrane network, it performs diversified and sometimes apparently opposing functions, such as cotranslation translocation of nascent polypeptides and retrotranslocation of misfolded proteins into the cytosol, or oxidation and reduction of cysteine residues of secretory and membrane proteins. To perform these various functions, the ER maintains morphologically and functionally different subdomains. Structurally, the rough ER (ribosome-attached), smooth ER (ribosome-free), and nuclear envelope are clearly distinguished (Voeltz et al. 2002). Constituents and contents of the ER, such as Ca2+ levels and lipid compositions, are also heterogeneous throughout (Rizzuto and Pozzan 2006; Pani and Singh 2009). Therefore, specialized regions will exist, such as the ER quality control (ERQC) compartment where ERAD machineries are enriched and N-glycans of misfolded glycoproteins are trimmed extensively for efficient degradation (Avezov et al. 2008; Lederkremer 2009). These specialized regions are also linked to other organelles, including mitochondria, the plasma membrane, peroxisomes, lysosomes, etc. (Lebiedzinska et al. 2009). In particular, a tight interaction between the ER and the mitochondria-associated membrane (MAM) is now emerging as a major cellular signaling hub (Fig. 3B)(Hayashi et al. 2009; Simmen et al. 2010). On the MAM, ER chaperones (CNX, BiP), oxidoreductases (ERp44, Ero1α), and Ca2+ channels/pumps (IP3R3, SERCA2b, and Sig-1R) are enriched, thereby creating an ideal environment for oxidative protein folding, as well as the regulation of Ca2+ flux (Gilady et al. 2010). On the MAM, IP3Rs, SERCA2b, and Sig-1R are regulated by ER oxidoreductases and/or chaperones, and ROS are produced as a by-product of oxidative protein folding. Both Ca2+ and ROS often function as signaling molecules. Thus, the MAM appears to be a processing hub that integrates signaling information derived from redox, Ca2+, and protein homeostasis. PAM (plasma membrane-associated membranes) is another example, albeit less well-characterized, in which the ER is located in the proximity of the plasma membrane (Lebiedzinska et al. 2009). This region is involved in cellular Ca2+ homeostasis, particularly capacitative Ca2+ entry (CCE), by facilitating interactions between STIMs and Orais. Elucidating the functions and mechanisms of these specialized compartments will broaden and integrate our understanding of proteostatic and metabolic regulation in the ER.
THERAPEUTIC PERSPECTIVES
Protein density in the ER is extremely high, around 100 mg/ml, together with concomitant protein manufacturing (Stevens and Argon 1999). Therefore, the elaborate proteostasis network described above is vital. Its absence inevitably leads to aberrant folding, degradation defects, and pathological consequences. For instance, the amounts and activities of ER chaperones and foldases decrease with age, resulting in a reduction in basal metabolism, which is responsible for a number of maladies, including neurodegenerative diseases, diabetes, cancer, and obesity (Douglas and Dillin 2010; Naidoo 2009). In addition, there are numerous inherited loss-of-function disorders caused by the mutation of specific genes related to ER homeostasis such as the cystic fibrosis trans-membrane conductance regulator (CFTR) and α 1-antitrypsin Z (ATZ) (Hebert and Molinari 2007).
For therapeutic purposes, chemical chaperones that stabilize mutant proteins and help polypeptides to achieve native structure have been extensively investigated (Lawrence et al. 2011; McLaughlin and Vandenbroeck 2010). 4-phenyl butyric acid (4-PBA) is a successful low-molecular weight compound that has been shown to have a beneficial effect on several misfolding-related diseases, including cystic fibrosis, α-1-antitrypsin (α1AT) deficiency, and type 2 diabetes mellitus (Ozcan et al. 2006; Hutt et al. 2009). The precise role of 4-PBA is not yet clear, but it has been determined that it has chaperone-like activities and may inhibit histone deacetylases (HDACs) at high concentrations (Powers et al. 2009). Boosting the capacity of the proteostasis network is another promising approach for therapy. For example, increasing calcium levels in the ER enhances the ER chaperone capacities and increases the production of misfolding-prone enzymes such as mutant variant of glucocerebrosidase (Ong et al. 2010). Also in the cytosol, the induction of Hsp70 or chaperonin CCT/TRiC may inhibit the formation of toxic oligomers, thereby preventing the onset of protein-folding diseases such as Huntington's disease (Behrends et al. 2006; Kitamura et al. 2006; Tam et al. 2006). On the other hand, inhibition of specific chaperones is also useful for the treatment of some diseases. Inhibitors of PDI were reported to suppress the toxicity of misfolded huntingtin in rat neuronal cells, presumably through inhibiting the proapoptotic function of PDI on the MAM (i.e., ROS production) (Hoffstrom et al. 2010). When it comes to in vivo case, vitamin A–coupled liposomes, which deliver small interfering RNA (siRNA) against collagen specific chaperone HSP47 to the hepatic stellate (HS) cell, showed the favorable therapeutic potential for suppressing the liver cirrhosis by reducing the accumulation of insoluble collagen in HS cells (Sato et al. 2008). In addition, stimulating the appropriate degradation of pathogenic proteins will also be beneficial for some cases. For example, the drug carbamazepine has been used to enhance the autophagic pathway, which results in a reduction of the hepatic load of ATZ (Hidvegi et al. 2010). Combination approach also could be used to synergize these effects (Mu et al. 2008).
CONCLUDING REMARKS
In the ER, protein homeostasis, redox, and calcium balances appear to be closely related to each other. We need to have a broader understanding on these processes, especially for complicated biological phenomena such as aging, disease chronicity, and neoplasm. The state-of-the-art “omics” technologies would be helpful to capture the holistic biological points of view (Chen et al. 2010; Churchman and Weissman 2011; Olzscha et al. 2011). On the other hand, established reductionistic approaches are also continuously needed for acquiring knowledge on fundamental biological processes such as the identification of components of the ERAD complex. The knowledge gained from these studies can be applied for the treatment of diseases and improving our health.
Footnotes
Editors: Richard I. Morimoto, Dennis Selkoe, and Jeff Kelly
Additional Perspectives on Protein Homeostasis available at www.cshperspectives.org
REFERENCES
- Aebi M, Bernasconi R, Clerc S, Molinari M 2010. N-glycan structures: Recognition and processing in the ER. Trends Biochem Sci 35: 74–82 [DOI] [PubMed] [Google Scholar]
- Anelli T, Sitia R 2008. Protein quality control in the early secretory pathway. EMBO J 27: 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appenzeller-Herzog C 2011. Glutathione- and non-glutathione-based oxidant control in the endoplasmic reticulum. J Cell Sci 124: 847–855 [DOI] [PubMed] [Google Scholar]
- Appenzeller-Herzog C, Ellgaard L 2008. The human PDI family: Versatility packed into a single fold. Biochim Biophys Acta 1783: 535–548 [DOI] [PubMed] [Google Scholar]
- Appenzeller-Herzog C, Riemer J, Zito E, Chin KT, Ron D, Spiess M, Ellgaard L 2010. Disulphide production by Ero1α-PDI relay is rapid and effectively regulated. EMBO J 29: 3318–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avezov E, Frenkel Z, Ehrlich M, Herscovics A, Lederkremer GZ 2008. Endoplasmic reticulum (ER) mannosidase I is compartmentalized and required for N-glycan trimming to Man5-6GlcNAc2 in glycoprotein ER-associated degradation. Mol Biol Cell 19: 216–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagola K, Mehnert M, Jarosch E, Sommer T 2011. Protein dislocation from the ER. Biochim Biophys Acta 1808: 925–936 [DOI] [PubMed] [Google Scholar]
- Ballar P, Zhong Y, Nagahama M, Tagaya M, Shen Y, Fang S 2007. Identification of SVIP as an endogenous inhibitor of endoplasmic reticulum-associated degradation. J Biol Chem 282: 33908–33914 [DOI] [PubMed] [Google Scholar]
- Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY 2001a. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nature Cell Biol 3: 24–29 [DOI] [PubMed] [Google Scholar]
- Bays NW, Wilhovsky SK, Goradia A, Hodgkiss-Harlow K, Hampton RY 2001b. HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol Biol Cell 12: 4114–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends C, Langer CA, Boteva R, Bottcher UM, Stemp MJ, Schaffar G, Rao BV, Giese A, Kretzschmar H, Siegers K, et al. 2006. Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Mol Cell 23: 887–897 [DOI] [PubMed] [Google Scholar]
- Bernasconi R, Molinari M 2011. ERAD and ERAD tuning: Disposal of cargo and of ERAD regulators from the mammalian ER. Curr Opin Cell Biol 23: 176–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi R, Galli C, Calanca V, Nakajima T, Molinari M 2010. Stringent requirement for HRD1, SEL1L, and OS-9/XTP3-B for disposal of ERAD-L S substrates. J Cell Biol 188: 223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasiole DA, Davis RA, Attie AD 2007. The physiological and molecular regulation of lipoprotein assembly and secretion. Mol Biosyst 3: 608–619 [DOI] [PubMed] [Google Scholar]
- Bordallo J, Plemper RK, Finger A, Wolf DH 1998. Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol Biol Cell 9: 209–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese N, Fasana E 2011. Targeting pathways of C-tail-anchored proteins. Biochim Biophys Acta 1808: 937–946 [DOI] [PubMed] [Google Scholar]
- Braakman I, Hoover-Litty H, Wagner KR, Helenius A 1991. Folding of influenza hemagglutinin in the endoplasmic reticulum. J Cell Biol 114: 401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun S, Matuschewski K, Rape M, Thoms S, Jentsch S 2002. Role of the ubiquitin-selective CDC48(UFD1/NPL4) chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J 21: 615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL 2007. The protective and destructive roles played by molecular chaperones during ERAD (endoplasmic-reticulum-associated degradation). Biochem J 404: 353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL 2010. The special delivery of a tail-anchored protein: Why it pays to use a dedicated courier. Mol Cell 40: 5–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr ML, Cano F, Svobodova S, Boyle LH, Boname JM, Lehner PJ 2011. HRD1 and UBE2J1 target misfolded MHC class I heavy chains for endoplasmic reticulum-associated degradation. Proc Natl Acad Sci 108: 2034–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bygrave FL, Benedetti A 1996. What is the concentration of calcium ions in the endoplasmic reticulum? Cell Calcium 19: 547–551 [DOI] [PubMed] [Google Scholar]
- Cabral WA, Chang W, Barnes AM, Weis M, Scott MA, Leikin S, Makareeva E, Kuznetsova NV, Rosenbaum KN, Tifft CJ, et al. 2007. Prolyl 3-hydroxylase 1 deficiency causes a recessive metabolic bone disorder resembling lethal/severe osteogenesis imperfecta. Nat Genet 39: 359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cali T, Galli C, Olivari S, Molinari M 2008. Segregation and rapid turnover of EDEM1 by an autophagy-like mechanism modulates standard ERAD and folding activities. Biochem Biophys Res Commun 371: 405–410 [DOI] [PubMed] [Google Scholar]
- Carvalho P, Goder V, Rapoport TA 2006. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell 126: 361–373 [DOI] [PubMed] [Google Scholar]
- Carvalho P, Stanley AM, Rapoport TA 2010. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell 143: 579–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman DC, Williams DB 2010. ER quality control in the biogenesis of MHC class I molecules. Semin Cell Dev Biol 21: 512–519 [DOI] [PubMed] [Google Scholar]
- Chapman E, Fry AN, Kang M 2010. The complexities of p97 function in health and disease. Mol Biosyst 7: 700–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Karnovsky A, Sans MD, Andrews PC, Williams JA 2010. Molecular characterization of the endoplasmic reticulum: Insights from proteomic studies. Proteomics 10: 4040–4052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JW, Sutor SL, Lindquist L, Evans GL, Madden BJ, Bergen HR III, Hefferan TE, Yaszemski MJ, Bram RJ 2009. Severe osteogenesis imperfecta in cyclophilin B-deficient mice. PLoS Genet 5: e1000750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JC, Shaler TA, Tyler RE, Kopito RR 2008. OS-9 and GRP94 deliver mutant α1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol 10: 272–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KT, Shen Y, Hendershot LM 2002. BAP, a mammalian BiP-associated protein, is a nucleotide exchange factor that regulates the ATPase activity of BiP. J Biol Chem 277: 47557–47563 [DOI] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS 2011. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469: 368–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessen JH, Mueller B, Spooner E, Pivorunas VL, Ploegh HL 2010. The transmembrane segment of a tail-anchored protein determines its degradative fate through dislocation from the endoplasmic reticulum. J Biol Chem 285: 20732–20739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc S, Hirsch C, Oggier DM, Deprez P, Jakob C, Sommer T, Aebi M 2009. Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J Cell Biol 184: 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe H, Michalak M 2009. Calcium binding chaperones of the endoplasmic reticulum. Gen Physiol Biophys 28: F96–F103 [PubMed] [Google Scholar]
- Coe H, Jung J, Groenendyk J, Prins D, Michalak M 2010. ERp57 modulates STAT3 signaling from the lumen of the endoplasmic reticulum. J Biol Chem 285: 6725–6738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppari S, Altieri F, Ferraro A, Chichiarelli S, Eufemi M, Turano C 2002. Nuclear localization and DNA interaction of protein disulfide isomerase ERp57 in mammalian cells. J Cell Biochem 85: 325–333 [DOI] [PubMed] [Google Scholar]
- Cortini M, Sitia R 2010. ERp44 and ERGIC-53 synergize in coupling efficiency and fidelity of IgM polymerization and secretion. Traffic 11: 651–659 [DOI] [PubMed] [Google Scholar]
- Csala M, Margittai E, Banhegyi G 2010. Redox control of endoplasmic reticulum function. Antioxid Redox Sign 13: 77–108 [DOI] [PubMed] [Google Scholar]
- D’Alessio C, Caramelo JJ, Parodi AJ 2010. UDP-GlC: Glycoprotein glucosyltransferase-glucosidase II, the ying-yang of the ER quality control. Semin Cell Dev Biol 21: 491–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancourt J, Barlowe C 2010. Protein sorting receptors in the early secretory pathway. Annu Rev Biochem 79: 777–802 [DOI] [PubMed] [Google Scholar]
- Denic V, Quan EM, Weissman JS 2006. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell 126: 349–359 [DOI] [PubMed] [Google Scholar]
- Deveraux Q, Ustrell V, Pickart C, Rechsteiner M 1994. A 26 S protease subunit that binds ubiquitin conjugates. J Biol Chem 269: 7059–7061 [PubMed] [Google Scholar]
- Dias-Gunasekara S, Gubbens J, van Lith M, Dunne C, Williams JA, Kataky R, Scoones D, Lapthorn A, Bulleid NJ, Benham AM 2005. Tissue-specific expression and dimerization of the endoplasmic reticulum oxidoreductase Ero1β. J Biol Chem 280: 33066–33075 [DOI] [PubMed] [Google Scholar]
- Ding WX, Yin XM 2008. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy 4: 141–150 [DOI] [PubMed] [Google Scholar]
- Dong M, Bridges JP, Apsley K, Xu Y, Weaver TE 2008. ERdj4 and ERdj5 are required for endoplasmic reticulum-associated protein degradation of misfolded surfactant protein C. Mol Biol Cell 19: 2620–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas PM, Dillin A 2010. Protein homeostasis and aging in neurodegeneration. J Cell Biol 190: 719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudek J, Benedix J, Cappel S, Greiner M, Jalal C, Muller L, Zimmermann R 2009. Functions and pathologies of BiP and its interaction partners. Cell Mol Life Sci 66: 1556–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eletto D, Dersh D, Argon Y 2010. GRP94 in ER quality control and stress responses. Semin Cell Dev Biol 21: 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L, Helenius A 2003. Quality control in the endoplasmic reticulum. Nature Rev Mol Cell Biol 4: 181–191 [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Ruddock LW 2005. The human protein disulphide isomerase family: Substrate interactions and functional properties. EMBO Rep 6: 28–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enyedi B, Varnai P, Geiszt M 2010. Redox state of the endoplasmic reticulum is controlled by Ero1L-α and intraluminal calcium. Antioxid Redox Sign 13: 721–729 [DOI] [PubMed] [Google Scholar]
- Ernst R, Mueller B, Ploegh HL, Schlieker C 2009. The otubain YOD1 is a deubiquitinating enzyme that associates with p97 to facilitate protein dislocation from the ER. Mol Cell 36: 28–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige MJ, Hendershot LM 2010. Disulfide bonds in ER protein folding and homeostasis. Curr Opin Cell Biol 23: 167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Liu G, Allen PD, Pessah IN 2000. Transmembrane redox sensor of ryanodine receptor complex. J Biol Chem 275: 35902–35907 [DOI] [PubMed] [Google Scholar]
- Finley D 2009. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem 78: 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. 2010. The Pfam protein families database. Nucleic Acids Res 38: D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frand AR, Kaiser CA 1998. The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol Cell 1: 161–170 [DOI] [PubMed] [Google Scholar]
- Fu L, Sztul E 2009. ER-associated complexes (ERACs) containing aggregated cystic fibrosis transmembrane conductance regulator (CFTR) are degraded by autophagy. Eur J Cell Biol 88: 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita E, Kouroku Y, Isoai A, Kumagai H, Misutani A, Matsuda C, Hayashi YK, Momoi T 2007. Two endoplasmic reticulum-associated degradation (ERAD) systems for the novel variant of the mutant dysferlin: Ubiquitin/proteasome ERAD(I) and autophagy/lysosome ERAD(II). Hum Mol Genet 16: 618–629 [DOI] [PubMed] [Google Scholar]
- Gale M Jr, Blakely CM, Hopkins DA, Melville MW, Wambach M, Romano PR, Katze MG 1998. Regulation of interferon-induced protein kinase PKR: Modulation of P58IPK inhibitory function by a novel protein, P52rIPK. Mol Cell Biol 18: 859–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbi N, Tanaka S, Momburg F, Hammerling GJ 2006. Impaired assembly of the major histocompatibility complex class I peptide-loading complex in mice deficient in the oxidoreductase ERp57. Nat Immunol 7: 93–102 [DOI] [PubMed] [Google Scholar]
- Gilady SY, Bui M, Lynes EM, Benson MD, Watts R, Vance JE, Simmen T 2010. Ero1α requires oxidizing and normoxic conditions to localize to the mitochondria-associated membrane (MAM). Cell Stress Chaperon 15: 619–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeckeler JL, Brodsky JL 2010. Molecular chaperones and substrate ubiquitination control the efficiency of endoplasmic reticulum-associated degradation. Diabetes Obes Metab 12: 32–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlach A, Klappa P, Kietzmann T 2006. The endoplasmic reticulum: Folding, calcium homeostasis, signaling, and redox control. Antioxid Redox Sign 8: 1391–1418 [DOI] [PubMed] [Google Scholar]
- Gorres KL, Raines RT 2010. Prolyl 4-hydroxylase. Crit Rev Biochem Mol Biol 45: 106–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinna LS, Robbins PW 1979. Glycoprotein biosynthesis. Rat liver microsomal glucosidases which process oligosaccharides. J Biol Chem 254: 8814–8818 [PubMed] [Google Scholar]
- Grove DE, Fan CY, Ren HY, Cyr DM 2010. The endoplasmic reticulum-associated Hsp40 DNAJB12 and Hsc70 cooperate to facilitate RMA1 E3-dependent degradation of nascent CFTRδF508. Mol Biol Cell 22: 301–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara M, Maegawa K, Suzuki M, Ushioda R, Araki K, Matsumoto Y, Hoseki J, Nagata K, Inaba K 2011. Structural basis of an ERAD pathway mediated by the ER-resident protein disulfide reductase ERdj5. Mol Cell 41: 432–444 [DOI] [PubMed] [Google Scholar]
- Hatahet F, Ruddock LW, Ahn K, Benham A, Craik D, Ellgaard L, Ferrari D, Ventura S 2009. Protein disulfide isomerase: A critical evaluation of its function in disulfide bond formation. Antioxid Redox Sign 11: 2807–2850 [DOI] [PubMed] [Google Scholar]
- Hayashi T, Su TP 2007. Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca2+ signaling and cell survival. Cell 131: 596–610 [DOI] [PubMed] [Google Scholar]
- Hayashi T, Rizzuto R, Hajnoczky G, Su TP 2009. MAM: More than just a housekeeper. Trends Cell Biol 19: 81–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A 2009. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol 11: 1433–1437 [DOI] [PubMed] [Google Scholar]
- Hebert DN, Molinari M 2007. In and out of the ER: Protein folding, quality control, degradation, and related human diseases. Physiol Rev 87: 1377–1408 [DOI] [PubMed] [Google Scholar]
- Hebert DN, Garman SC, Molinari M 2005. The glycan code of the endoplasmic reticulum: Asparagine-linked carbohydrates as protein maturation and quality-control tags. Trends Cell Biol 15: 364–370 [DOI] [PubMed] [Google Scholar]
- Hegde RS, Kang SW 2008. The concept of translocational regulation. J Cell Biol 182: 225–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde RS, Ploegh HL 2010. Quality and quantity control at the endoplasmic reticulum. Curr Opin Cell Biol 22: 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A 1994. How N-linked oligosaccharides affect glycoprotein folding in the endoplasmic reticulum. Mol Biol Cell 5: 253–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendershot LM 2004. The ER function BiP is a master regulator of ER function. Mt Sinai J Med 71: 289–297 [PubMed] [Google Scholar]
- Hidvegi T, Ewing M, Hale P, Dippold C, Beckett C, Kemp C, Maurice N, Mukherjee A, Goldbach C, Watkins S, et al. 2010. An autophagy-enhancing drug promotes degradation of mutant α1-antitrypsin Z and reduces hepatic fibrosis. Science 329: 229–232 [DOI] [PubMed] [Google Scholar]
- Higo T, Hattori M, Nakamura T, Natsume T, Michikawa T, Mikoshiba K 2005. Subtype-specific and ER lumenal environment-dependent regulation of inositol 1,4,5-triphosphate receptor type 1 by ERp44. Cell 120: 85–98 [DOI] [PubMed] [Google Scholar]
- Hirao K, Natsuka Y, Tamura T, Wada I, Morito D, Natsuka S, Romero P, Sleno B, Tremblay LO, Herscovics A, et al. 2006. EDEM3, a soluble EDEM homolog, enhances glycoprotein endoplasmic reticulum-associated degradation and mannose trimming. J Biol Chem 281: 9650–9658 [DOI] [PubMed] [Google Scholar]
- Hirsch C, Gauss R, Horn SC, Neuber O, Sommer T 2009. The ubiquitylation machinery of the endoplasmic reticulum. Nature 458: 453–460 [DOI] [PubMed] [Google Scholar]
- Hoffstrom BG, Kaplan A, Letso R, Schmid RS, Turmel GJ, Lo DC, Stockwell BR 2010. Inhibitors of protein disulfide isomerase suppress apoptosis induced by misfolded proteins. Nat Chem Biol 6: 900–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoseki J, Ushioda R, Nagata K 2010. Mechanism and components of endoplasmic reticulum-associated degradation. J Biochem 147: 19–25 [DOI] [PubMed] [Google Scholar]
- Hosoda A, Tokuda M, Akai R, Kohno K, Iwawaki T 2010. Positive contribution of ERdj5/JPDI to endoplasmic reticulum protein quality control in the salivary gland. Biochem J 425: 117–125 [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Wada I, Hasegawa K, Yorihuzi T, Tremblay LO, Herscovics A, Nagata K 2001. A novel ER α-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep 2: 415–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Wada I, Natsuka Y, Nagata K 2006. EDEM accelerates ERAD by preventing aberrant dimer formation of misfolded α1-antitrypsin. Genes Cells 11:465–476 [DOI] [PubMed] [Google Scholar]
- Hosokawa N, You Z, Tremblay LO, Nagata K, Herscovics A 2007. Stimulation of ERAD of misfolded null Hong Kong α1-antitrypsin by Golgi α1,2-mannosidases. Biochem Biophys Res Commun 362: 626–632 [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Wada I, Nagasawa K, Moriyama T, Okawa K, Nagata K 2008. Human XTP3-B forms an endoplasmic reticulum quality control scaffold with the HRD1-SEL1L ubiquitin ligase complex and BiP. J Biol Chem 283: 20914–20924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Kamiya Y, Kato K 2010a. The role of MRH domain-containing lectins in ERAD. Glycobiology 20: 651–660 [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Tremblay LO, Sleno B, Kamiya Y, Wada I, Nagata K, Kato K, Herscovics A 2010b. EDEM1 accelerates the trimming of α1,2-linked mannose on the C branch of N-glycans. Glycobiology 20: 567–575 [DOI] [PubMed] [Google Scholar]
- Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I 2008. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453: 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutt D, Balch WE 2010. Cell biology. The proteome in balance. Science 329: 766–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutt DM, Powers ET, Balch WE 2009. The proteostasis boundary in misfolding diseases of membrane traffic. FEBS Lett 583: 2639–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang C, Sinskey AJ, Lodish HF 1992. Oxidized redox state of glutathione in the endoplasmic reticulum. Science 257: 1496–1502 [DOI] [PubMed] [Google Scholar]
- Inaba K, Masui S, Iida H, Vavassori S, Sitia R, Suzuki M 2010. Crystal structures of human Ero1α reveal the mechanisms of regulated and targeted oxidation of PDI. EMBO J 29: 3330–3343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida Y, Yamamoto A, Kitamura A, Lamande SR, Yoshimori T, Bateman JF, Kubota H, Nagata K 2009. Autophagic elimination of misfolded procollagen aggregates in the endoplasmic reticulum as a means of cell protection. Mol Biol Cell 20: 2744–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa Y, Wirz J, Vranka JA, Nagata K, Bachinger HP 2009. Biochemical characterization of the prolyl 3-hydroxylase 1.cartilage-associated protein.cyclophilin B complex. J Biol Chem 284: 17641–17647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikura S, Weissman AM, Bonifacino JS 2010. Serine residues in the cytosolic tail of the T-cell antigen receptor α-chain mediate ubiquitination and endoplasmic reticulum-associated degradation of the unassembled protein. J Biol Chem 285: 23916–23924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi Y, Okada F, Tsunoda S, Kibe N, Shirasawa N, Ikawa M, Okabe M, Ikeda Y, Fujii J 2009. Peroxiredoxin 4 knockout results in elevated spermatogenic cell death via oxidative stress. Biochem J 419: 149–158 [DOI] [PubMed] [Google Scholar]
- Jarosch E 2002. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat Cell Biol 4: 134–139 [DOI] [PubMed] [Google Scholar]
- Jessop CE, Watkins RH, Simmons JJ, Tasab M, Bulleid NJ 2009. Protein disulphide isomerase family members show distinct substrate specificity: P5 is targeted to BiP client proteins. J Cell Sci 122: 4287–4295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Awad W, Petrova K, Hendershot LM 2008. Regulated release of ERdj3 from unfolded proteins by BiP. EMBO J 27: 2873–2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo Y, Debose-Boyd RA 2010. Control of cholesterol synthesis through regulated ER-associated degradation of HMG CoA reductase. Crit Rev Biochem Mol Biol 45: 185–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John LM, Lechleiter JD, Camacho P 1998. Differential modulation of SERCA2 isoforms by calreticulin. J Cell Biol 142: 963–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AE, Van Waes MA 1999. The translocon: A dynamic gateway at the ER membrane. Annu Rev Cell Dev Biol 15: 799–842 [DOI] [PubMed] [Google Scholar]
- Kamiya Y, Kamiya D, Yamamoto K, Nyfeler B, Hauri HP, Kato K 2008. Molecular basis of sugar recognition by the human L-type lectins ERGIC-53, VIPL, and VIP36. J Biol Chem 283: 1857–1861 [DOI] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA 2010. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol 11: 579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehara K, Kawaguchi S, Ng DT 2007. The EDEM and Yos9p families of lectin-like ERAD factors. Semin Cell Dev Biol 18: 743–750 [DOI] [PubMed] [Google Scholar]
- Karala AR, Lappi AK, Saaranen MJ, Ruddock LW 2009. Efficient peroxide-mediated oxidative refolding of a protein at physiological pH and implications for oxidative folding in the endoplasmic reticulum. Antioxid Redox Sign 11: 963–970 [DOI] [PubMed] [Google Scholar]
- Kawaguchi S, Ng DT 2007. SnapShot: ER-associated protein degradation pathways. Cell 129: 1230. [DOI] [PubMed] [Google Scholar]
- Kim I, Rodriguez-Enriquez S, Lemasters JJ 2007. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys 462: 245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Horibe T, Sakamoto C, Shitara Y, Fujiwara F, Komiya T, Yamamoto A, Hayano T, Takahashi N, Kikuchi M 2008. Evidence for mitochondrial localization of P5, a member of the protein disulphide isomerase family. J Biochem 144: 187–196 [DOI] [PubMed] [Google Scholar]
- Kitamura A, Kubota H, Pack CG, Matsumoto G, Hirayama S, Takahashi Y, Kimura H, Kinjo M, Morimoto RI, Nagata K 2006. Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nat Cell Biol 8: 1163–1170 [DOI] [PubMed] [Google Scholar]
- Kjeldsen T, Ludvigsen S, Diers I, Balschmidt P, Sorensen AR, Kaarsholm NC 2002. Engineering-enhanced protein secretory expression in yeast with application to insulin. J Biol Chem 277: 18245–18248 [DOI] [PubMed] [Google Scholar]
- Knop M, Finger A, Braun T, Hellmuth K, Wolf DH 1996. Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J 15: 753–763 [PMC free article] [PubMed] [Google Scholar]
- Kopito RR 1999. Biosynthesis and degradation of CFTR. Physiol Rev 79 (Suppl): S167–S173 [DOI] [PubMed] [Google Scholar]
- Korotkov KV, Kumaraswamy E, Zhou Y, Hatfield DL, Gladyshev VN 2001. Association between the 15-kDa selenoprotein and UDP-glucose:glycoprotein glucosyltransferase in the endoplasmic reticulum of mammalian cells. J Biol Chem 276: 15330–15536 [DOI] [PubMed] [Google Scholar]
- Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, Ogawa S, Kaufman RJ, Kominami E, Momoi T 2007. ER stress (PERK/eIF2α phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ 14: 230–239 [DOI] [PubMed] [Google Scholar]
- Kowalski JM, Parekh RN, Mao J, Wittrup KD 1998. Protein folding stability can determine the efficiency of escape from endoplasmic reticulum quality control. J Biol Chem 273: 19453–19458 [DOI] [PubMed] [Google Scholar]
- Kroeger H, Miranda E, MacLeod I, Perez J, Crowther DC, Marciniak SJ, Lomas DA 2009. Endoplasmic reticulum-associated degradation (ERAD) and autophagy cooperate to degrade polymerogenic mutant serpins. J Biol Chem 284: 22793–22802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YA, Lawson TG, Velayutham M, Zweier JL, Pickart CM 2002. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature 416: 763–767 [DOI] [PubMed] [Google Scholar]
- Lawrence JM, Samways MJ, Henwood J, Kelly J 2011. Effect of an invasive ant and its chemical control on a threatened endemic Seychelles millipede. Ecotoxicology [DOI] [PubMed] [Google Scholar]
- Le Fourn V, Gaplovska-Kysela K, Guhl B, Santimaria R, Zuber C, Roth J 2009. Basal autophagy is involved in the degradation of the ERAD component EDEM1. Cell Mol Life Sci 66: 1434–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebiedzinska M, Szabadkai G, Jones AW, Duszynski J, Wieckowski MR 2009. Interactions between the endoplasmic reticulum, mitochondria, plasma membrane and other subcellular organelles. Int J Biochem Cell Biol 41: 1805–1816 [DOI] [PubMed] [Google Scholar]
- Lederkremer GZ 2009. Glycoprotein folding, quality control and ER-associated degradation. Curr Opin Struct Biol 19: 515–523 [DOI] [PubMed] [Google Scholar]
- Lee SO, Cho K, Cho S, Kim I, Oh C, Ahn K 2010. Protein disulphide isomerase is required for signal peptide peptidase-mediated protein degradation. EMBO J 29: 363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P 2009. SMART 6: Recent updates and new developments. Nucleic Acids Res 37: D229–D232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Camacho P 2004. Ca2+-dependent redox modulation of SERCA 2b by ERp57. J Cell Biol 164: 35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Mongillo M, Chin KT, Harding H, Ron D, Marks AR, Tabas I 2009. Role of ERO1-α-mediated stimulation of inositol 1,4,5-triphosphate receptor activity in endoplasmic reticulum stress-induced apoptosis. J Cell Biol 186: 783–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Yin C, Doong H, Fang S, Peterhoff C, Nixon RA, Monteiro MJ 2006. Characterization of erasin (UBXD2): A new ER protein that promotes ER-associated protein degradation. J Cell Sci 119: 4011–4024 [DOI] [PubMed] [Google Scholar]
- Lilley BN, Ploegh HL 2004. A membrane protein required for dislocation of misfolded proteins from the ER. Nature 429: 834–840 [DOI] [PubMed] [Google Scholar]
- Lim PJ, Danner R, Liang J, Doong H, Harman C, Srinivasan D, Rothenberg C, Wang H, Ye Y, Fang S, et al. 2009. Ubiquilin and p97/VCP bind erasin, forming a complex involved in ERAD. J Cell Biol 187: 201–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou J, Kim ML, Heo WD, Jones JT, Myers JW, Ferrell JE Jr, Meyer T 2005. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol 15: 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loureiro J, Lilley BN, Spooner E, Noriega V, Tortorella D, Ploegh HL 2006. Signal peptide peptidase is required for dislocation from the endoplasmic reticulum. Nature 441: 894–897 [DOI] [PubMed] [Google Scholar]
- Manjithaya R, Nazarko TY, Farre JC, Subramani S 2010. Molecular mechanism and physiological role of pexophagy. FEBS Lett 584: 1367–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margittai E, Banhegyi G 2010. Oxidative folding in the endoplasmic reticulum: Towards a multiple oxidant hypothesis? FEBS Lett 584: 2995–2998 [DOI] [PubMed] [Google Scholar]
- Mariappan M, Li X, Stefanovic S, Sharma A, Mateja A, Keenan RJ, Hegde RS 2010. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature 466: 1120–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mast SW, Diekman K, Karaveg K, Davis A, Sifers RN, Moremen KW 2005. Human EDEM2, a novel homolog of family 47 glycosidases, is involved in ER-associated degradation of glycoproteins. Glycobiology 15: 421–436 [DOI] [PubMed] [Google Scholar]
- Mateja A, Szlachcic A, Downing ME, Dobosz M, Mariappan M, Hegde RS, Keenan RJ 2009. The structural basis of tail-anchored membrane protein recognition by Get3. Nature 461: 361–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin M, Vandenbroeck K 2010. The endoplasmic reticulum protein folding factory and its chaperones: New targets for drug discovery? Br J Pharmacol 162: 328–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier L, Usherwood YK, Tae Chung K, Hendershot LM 2002. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol Biol Cell 13: 4456–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak M, Groenendyk J, Szabo E, Gold LI, Opas M 2009. Calreticulin, a multi-process calcium-buffering chaperone of the endoplasmic reticulum. Biochem J 417: 651–666 [DOI] [PubMed] [Google Scholar]
- Molinari M 2007. N-glycan structure dictates extension of protein folding or onset of disposal. Nat Chem Biol 3: 313–320 [DOI] [PubMed] [Google Scholar]
- Molinari M, Calanca V, Galli C, Lucca P, Paganetti P 2003. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science 299: 1397–1400 [DOI] [PubMed] [Google Scholar]
- Morello R, Rauch F 2010. Role of cartilage-associated protein in skeletal development. Curr Osteoporos Rep 8: 77–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morello R, Bertin TK, Chen Y, Hicks J, Tonachini L, Monticone M, Castagnola P, Rauch F, Glorieux FH, Vranka J, et al. 2006. CRTAP is required for prolyl 3- hydroxylation and mutations cause recessive osteogenesis imperfecta. Cell 127: 291–304 [DOI] [PubMed] [Google Scholar]
- Morito D, Hirao K, Oda Y, Hosokawa N, Tokunaga F, Cyr DM, Tanaka K, Iwai K, Nagata K 2008. Gp78 cooperates with RMA1 in endoplasmic reticulum-associated degradation of CFTRΔ508. Mol Biol Cell 19: 1328–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu TW, Ong DS, Wang YJ, Balch WE, Yates JR III, Segatori L, Kelly JW 2008. Chemical and biological approaches synergize to ameliorate protein-folding diseases. Cell 134: 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller B, Klemm EJ, Spooner E, Claessen JH, Ploegh HL 2008. SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc Natl Acad Sci 105: 12325–12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myhill N, Lynes EM, Nanji JA, Blagoveshchenskaya AD, Fei H, Carmine Simmen K, Cooper TJ, Thomas G, Simmen T 2008. The subcellular distribution of calnexin is mediated by PACS-2. Mol Biol Cell 19: 2777–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai N, Hosokawa M, Itohara S, Adachi E, Matsushita T, Hosokawa N, Nagata K 2000. Embryonic lethality of molecular chaperone hsp47 knockout mice is associated with defects in collagen biosynthesis. J Cell Biol 150: 1499–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa K, Higashi T, Hosokawa N, Kaufman RJ, Nagata K 2007. Simultaneous induction of the four subunits of the TRAP complex by ER stress accelerates ER degradation. EMBO Rep 8: 483–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K 2003. HSP47 as a collagen-specific molecular chaperone: Function and expression in normal mouse development. Semin Cell Dev Biol 14: 275–282 [DOI] [PubMed] [Google Scholar]
- Naidoo N 2009. ER and aging-Protein folding and the ER stress response. Ageing Res Rev 8: 150–159 [DOI] [PubMed] [Google Scholar]
- Neutzner A, Neutzner M, Benischke AS, Ryu SW, Frank S, Youle RJ, Karbowski M 2011. A systematic search for ER membrane-associated RING finger proteins identifies Nixin/ZNRF4 as a regulator of calnexin stability and ER homeostasis. J Biol Chem 286: 8633–8643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng W, Sergeyenko T, Zeng N, Brown JD, Romisch K 2007. Characterization of the proteasome interaction with the Sec61 channel in the endoplasmic reticulum. J Cell Sci 120: 682–691 [DOI] [PubMed] [Google Scholar]
- Oda Y, Hosokawa N, Wada I, Nagata K 2003. EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science 299: 1394–1397 [DOI] [PubMed] [Google Scholar]
- Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, et al. 2006. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol 26: 9220–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda-Shimizu Y, Hendershot LM 2007. Characterization of an ERAD pathway for nonglycosylated BiP substrates, which require Herp. Mol Cell 28: 544–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivari S, Galli C, Alanen H, Ruddock L, Molinari M 2005. A novel stress-induced EDEM variant regulating endoplasmic reticulum-associated glycoprotein degradation. J Biol Chem 280: 2424–2428 [DOI] [PubMed] [Google Scholar]
- Olivari S, Cali T, Salo KEH, Paganetti P, Ruddock LW, Molinari M 2006. EDEM1 regulates ER-associated degradation by accelerating de-mannosylation of folding-defective polypeptides and by inhibiting their covalent aggregation. Biochem Biophys Res Commun 349: 1278–1284 [DOI] [PubMed] [Google Scholar]
- Oliver JD, Roderick HL, Llewellyn DH, High S 1999. ERp57 functions as a subunit of specific complexes formed with the ER lectins calreticulin and calnexin. Mol Biol Cell 10: 2573–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzscha H, Schermann SM, Woerner AC, Pinkert S, Hecht MH, Tartaglia GG, Vendruscolo M, Hayer-Hartl M, Hartl FU, Vabulas RM 2011. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144: 67–78 [DOI] [PubMed] [Google Scholar]
- Ong DS, Mu TW, Palmer AE, Kelly JW 2010. Endoplasmic reticulum Ca2+ increases enhance mutant glucocerebrosidase proteostasis. Nat Chem Biol 6: 424–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando RA 2004. The low-density lipoprotein receptor-related protein associates with calnexin, calreticulin, and protein disulfide isomerase in receptor-associated-protein-deficient fibroblasts. Exp Cell Res 294: 244–253 [DOI] [PubMed] [Google Scholar]
- Otero JH, Lizak B, Hendershot LM 2010. Life and death of a BiP substrate. Semin Cell Dev Biol 21: 472–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan U, Yilmaz E, Ozcan L, Furuhashi M, Vaillancourt E, Smith RO, Gorgun CZ, Hotamisligil GS 2006. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science 313: 1137–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paetzel M, Karla A, Strynadka NC, Dalbey RE 2002. Signal peptidases. Chem Rev 102: 4549–4580 [DOI] [PubMed] [Google Scholar]
- Pani B, Singh BB 2009. Lipid rafts/caveolae as microdomains of calcium signaling. Cell Calcium 45: 625–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B, Lee S, Kim E, Cho K, Riddell SR, Cho S, Ahn K 2006. Redox regulation facilitates optimal peptide selection by MHC class I during antigen processing. Cell 127: 369–382 [DOI] [PubMed] [Google Scholar]
- Park CY, Shcheglovitov A, Dolmetsch R 2010. The CRAC channel activator STIM1 binds and inhibits L-type voltage-gated calcium channels. Science 330: 101–105 [DOI] [PubMed] [Google Scholar]
- Petrova K, Oyadomari S, Hendershot LM, Ron D 2008. Regulated association of misfolded endoplasmic reticulum lumenal proteins with P58/DNAJc3. EMBO J 27: 2862–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon M, Schekman R, Romisch K 1997. Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J 16: 4540–4548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard MG, Travers KJ, Weissman JS 1998. Ero1p: A novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol Cell 1: 171–182 [DOI] [PubMed] [Google Scholar]
- Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE 2009. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem 78: 959–991 [DOI] [PubMed] [Google Scholar]
- Raasi S, Wolf DH 2007. Ubiquitin receptors and ERAD: A network of pathways to the proteasome. Semin Cell Dev Biol 18: 780–791 [DOI] [PubMed] [Google Scholar]
- Rabinovich E, Kerem A, Froehlich KU, Diamant N, Bar-Nun S 2002. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol Cell Biol 22: 626–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabu C, Schmid V, Schwappach B, High S 2009. Biogenesis of tail-anchored proteins: The beginning for the end? J Cell Sci 122: 3605–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Monastyrska I, Verheije MH, Cali T, Ulasli M, Bianchi S, Bernasconi R, de Haan CA, Molinari M 2010. Coronaviruses hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe 7: 500–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemer J, Appenzeller-Herzog C, Johansson L, Bodenmiller B, Hartmann-Petersen R, Ellgaard L 2009. A luminal flavoprotein in endoplasmic reticulum-associated degradation. Proc Natl Acad Sci 106: 14831–14836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto R, Pozzan T 2006. Microdomains of intracellular Ca2+: Molecular determinants and functional consequences. Physiol Rev 86: 369–408 [DOI] [PubMed] [Google Scholar]
- Rizzuto R, Marchi S, Bonora M, Aguiari P, Bononi A, De Stefani D, Giorgi C, Leo S, Rimessi A, Siviero R, et al. 2009. Ca2+ transfer from the ER to mitochondria: When, how and why. Biochim Biophys Acta 1787: 1342–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Canada C, Kelleher DJ, Gilmore R 2009. Cotranslational and posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms. Cell 136: 272–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumpf S, Jentsch S 2006. Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone. Mol Cell 21: 261–269 [DOI] [PubMed] [Google Scholar]
- Rutkevich LA, Williams DB 2010. Participation of lectin chaperones and thiol oxidoreductases in protein folding within the endoplasmic reticulum. Curr Opin Cell Biol 23: 157–166 [DOI] [PubMed] [Google Scholar]
- Rutkevich LA, Cohen-Doyle MF, Brockmeier U, Williams DB 2010. Functional relationship between protein disulfide isomerase family members during the oxidative folding of human secretory proteins. Mol Biol Cell 21: 3093–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski DT, Kang SW, Goodman AG, Garrison JL, Taunton J, Katze MG, Kaufman RJ, Hegde RS 2007. The role of p58IPK in protecting the stressed endoplasmic reticulum. Mol Biol Cell 18: 3681–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders CR, Myers JK 2004. Disease-related misassembly of membrane proteins. Annu Rev Biophys Biomol Struct 33: 25–51 [DOI] [PubMed] [Google Scholar]
- Saraogi I, Shan SO 2011. Molecular mechanism of co-translational protein targeting by the signal recognition particle. Traffic 12: 535–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Murase K, Kato J, Kobune M, Sato T, Kawano Y, Takimoto R, Takada K, Miyanishi K, Matsunaga T, et al. 2008. Resolution of liver cirrhosis using vitamin A-coupled liposomes to deliver siRNA against a collagen-specific chaperone. Nat Biotechnol 26: 431–442 [DOI] [PubMed] [Google Scholar]
- Satoh T, Chen Y, Hu D, Hanashima S, Yamamoto K, Yamaguchi Y 2010. Structural basis for oligosaccharide recognition of misfolded glycoproteins by OS-9 in ER-associated degradation. Mol Cell 40: 905–916 [DOI] [PubMed] [Google Scholar]
- Schafer A, Wolf DH 2009. Sec61p is part of the endoplasmic reticulum-associated degradation machinery. EMBO J 28: 2874–2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuberth C, Buchberger A 2008. UBX domain proteins: Major regulators of the AAA ATPase Cdc48/p97. Cell Mol Life Sci 65: 2360–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekijima Y, Wiseman RL, Matteson J, Hammarstrom P, Miller SR, Sawkar AR, Balch WE, Kelly JW 2005. The biological and chemical basis for tissue-selective amyloid disease. Cell 121: 73–85 [DOI] [PubMed] [Google Scholar]
- Shen Y, Hendershot LM 2005. ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a cofactor for BiP’s interactions with unfolded substrates. Mol Biol Cell 16: 40–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibatani T, David LL, McCormack AL, Frueh K, Skach WR 2005. Proteomic analysis of mammalian oligosaccharyltransferase reveals multiple subcomplexes that contain Sec61, TRAP, and two potential new subunits. Biochemistry 44: 5982–5992 [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Okuda-Shimizu Y, Hendershot LM 2010. Ubiquitylation of an ERAD substrate occurs on multiple types of amino acids. Mol Cell 40: 917–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmen T, Lynes EM, Gesson K, Thomas G 2010. Oxidative protein folding in the endoplasmic reticulum: Tight links to the mitochondria-associated membrane (MAM). Biochim Biophys Acta 1798: 1465–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solda T, Galli C, Kaufman RJ, Molinari M 2007. Substrate-specific requirements for UGT1-dependent release from calnexin. Mol Cell 27: 238–249 [DOI] [PubMed] [Google Scholar]
- Stevens FJ, Argon Y 1999. Protein folding in the ER. Semin Cell Dev Biol 10: 443–454 [DOI] [PubMed] [Google Scholar]
- Swanson R, Locher M, Hochstrasser M 2001. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matα2 repressor degradation. Genes Dev 15: 2660–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam S, Geller R, Spiess C, Frydman J 2006. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nat Cell Biol 8: 1155–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe K, Lennarz WJ, Suzuki T 2006. A cytoplasmic peptide: N-glycanase. Meth Enzymol 415: 46–55 [DOI] [PubMed] [Google Scholar]
- Tavender TJ, Bulleid NJ 2010a. Molecular mechanisms regulating oxidative activity of the Ero1 family in the endoplasmic reticulum. Antioxid Redox Sign 13: 1177–1187 [DOI] [PubMed] [Google Scholar]
- Tavender TJ, Bulleid NJ 2010b. Peroxiredoxin IV protects cells from oxidative stress by removing H2O2 produced during disulphide formation. J Cell Sci 123: 2672–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavender TJ, Springate JJ, Bulleid NJ 2010. Recycling of peroxiredoxin IV provides a novel pathway for disulphide formation in the endoplasmic reticulum. EMBO J 29: 4185–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxis C, Hitt R, Park SH, Deak PM, Kostova Z, Wolf DH 2003. Use of modular substrates demonstrates mechanistic diversity and reveals differences in chaperone requirement of ERAD. J Biol Chem 278: 35903–35913 [DOI] [PubMed] [Google Scholar]
- Tien AC, Rajan A, Schulze KL, Ryoo HD, Acar M, Steller H, Bellen HJ 2008. Ero1L, a thiol oxidase, is required for Notch signaling through cysteine bridge formation of the Lin12-Notch repeats in Drosophila melanogaster. J Cell Biol 182: 1113–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmins JM, Ozcan L, Seimon TA, Li G, Malagelada C, Backs J, Backs T, Bassel-Duby R, Olson EN, Anderson ME, et al. 2009. Calcium/calmodulin-dependent protein kinase II links ER stress with Fas and mitochondrial apoptosis pathways. J Clin Invest 119: 2925–2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todde V, Veenhuis M, van der Klei IJ 2009. Autophagy: Principles and significance in health and disease. Biochim Biophys Acta 1792: 3–13 [DOI] [PubMed] [Google Scholar]
- Tsai YC, Weissman AM 2010. The unfolded protein response, degradation from endoplasmic reticulum and cancer. Genes Cancer 1: 764–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushioda R, Nagata K 2011. The endoplasmic reticulum-associated degradation and disulfide reductase ERdj5. Meth Enzymol 490: 235–258 [DOI] [PubMed] [Google Scholar]
- Ushioda R, Hoseki J, Araki K, Jansen G, Thomas DY, Nagata K 2008. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science 321: 569–572 [DOI] [PubMed] [Google Scholar]
- Vashist S, Ng DT 2004. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J Cell Biol 165: 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Rolls MM, Rapoport TA 2002. Structural organization of the endoplasmic reticulum. EMBO Rep 3: 944–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlman J 2007. Real-time fluorescence detection of ERAD substrate retrotranslocation in a mammalian in vitro system. Cell 129: 943–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Heath-Engel H, Zhang D, Nguyen N, Thomas DY, Hanrahan JW, Shore GC 2008. BAP31 interacts with Sec61 translocons and promotes retrotranslocation of CFTRδF508 via the derlin-1 complex. Cell 133: 1080–1092 [DOI] [PubMed] [Google Scholar]
- Wang X, Herr RA, Rabelink M, Hoeben RC, Wiertz EJ, Hansen TH 2009. Ube2j2 ubiquitinates hydroxylated amino acids on ER-associated degradation substrates. J Cell Biol 187:655–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Brown EC, Mak G, Zhuang J, Denic V 2010a. A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol Cell 40: 159–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Deng X, Mancarella S, Hendron E, Eguchi S, Soboloff J, Tang XD, Gill DL 2010b. The calcium store sensor, STIM1, reciprocally controls Orai and CaV1.2 channels. Science 330: 105–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhu L, Wang CC 2011. The endoplasmic reticulum sulfhydryl oxidase Ero1β drives efficient oxidative protein folding with loose regulation. Biochem J 434: 113–121 [DOI] [PubMed] [Google Scholar]
- Wiertz EJ 1996. The human cytomegalovirus us11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84: 769–779 [DOI] [PubMed] [Google Scholar]
- Wiertz EJ, Tortorella D, Bogyo M, Yu J, Mothes W, Jones TR, Rapoport TA, Ploegh HL 1996. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 384: 432–438 [DOI] [PubMed] [Google Scholar]
- Willer M, Forte GMA, Stirling CJ 2008. Sec61p is required for ERAD-L: Genetic dissection of the translocation and ERAD-L functions of Sec61P using novel derivatives of CPY. J Biol Chem 283: 33883–33888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DB 2006. Beyond lectins: The calnexin/calreticulin chaperone system of the endoplasmic reticulum. J Cell Sci 119: 615–623 [DOI] [PubMed] [Google Scholar]
- Wiseman RL, Powers ET, Buxbaum JN, Kelly JW, Balch WE 2007. An adaptable standard for protein export from the endoplasmic reticulum. Cell 131: 809–821 [DOI] [PubMed] [Google Scholar]
- Wong E, Cuervo AM 2010. Integration of clearance mechanisms: The proteasome and autophagy. Cold Spring Harb Perspect Biol 2: a006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto YH, Kimura T, Momohara S, Takeuchi M, Tani T, Kimata Y, Kadokura H, Kohno K 2010. A novel ER J-protein DNAJB12 accelerates ER-associated degradation of membrane proteins including CFTR. Cell Struct Funct 35: 107–116 [DOI] [PubMed] [Google Scholar]
- Yan W, Frank CL, Korth MJ, Sopher BL, Novoa I, Ron D, Katze MG 2002. Control of PERK eIF2α kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proc Natl Acad Sci 99: 15920–15925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA 2004. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature 429: 841–847 [DOI] [PubMed] [Google Scholar]
- Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL 2009. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 5: 1180–1185 [DOI] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ 2007a. Eating the endoplasmic reticulum: Quality control by autophagy. Trends Cell Biol 17: 279–285 [DOI] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ 2007b. Endoplasmic reticulum stress: A new pathway to induce autophagy. Autophagy 3: 160–162 [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Tanaka K 2010. Lectin-like ERAD players in ER and cytosol. Biochim Biophys Acta 1800: 172–180 [DOI] [PubMed] [Google Scholar]
- Younger JM, Chen L, Ren HY, Rosser MF, Turnbull EL, Fan CY, Patterson C, Cyr DM 2006. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell 126: 571–582 [DOI] [PubMed] [Google Scholar]
- Zable AC, Favero TG, Abramson JJ 1997. Glutathione modulates ryanodine receptor from skeletal muscle sarcoplasmic reticulum. Evidence for redox, regulation of the Ca2+ release mechanism. J Biol Chem 272: 7069–7077 [DOI] [PubMed] [Google Scholar]
- Zahedi RP, Volzing C, Schmitt A, Frien M, Jung M, Dudek J, Wortelkamp S, Sickmann A, Zimmermann R 2009. Analysis of the membrane proteome of canine pancreatic rough microsomes identifies a novel Hsp40, termed ERj7. Proteomics 9: 3463–3473 [DOI] [PubMed] [Google Scholar]
- Zavacki AM, Arrojo EDR, Freitas BC, Chung M, Harney JW, Egri P, Wittmann G, Fekete C, Gereben B, Bianco AC 2009. The E3 ubiquitin ligase TEB4 mediates degradation of type 2 iodothyronine deiodinase. Mol Cell Biol 29: 5339–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann R, Eyrisch S, Ahmad M, Helms V 2010. Protein translocation across the ER membrane. Biochim Biophys Acta 1808: 912–924 [DOI] [PubMed] [Google Scholar]
- Zito E, Chin KT, Blais J, Harding HP, Ron D 2010a. ERO1-β, a pancreas-specific disulfide oxidase, promotes insulin biogenesis and glucose homeostasis. J Cell Biol 188: 821–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito E, Melo EP, Yang Y, Wahlander A, Neubert TA, Ron D 2010b. Oxidative protein folding by an endoplasmic reticulum-localized peroxiredoxin. Mol Cell 40: 787–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber C, Cormier JH, Guhl B, Santimaria R, Hebert DN, Roth J 2007. EDEM1 reveals a quality control vesicular transport pathway out of the endoplasmic reticulum not involving the COPII exit sites. Proc Natl Acad Sci 104: 4407–4412 [DOI] [PMC free article] [PubMed] [Google Scholar]