Abstract
The crystal structures of ribosomes that have been obtained since 2000 have transformed our understanding of protein synthesis. In addition to proving that RNA is responsible for catalyzing peptide bond formation, these structures have provided important insights into the mechanistic details of how the ribosome functions. This review emphasizes what has been learned about the mechanism of peptide bond formation, the antibiotics that inhibit ribosome function, and the fidelity of decoding.
Crystal structures of the ribosome reveal much about the mechanism of protein synthesis, showing how RNA catalyses peptide bond formation and how antibiotics inhibit this.
1. INTRODUCTION
In December, 1962, James Watson delivered a Nobel Prize lecture in Stockholm entitled “Involvement of RNA in the Synthesis of Protein” (Watson 1963) in which he described what was then known about protein synthesis. When one reads that text today, one is struck by how much had already been learned; the picture Watson painted for his audience was correct in its essence. However, there were still many missing pieces. For example, in 1962 no one knew how many tRNA binding sites there are on the ribosome, nor even whether all the ribosomes in a cell are the same.
In 1962, only three kinds of RNA were known: transfer RNAs, which were then called soluble RNAs, messenger RNAs, which had just been discovered, and ribosomal RNAs. It was understood that mRNAs convey sequence information from the genome to the protein synthetic apparatus, and that tRNAs are carrier molecules for amino acids that perform two functions. First, the esterification of amino acids to tRNAs, which is driven by ATP hydrolysis, activates them; the formation of polypeptides from aminoacyl tRNAs is spontaneous under intracellular conditions, but the formation of polypeptides from free amino acids is not. Second, tRNAs are adaptor molecules. There is at least one tRNA for every amino acid the cell uses for protein synthesis, and an enzyme that specifically aminoacylates each tRNA with its cognate amino acid. Furthermore, base pairing interactions between tRNA bases, i.e., between tRNA anticodons and mRNA codons, determine protein sequences. The RNAs whose role(s) in protein synthesis Watson could not explain were the ribosomal RNAs. No one knew why the ribosome, the enzyme that catalyzes peptide bonds formation, unlike any other enzyme then known, is made primarily of RNA.
The reason for writing an essay on the role of RNA in protein synthesis today is we can now answer the one major question that Watson could not. Ribosomes contain rRNA because the ribosome is a ribozyme. Even before the first ribozymes were discovered in the early 1980s, it had been suspected that rRNA might be the active principle in the ribosome (Crick 1968; see Noller 1991). The validity of that surmise was proven by the atomic resolution crystal structures of ribosomes that began appearing in the summer of 2000 (Fig. 1).
Figure 1.
Spacefilling model of the 70S ribosome. The structure of the Hma large subunit (Ban et al. 2000) and that of the T. thermophilus small subunit (Wimberly et al. 2000) were docked using the rRNA from the Noller model (Yusupov et al. 2001) of the 70S as well as the A, P, and E site tRNAs from that model. The 23S rRNA and 5S r RNA are in pink and white whereas the 16S rRNA is in light and dark yellow. rProteins of the large subunit are blue and those of the small subunit are green. The A-site tRNA with its 3' end extending into the peptidyl transferase cavity is red and the P-site tRNA is yellow.
Here we review the crystallographic information on which the conclusion that the ribosome is a ribozyme is based, stressing the role of rRNA as a mediator of both peptide bond formation, and mRNA/tRNA interactions, the two aspects of ribosome function now best understood. In addition we will comment on some of the crystal structures that have been obtained of ribosomes with factors bound, and summarize what has been learned about the antibiotic inhibitors of large subunit function. Rather than discussing all of the ribosome crystal structures now available, we will concentrate on those obtained using the large ribosomal subunit of Haloarcula marismortui (Hma), and both the 70S ribosome and the small ribosomal subunit of Thermus thermophilus (Tth). Those interested in what has been learned from other ribosomal crystal structures should consult the appropriate reviews (e.g., Yonath and Bashan 2004; Berk and Cate 2007).
2. THE ROLE OF THE RIBOSOME IN PROTEIN SYNTHESIS
In all organisms, the ribosome consists of two ribonucleoprotein subunits, one about twice the size of the other. Prokaryotic large ribosomal subunits sediment at ∼50S, and have masses of ∼1.5 megadaltons. In prokaryotes, the small ribosomal subunit sediments at ∼30S, and has a mass of ∼0.8 megadaltons. The object that catalyzes protein synthesis is a 1:1 complex of the two subunits that sediments at ∼70S, and it is about two-thirds RNA by weight. The large subunit contains 34 proteins and two RNAs, 23S rRNA and 5S rRNA, whereas the small subunit consists of 21 proteins and a single RNA, 16S rRNA. With the exception of a single large subunit protein, L7/L12, which is present in four copies per ribosome, the ribosome is a 1:1 complex of all its components (Wittmann-Liebold 1986). The cytoplasmic ribosomes in eukaryotes are homologous to prokaryotic ribosomes, but are bigger and have more components (Wool 1986; Wool et al. 1995).
The roles of the two subunits of the ribosome in protein synthesis are different (Green and Noller 1997). The small ribosomal subunit mediates the interactions between mRNA codons and tRNA anticodons that determine the order in which amino acids are assembled into protein. The large ribosomal subunit contains the peptidyl transferase center, the site where peptide bond formation occurs. In isolation, both subunits can perform functions closely related to those they play in the intact 70S ribosome. By itself, the small subunit binds mRNA, and when mRNA is bound, it will bind tRNAs in a codon-specific manner. By itself, the large subunit will catalyze the formation of peptide bonds between aminoacyl tRNA-like substrates.
Both subunits have three tRNA binding sites: an A site that binds aminoacyl-tRNAs, a P site that interacts with peptidyl-tRNAs, and an E site through which deacylated tRNAs pass as they are discharged from the ribosome. The tRNA sites in the large subunit interact primarily with the CCA terminus of tRNAs, but also with their acceptor stems. The anticodon stems of tRNAs interact with the A, P, and E sites of the small ribosomal subunit. In a functioning ribosome, mRNA also binds to the A and P sites of the small subunit, and the interaction between mRNA codons and tRNA anticodons in the A site ensure that the correct aminoacyl tRNA is selected from the cytoplasmic pool at each step in chain elongation.
The two aspects of the mechanism of ribosome function of primary interest here, peptide bond formation and mRNA decoding, are critical for the elongation phase of protein synthesis, i.e., the stage in protein synthesis during which nascent polypeptides are elongated amino-terminal to carboxy-terminal, one amino acid at a time. Elongation is promoted by two protein factors. One facilitates the delivery of aminoacyl-tRNAs to the ribosome, and the other promotes translocation, the complex process that ultimately enables the ribosome to read the next codon of the mRNA bound to it. In prokaryotes, the protein factor responsible for tRNA delivery is called EF-Tu, and the protein that promotes translocation is called EF-G. It is important to note that ribosomes can catalyze mRNA-dependent polypeptide synthesis without the assistance of factors, but factor-free translation is slow, and inefficient (Pestka 1974; Gavrilova et al. 1976; Southworth et al. 2002).
The elongation cycle starts with a ribosome that has a mRNA bound to its small subunit, a peptidyl tRNA bound to the P site of both the large and small ribosomal subunits that makes the expected codon–anticodon interaction with the mRNA triplet exposed in the P site, an A site that has the mRNA codon 3' to the one in the P site bound to it, but no tRNA, and an E site that contains a deacylated tRNA. Ribosomes in this state interact preferentially with aminoacyl tRNA ternary complexes, which is to say aminoacyl tRNA·EF-Tu·GTP complexes. Base pairing interactions between the A-site mRNA codon and ternary complex anticodons determine which ternary complex will be selected from the mixture present in the cell. When a cognate ternary complex has been selected, the anticodon stem of its tRNA occupies the A-site of the small subunit, but not the A site of the large subunit. Hydrolysis of the GTP in the ternary complex ensues, EF-Tu·GDP is released from the ribosome, and the bound aminoacyl tRNA reorients so that its aminoacyl-CCA end can enter the large subunit's A site. This reorientation is called “accommodation.” (At about this stage, the deacylated tRNA in the E site leaves the ribosome.) The peptidyl transferase reaction ensues quickly thereafter; the nascent peptide that was esterified to the 3' terminal ribose of the tRNA in the P site is transferred to the amino group of the aminoacyl tRNA bound to the A site, which elongates it by one amino acid. The tRNA in the P site is left deacylated. In the final step of the cycle, EF-G·GTP binds to the same site on the ribosome where EF-Tu binds, and facilitates translocation. The deacylated tRNA in the P site moves to the E site, the peptidyl tRNA in the A site moves to the P site, and the ribosome advances along its mRNA by 1 codon. Hydrolysis of the GTP bound to EF-G follows, and EF-G·GDP leaves the ribosome, returning it to its initial state.
3. RIBOSOME CRYSTAL STRUCTURES
No single experimental technique can ever provide all the information needed to understand the mechanism of a complex biological process, but history shows that mechanisms cannot be understood unless the structures of the participating macromolecules are known at atomic resolution. Fortunately, in 2000, the ribosome community began to obtain the high-resolution structural information about the ribosome it had needed for so long, and not surprisingly, the field has been utterly transformed by it.
A monograph could be written on the history of ribosome crystallography, and it would be a good read because the scientific hurdles that had to be overcome were substantial, and the personalities involved were/are colorful. In the end, once a strategy had been devised for phasing ribosome diffraction patterns (Ban et al. 1998), a burst of atomic resolution structures appeared. A 2.4 Å resolution structure of the large ribosomal subunit from Hma (Ban et al. 2000) appeared in August, 2000. A month later, a 3.05 Å resolution structure appeared of the 30S subunit from Tth (Wimberly et al. 2000), and it followed by only a week or two the publication of an independently determined, but less accurate, and lower resolution (3.4 Å) version of the same structure (Schluenzen et al. 2000), the faults of which were later corrected (Pioletti et al. 2001). The big surprise for the cognoscenti was the 3.0 Å resolution crystal structure of the Deinococcus radiodurans large ribosomal subunit, which appeared at the end of 2001 (Harms et al. 2001). The paper describing that structure was also the paper in which crystallization of those ribosomes was first announced. Implicit in this “coincidence” was the message that the technical problems that for so long plagued ribosome crystallography had been overcome.
Although the structures of the large and small subunits mentioned earlier were extraordinarily illuminating, they left unanswered a host of questions related to the way subunits interact in the 70S ribosome, but the crystal structures needed to address those issues began appearing not long thereafter. The first atomic model of the structure of 70S ribosome published was derived from a 5.5 Å resolution electron density map of the ribosome from T. thermophilus, which was interpreted using the structures of the two subunits that had already been determined at higher resolution (Yusupov et al. 2001). The principle drawback of such models is that they are least informative where information is most needed, namely in those regions where structures used to guide interpretation do not explain electron density. Fortunately, crystal structures of 70S ribosomes are now available that have resolutions high enough to eliminate such ambiguities. In 2005 Schuwirth et al. produced a 3.5 Å resolution structure for the 70S ribosome from Escherichia coli. A year later a 2.8 Å resolution structure was obtained for a new crystal form of the T. thermophilus 70S ribosome (Selmer et al. 2006), and shortly thereafter a higher resolution (3.7 Å) version of the original Yusupov structure appeared (Korostelev et al. 2006). These crystal structures have been the “parent structures” for scores of additional structures that show ribosomes and ribosomal subunits bound to substrates, substrate analogs, proteins factors, and antibiotic inhibitors.
4. THE PTC IS MADE OF RNA
The first atomic resolution structures of the large ribosomal subunit and its substrate complexes proved that rRNA is responsible for peptidyl transferase activity of the ribosome (Nissen et al. 2000). The evidence is straightforward. It has long been known that large subunits catalyze the formation of peptide bonds in the absence of mRNA, protein factors, or small ribosomal subunits when provided with low molecular weight analogs of the CCA-ends of peptidyl tRNA and aminoacyl tRNA (Monro et al. 1969). For that reason, structures were obtained of the complexes the Hma large subunit forms both with an A-site analog, and with a putative transition state analog (Nissen et al. 2000). They revealed that peptidyl tRNA mimics bind to the large subunit with their C74 and C75 analogs base-paired with two G residues belonging to the P-loop of 23S rRNA, and that aminoacyl tRNA mimics bind with their C75 analogs base-paired with a G residue belonging to the A-loop of 23S rRNA, as earlier biochemical and genetic experiments had indicated they should (Samaha et al. 1995; Kim and Green 1999). Thus there could be no doubt that these analog structures identified the site where peptide bond formation occurs in the ribosome, i.e., the peptidyl transferase center (PTC). Inspection of the surrounding region showed that there is no protein in the vicinity, and hence that protein cannot be directly involved in peptide bond formation. Although it was subsequently discovered that there is some poorly ordered protein in the vicinity of the PTC that had not been taken into account in these first large subunit crystal structures (see Klein et al. 2004), the conclusions still stands.
5. WHAT DOES THE RNA IN THE PTC DO TO PROMOTE PEPTIDE BOND FORMATION?
Enzymes can facilitate chemical reactions in three general ways: (1) substrate orientation, (2) specific chemical catalysis, and (3) transition state stabilization. Substrate orientation is a major contributor to the catalytic power of most enzymes. By binding substrates with their orbitals oriented properly for reaction, enzymes reduce the entropic barrier to reaction. Page and Jencks (1971) concluded that the rate enhancement caused by orientation can exceed 107-fold. As expected, the PTC avails itself of this important source of catalytic power by positioning the α-amino group of the aminoacyl moiety of the aminoacyl tRNA bound to the A site so that it is close to and pointed toward the carbonyl carbon of the ester that links the peptidyl moiety to the CCA portion of the pepetidyl tRNA bound in the P site. Thus, substrate orientation makes a major contribution to the ribosome's catalytic power, as pointed out by Nissen et al. (2000). Indeed, it was proposed some two decades ago that substrate orientation alone might accelerate the rate of peptide bond formation sufficiently to account for the catalytic activity of the ribosome (Nierhaus et al. 1980), and similar arguments have been advanced more recently (Sievers et al. 2004). Nevertheless, whether and to what extent the ribosome uses additional mechanisms to enhance the rate of peptide bond formation was still an open question. Does it use RNA, with or without the assistance of metal ions, to assist catalysis chemically, and does the ribosome stabilize the oxyanion intermediate in the synthesis reaction as proteases do when catalyzing the peptide hydrolysis reaction? These questions have now been largely answered.
Initial insights into the substrate complexes that form in the PTC were obtained from the structures of separate complexes with A-site analogs, e.g., C puromycin (Cpmn), and the P-site substrates analog, CCA-phe-caproic acid-biotin (CCApcb) plus sparsomycin (Hansen et al. 2002). An approximation to the structure of the large subunit containing both substrates was achieved initially by superimposing the structures of these separately determined A-site and P-site substrate complexes. In this hypothetical, two-substrate complex the α-amino group of the A-site amino acid is adjacent to the ester linked carbonyl carbon of the peptidyl-tRNA it is to attack. The orientations of the two single-stranded CCA sequences bound in these two sites are related by a two-fold rotation axis despite the fact that the tRNA molecules to which they are attached are related to each other by a translation. The proposal that this difference in the orientations of the 3′ termini of the two tRNA molecules may facilitate their translocation after peptide bond formation is as yet untested (Nissen et al. 2000). In any case the relative orientation of the aminoacyl and peptidyl CCAs correctly positions the attacking αNH2 and the ester linked carbonyl carbon, more or less.
6. COMPLEXES WITH BOTH A- AND P-SITE SUBSTRATES BOUND
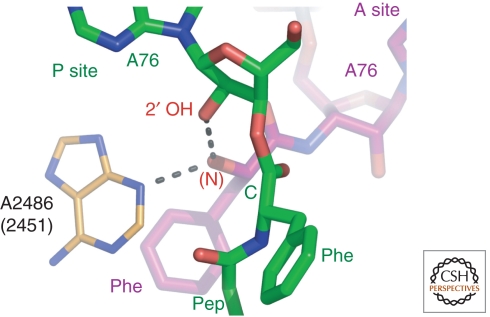
The structures obtained of Hma large subunit complexes with analogs of A- and P-site substrates bound simultaneously (Fig. 2) both limited the number of ways RNA might promote peptide bond formation, and showed that premature peptidyl-tRNA hydrolysis is suppressed by an induced-fit mechanism. Two A-site substrates were prepared that differed in whether or not a C74 mimic was included: CC-hydroxypuromycin (CChPmn) or CPmn. Both were studied in combination with a P-site substrate, CCApcb. The structures of these complexes confirmed that only the N3 of A2486 (2451 E. coli) and the 2′OH of A76 of the P-site substrate contact the attacking α-amino group of the aminoacyl-tRNA, as had earlier been concluded (Hansen et al. 2002), and thus only they could possibly play a direct, chemical role in catalysis (Fig. 2).
Figure 2.
Preattack conformation of the substrates of the peptidyl transferase reaction. The hydroxyl group representing the α-amino group of the A-site substrate, CChPmn (purple) is in position to attack the ester group of the P-site substrate CCApcb (green). It is within hydrogen bonding distance of the N3 of A2486 (2451), and the 2′ hydroxyl group of the P-site substrate. In the ground state, the two reactive groups are 3.7 Å apart.
The possibility that the N3 of A2486 (2451) might serve as a general base to activate the attacking αNH2, despite its normally low pKa (Nissen et al. 2000), was ruled out by the genetic and biochemical experiments reported by Green and coworkers, who showed that mutation of A2486 to any other base has no effect on the rate of peptide bond formation when full tRNA substrates are used with the 70s ribosome; these mutations do, however, inhibit the termination step, accounting for their dominant lethal phenotype (Youngman et al. 2004).
In contrast, the 2′OH of the P-site A76 remains an interesting candidate for catalytic involvement as both a general acid and general base; numerous biochemical experiments employing a deoxA76 in the P-site substrate have suggested that the 2′OH of A76 plays an important role in the PTC. Barta and colleagues proposed a mechanism in which the 2′OH serves simultaneously as a general base to activate the nucleophilic attack of the α-amino group and as a general acid to facilitate the leaving of the 3′ linked peptide ester (Dorner et al. 2003). Their proposal was based on experiments which showed that when acetylated aminoacyl deoxA is used as a P-site substrate and Pmn employed as an A-site substrate, the rate of peptide bond formation is reduced at least 100-fold, and on the structure-based model of Hansen et al. (2002), which suggested an interaction between this 2′OH and the α-NH2 group. A more quantitative assessment of the magnitude of the effect of a deoxy-A76 on the rate of peptide bond formation was obtained using full-length tRNA substrates by Strobel and coworkers who found that the rate of peptide bond formation for peptidyl-tRNAs containing a 2′ deoxy A76 is at least 106-fold slower than normal (Weinger et al. 2004). This rate reduction also occurs if a 2′ fluoro analog rather than a 2′ deoxy analog is used. However, it is not obvious that this entire rate enhancement should be credited to the 2′OH group. A 100-fold rate enhancement would result even if the reaction were uncatalyzed simply because of the vicinal effect of the 2′OH. Also, in the substrate complex, water is sterically excluded from donating protons to the 3′ ester-linked hydroxyl (Schmeing et al. 2005a).
7. THE TRANSITION STATE INTERMEDIATE AND ITS POSSIBLE STABILIZATION
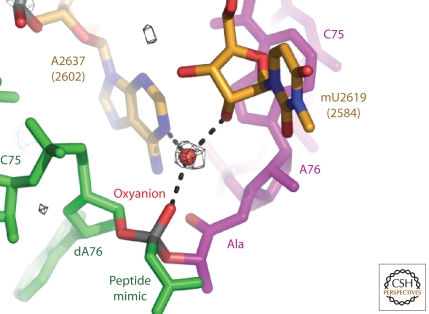
Structures have been obtained of several of the complexes the Hma large subunit forms with analogs of the expected intermediate in the peptide synthesis reaction in which either a phosphodiester or triester is used to mimic the tetrahedral carbon transition state (Fig. 3). The first such transition state analog (TSA) used to examine the structure of a complex was CCA-phosphate puromycin (Nissen et al. 2000). A 3.2 Å resolution map was interpreted as indicating that one of its nonbridging phosphoryl oxygens was interacting with the N3 of A2486 (2451) and represented the oxyanion mimic of the tetrahedral carbon. However, subsequent modeling of the tetrahedral intermediate from the structures of the A- and P-site substrate complexes implied that the oxyanion points away from A2486, and the pH dependence of the TSA binding was not consistent with the proposed interaction with A2486 (Parnell et al. 2002). The structures of numerous additional analogs of the transition state intermediate, which were determined at resolutions between 2.4 Å and 2.8 Å, clearly showed that the oxyanion of the intermediate points away from A2486 (2451), and interacts with a water molecule that is positioned by its interactions with several adjacent nucleotides. The polarity of this water molecule may increase the rate of peptide bond formation by stabilizing the oxyanion, but the magnitude of its contribution is difficult to estimate.
Figure 5.
Proteins in the vicinity of the E site of the large ribosomal subunit. Oligonucleotide analogs of the acceptor stem of a tRNA (orange) will bind to the E site of the large ribosomal subunit from H. marismortui. The proteins in the immediate neighborhood, L44e (green) and L15e (blue), have no homologs in the eubacterial ribosome. When the structure of the large ribosomal subunit from D. radiodurans (Harms et al. 2001) is superimposed on that of H. marismortui, it is found that L33 (red) and L31 (yellow) occupy suggestively similar positions. (Reproduced with permission from Schmeing et al. (2003).)
Figure 3.
Structure of an analog of a transition site intermediate in the peptidyl transferase reaction showing the oxyanion bound to a water molecule and pointing away from A2486.
8. METAL IONS IN THE PTC?
The observation that the 2′OH of the A76 of the P-site bound tRNA enhances the rate of peptidyl-transferase activity by 106-fold or more raises the question of why it is so effective. Difference electron density maps of TSA complexes showed density peaks near this 2′OH, opening the possibility that there might be a Mg++ or Na+ ion present, even though the peak was 2.8 Å from the A76 2′OH. The possibility that a Mg++ ion might be binding to this 2′OH, or anywhere else at the site of reaction, was eliminated by examining crystals of TSA complexes with the large subunit in which the Mg++ in the crystals was exchanged for Mn++, because no peaks appeared in this region in anomalous difference Fourier maps. Likewise, the possibility that there might be a monovalent metal ion binding in the vicinity was eliminated by examining crystals containing 1.5-M potassium. There is no structural or biochemical evidence that metal ions play a direct role in peptide bond formation.
The analysis of the mechanism of peptide bond formation presented here, which derives from the structures of complexes of the large ribosomal subunit from Hma with substrate analogs, as well as numerous biochemical experiments, has not gone unchallenged. For example, Korostelev et al. (2006) concluded from their 3.7 Å resolution structure of the Tth 70S ribosome with tRNAs bound in both the P- and E-sites that the structure of the PTC in isolated 50S subunits is not the same as it is in 70S ribosomes. However, subsequent recalculations of the Korostelev electron density map (Simonovic and Steitz 2008), and some recently reported crystal structures of 70S ribosomes with tRNAs bound in both the A- and P-sites (Voorhees et al. 2009; Blaha et al. 2009) indicate that this is not the case. These new structures support the conclusions about the mechanism of peptide bond formation that were reached using Hma structures. Nevertheless, anyone concerned that peace is about to break out should consult (Gindulyte et al. 2006) and (Korostelev et al. 2009).
9. PROTECTION OF PEPTIDYL-tRNA FROM HYDROLYSIS AND INDUCED-FIT ACTIVATION OF PEPTIDE SYNTHESIS
Another long-standing question that can now be answered is how the ribosome prevents the premature hydrolysis of peptidyl-tRNAs. If the PTC can activate α-NH2 groups for reaction, why does it not activate water molecules when the A-site is empty? The reason is that the ester-linked carbonyl carbon to be attacked is sterically protected by the PTC until the CCA of an aminoacyl-tRNA binds to the A-site and induces a (modest) conformational change in the PTC rRNA, which repositions the carbonyl group so that it can be attacked by an appropriately positioned α-amino group (Schmeing et al. 2005b). In short, this property of the PTC depends on induced fit, a mechanism responsible for enhancing the substrate specificity of many protein enzymes, that was first advanced by Koshland (1959) to explain why hexokinase does not hydrolyze ATP in the absence of glucose, and then proven to be correct by Bennett and Steitz (1976).
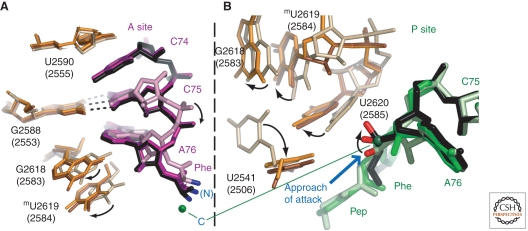
The structural evidence for an induced fit conformational change in the large ribosomal subunit derives from structures of the Hma 50S subunit complexed with CCApcb in the P-site, and simultaneously with either CChPmn or ChPmn in the A-site (Schmeing et al. 2005b). When ChPmn is in the A-site the ester group of CCApcb is both positioned and sequestered by the bases of U2620 (2585) on one side and A2486 (2451) and C2104 (2063) on the other so that the carbonyl oxygen of its ester group points toward the α-amino group of the A-site substrate, and in this orientation hydrolysis is prevented because water molecules cannot access its carbonyl carbon. However, when CChPmn binds to the A-site, it induces a series of conformational changes in the bases forming the PTC; the base of U2620 (2582) moves away from the ester group of the peptidyl-tRNA mimic, allowing its carbonyl group to reorient in a manner that is favorable for nucleophilic attack by the nearby α-hydroxl or α-amino group (Fig. 4). These conformational changes also explain why the rate of hydrolysis of peptidyl tRNAs is 100 times faster in the presence of CCA than it is in the presence of CA (Caskey et al. 1971).
Figure 4.
Movements of rRNA and peptidyl-tRNA are induced by proper binding of the A-site substrate. (A) A view of the A-site of the three complexes. Without C74, ChPmn (pink, with rRNA colored wheat) is positioned higher in the A-site. In substrates with C74, both CChPmn (purple, with orange rRNA) and the TSA (black, with brown rRNA), C74 stacks with rRNA base U2590 (2555), shifting the substrates down, and the α-amino group closer to the ester carbon of the P-site substrate (green). rRNA base G2618 (2583) shifts to maintain the A-minor interaction, causing methylU2619 (2584) to also move. (B) A view of the P-site of the same three complexes. The movement induced in G2618 (2583) by A-site substrate binding breaks its G-U wobble pair with U2451 (2506), which swings 90o. MethylU2619 (2584) and U2620 (2585) also shift to allow the ester group to move from the position it occupies when CCApcb (light green) is bound together with ChPmn, to that when CCApcb (medium green) is bound with CChPmn, and finally to that when it has been attacked by the A-site substrate, as shown with the TSA (black).
10. RELEASE FACTORS ARE tRNA MIMICS
The synthesis of nascent polypeptide chains ends when translocation brings the stop codon of a mRNA into the A site of the ribosome, and the reason synthesis ceases at that junction is that cells do not (normally) contain tRNAs that are cognate to stop codons. Based on what has been said so far one might think that ribosomal complexes of this sort would be very stable because in the absence of A-site bound tRNAs, the conformation adopted by the PTC protects the ester bonds that link nascent polypeptides to P-site bound tRNAs from hydrolysis. However, in vivo, polypeptides are rapidly released from these complexes because of the action of proteins called release factors, of which there are two in bacteria, RF1 and RF2. RF1 is specific for 70S ribosomes carrying nascent polypeptides that have stopped synthesis at UAG or UAA codons, and RF2 is specific for UGA and UAA codons. Both recognize stop codons when they are exposed in the decoding centers of ribosomes, and make the PTC catalyze a reaction it otherwise promotes very inefficiently, namely the hydrolysis of the ester bonds that link peptides to tRNAs bound in the P site.
Atomic resolution structures have recently been obtained of 70S ribosome/tRNA complexes with RF1 bound (Lauerberg et al. 2008), or RF2 bound (Korostelev et al. 2008; Weixlbaumer et al. 2008). These structures show that RFs bind to the ribosome in much the same way that aminoacyl tRNAs bind before peptide bond formation. Specific interactions between the ends of RFs that bind to the A site of the small subunit decoding center and stop codons stabilize RFs in a conformation which ensures that their far ends will insert into PTC, where they occupy the same space as the CCA end of a A-site bound tRNA. The conformation of the PTC changes in response to RF binding in much the same way it changes when aminoacyl tRNAs bind to the A site, rendering the ester bond in the P-site of the PTC accessible to water. The GGQ sequence found in all RFs is positioned so that it can assist with the attack of a water molecule on that ester bond.
11. BINDING OF THE CCA END OF tRNA TO E SITE OF THE LARGE SUBUNIT
The exit site, or E-site, binds deacylated tRNAs as they transit from the P-site, after peptide bond formation, back to solution. Although proteins are found in the present day E-site of archaea (L44e and L15e) and eubacteria (L33), it appears that prior to the divergence of archaea and eubacteria, the binding site for CCA in the E-site on the large subunit was entirely rRNA. Not only are the sequence-specific interactions that ensure that only deacylated tRNAs bind to the E-site mediated by RNA, but the proteins that contact the CCA in eubacteria and archebacteria are not homologous, implying that they were added to the ribosome subsequent to the divergence of the two kingdoms (Schmeing et al. 2003) (Fig. 5). Furthermore, the conformation of the CCA end of a tRNA bound to the E site of an archaeal ribosome is not the same as it is when it is bound to the E site of a eubacterial ribosome (Schmeing et al. 2003; Selmer et al. 2006). Only A76 binds the same way to the E site of both types of ribosome, where it interacts with universally conserved bases.
12. INHIBITION OF THE PEPTIDYL TRANSFERASE REACTION BY ANTIBIOTICS
Microorganisms synthesize a wide variety of secondary metabolites that seem to function at low concentrations as cell-signaling molecules (Yim et al. 2007). At higher concentrations, some of these compounds are antibiotics because they inhibit ribosome function. The species specificities of these ribosome inhibitors vary a lot, but some are sufficiently specific for eubacterial ribosomes to be useful for treating human diseases.
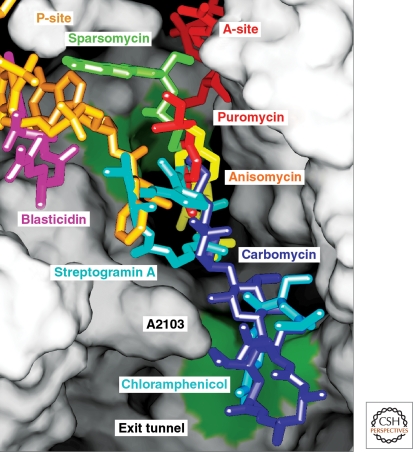
Crystal structures have been obtained of dozens of the complexes antibiotics and other inhibitors of protein synthesis form with the large ribosomal subunit (e.g., Fig. 6). So far, almost all of the anti-ribosomal antibiotics studied crystallographically bind to sites that are composed entirely of RNA, and most of those that target the large ribosomal subunit inhibit its activity in two different ways (Schleunzen et al. 2001; Hansen et al. 2002; Hansen et al. 2003). One class, the macrolides (e.g., erythromycin, tylosin, and azythromycin), bind to a site in the proximal part of the polypeptide exit tunnel adjacent to the peptidyl-transferase center, and although some macrolides also block PTC activity sterically, they all inhibit protein synthesis by interfering with the passage of nascent polypeptide down the exit tunnel. In the presence of macrolides ribosomes tend to synthesize short peptides rather than complete proteins (Tenson et al. 2003). The hypothesis that macrolides occlude the tunnel like the stopper in a bottle, which explains a lot of the phenomenology of macrolides, may be an oversimplification (Tu et al. 2005; Voss et al. 2006), and there is abundant evidence that the efficiency with which nascent polypeptides pass through the exit tunnel, which is the property of the protein synthesis system that macrolides alter, is affected by amino acid sequence (Nakatogawa and Ito 2004). In any case, all the macrolides studied so far bind to the H. marismortui 50S subunit (Hansen et al. 2003) in the same general location that macrolides bind to the large ribosomal subunit from the eubacterium D. radiodurans (Schleunzen et al. 2001).
Figure 6.
The positions of seven antibiotics and A-site (red) plus P-site (yellow) substrates bound to the peptidyl transferase center. The ribosome has been split open to reveal the lumen of the exit tunnel and adjacent regions of the peptidyl transferase site. Ribosomal components are depicted as a continuous surface that is colored green at two positions where splayed out bases provide hydrophobic binding sites for small molecules. Seven independently determined cocrystal structures have been aligned by superimposing the 23S rRNA in each complex.
The second set of antibiotics we will discuss is much more diverse chemically. Its members bind to either the A-site or the P-site of the PTC, and appear to obstruct the binding of either the A-site or P-site tRNA to the PTC, or both, consistent with their being competitive inhibitors of peptide bond formation (Fig. 6). For example, anisomycin bound to Hma large subunit (Hansen et al. 2003) and chloramphenicol bound to D. radiodurans 50S subunit (Schleunzen et al. 2001) both occupy a hydrophobic crevice formed by two splayed-out bases that also provides the binding site for the tyrosine side chain of an A-site bound substrate analog. Linezolid, a completely synthetic protein synthesis inhibitor, binds to ribosomes the same way (Ippolito et al. 2008). (At mM concentrations chloramphenicol binds to a second hydrophobic crevice in Hma subunits where macrolide antibiotics also bind.) Virginiamycin M occupies portions of both the A-site and P-site whereas blastocydin S exploits another strategy by mimicking C74 and C75 of the P-site bound tRNA and base-pairing with the P-loop.
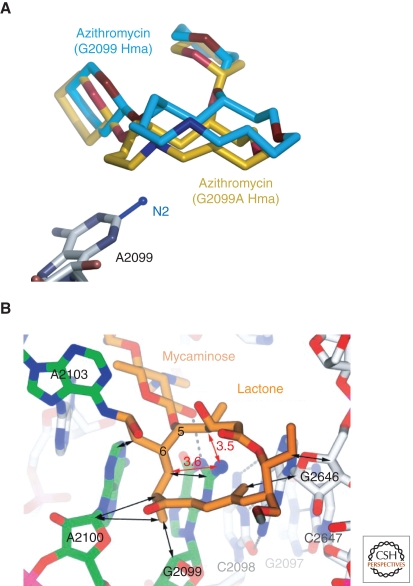
Recent structural studies of mutant Hma large subunit have provided insights into the structural basis of the resistance that mutations confer to the macrolides, and to anisomycin (Blaha et al. 2008; Gurel et al. 2009). It has been known for years that the mutation A2058G (E. coli) in 23S rRNA makes eubacterial ribosomes highly resistant to macrolides. The nucleotide at that location in the Hma ribosome is a G, and as expected, erythromycin does not bind to crystals of the Hma large subunit, even at a concentration of 3 mM, and other macrolides, e.g., azithromycin, bind only weakly. However, when that G is mutated to an A, the affinity of the Hma ribosome for erythromycin increases by >104-fold, and it binds at <1 µM concentrations (Tu et al. 2005). Azythromycin also binds more tightly to the mutant, but in much the same orientation that it binds to wild-type ribosomes (Fig. 7A). The reduction in binding constant caused by the A2058G mutation in eubacterial ribosomes reflects the energetic cost of desolvating and burying the N2 of that G, and the less snug packing of the macrolide ring on the tunnel wall its presence requires (Fig. 7B). Dimethylation of the N6 of A2058 would also sterically interfere with macrolide binding. Erythromycin, telithromycin and azythromycin bind identically to the G2099A mutant subunit, and very similarly to the way 16-membered macrolides bind to wild-type 50S subunit. (Initial reports indicated that the orientation of the macrolide ring of erythromycin bound to the Hma 50S subunit is orthogonal to that observed when it binds to the large subunit from Dra (Schleunzen et al. 2001), but subsequent studies performed with Dra have indicated that this not the case (Wilson et al. 2005).)
Figure 7.
Macrolide interactions with the large ribosomal subunit of H. marismortui. (A) Comparison of the location and conformation of azithromycin bound to wild-type H. marismortui large ribosomal subunits, and G2099A mutant large ribosomal subunits from the same organism. Structures of both forms of the subunit with the drug bound have been superimposed. The drug bound to G2099A subunits is shown in brown, and the drug bound to wild-type is in cyan. The location that would be occupied by the N2 of a G residue at position 2099 is modeled in. (Reproduced with permission from Tu et al. 2005 [© Elsevier].) (B) Intrusion of the N2 of G2099 into the lactone ring of macrolide antibiotics. The structure of the 16-membered macrolide tylosin is shown bound to wild-type H. marismortui large ribosomal subunits. The N2 of G2099 lies under the hydrophobic surface of the lactone ring of the drug requiring it to move away from the base where it would otherwise make a number of close contacts that would be unfavorable for binding. In addition, when the drug is bound, the N2 of G2099 is desolvated and unable to make any hydrogen bonds. (Reproduced with permission from Hansen et al. 2002 [© Elsevier].)
Anisomycin is one of three low molecular weight antibiotics that inhibit the large ribosomal subunit by binding to its A-site cleft. These compounds are interesting because even though their binding site, i.e., the A-site cleft and its immediate surround, is highly conserved and their chemical structures quite similar, they are species-specific. Two members of this group, chloramphenicol and linezolid, are specific for eubacteria, whereas the third, anisomycin, is much more toxic to eukaryotes and archaea than it is to eubacteria. What is the structural basis for this difference?
No single mutation makes Hma highly resistant to anisomycin the way A2058G (E. coli) makes eubacterial highly resistant to macrolides. Nevertheless, when the structures of the Hma large ribosomal subunits containing mutations that confer comparatively weak resistance to anisomycin are considered as a group, an interesting picture of the conformational properties of the PTC emerges that may do more than explain the phenotypes of those mutations.
It appears likely that there are several different conformations possible for the PTC region of the ribosome that differ modestly in RNA backbone trajectory, but more conspicuously in the orientations of their nucleotides bases. These conformational states are in equilibrium with each other, and may vary in free energy by as much as 3 or 4 kcal/mol at 37oC. Any given substrates, substrate analog, or inhibitor of the PTC binds only when it is in a compatible conformational state, and thus that conformation will dominate when the ligand in question is present. For example, when anisomycin is bound to the Hma ribosome the conformation of its PTC is not the same as it is in the absence of anisomycin (Blaha et al. 2008). The conformational change that explains why the PTC catalyzes nucleophilic attacks on the peptidyl tRNA only when proper A-site substrates are present is undoubtedly a manifestation of this same phenomenon (Schmeing et al. 2005b).
The structural data support the hypothesis that mutations in Hma that cause resistance to anisomycin either stabilize the apo- conformation of the PTC or destabilize its anisomycin-bound conformation (Blaha et al. 2008); thermodynamically it does not matter which happens. Because the conformation of the center of the PTC, which includes the A-site cleft, is stabilized by its interactions with more peripheral nucleotides, mutations in nucleotides that do not contact anisomycin when it is bound to the ribosome can alter its binding constant, as observed. They also determine which conformations of the PTC will be favored in the ribosomes from any given species in the absence of PTC ligands. It follows that the conformation of the PTC in apo- structures of the ribosome should vary somewhat between species, and they do.
Comparison of the structures of large ribosomal subunits from several species, with and without A-site cleft antibiotics of several kinds bound strongly suggests that the position of a single, highly conserved nucleotide that neighbors the A-site cleft (U2504 in E. coli) controls the response of the ribosome to A-site cleft antibiotics (Gurel et al. 2009). In eubacteria its placement restricts the volume antibiotics can occupy on one side of the A-site cleft. In Hma, and one would surmise eukaryotes as well, its placement is more permissive allowing larger molecules can bind to the cleft. Because the position of this nucleotide is determined by its interactions with nucleotides that are even further removed from the center of the PTC and the identities of which vary from one group of organisms to the next, its conformation provides a satisfying explanation for species specificities of drugs of this sort.
Mutations in ribosomal proteins can also cause antibiotic resistance. A deletion of three residues in a long, antiparallel protein loop of L22 causes resistance to erythromycin, but does not prevent the drug from binding. The structure of Hma large subunit containing such a deletion mutation in protein L22 shows that the protein loop that lies on the wall of the wild-type ribosomal tunnel has moved from that position, making the tunnel wider by several Å just below the macrolide binding site (Tu et al. 2005). The resistance mechanism is less clear in this case, but may be related to the tunnel widening. Weak resistance to anisomycin also results from mutations in ribosomal protein L3 (Gurel, G., unpublished data).
13. TRANSLATIONAL FIDELITY
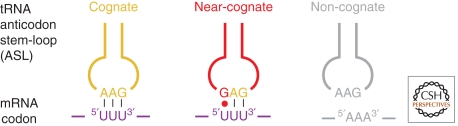
No aspect of protein synthesis has been more intensively studied than the fidelity with which mRNA sequences are translated into protein sequences. Fidelity is interesting because it cannot be explained by the energetics of the base pairing interactions on which it depends. The free energy of forming a three-base pair helix between an mRNA codon and the anticodon of an aminoacyl tRNA that are perfectly complementary, i.e., that are cognate, is in many instances only slightly more favorable than the free energy of the pairing of the same codon with tRNA codons that are not complementary at a single position, i.e., that are near cognate (Fig. 8). Thus if tRNA-mRNA interactions were all that counted, the error rate of protein synthesis might exceed one wrong amino acid for every 100 amino acids incorporated into protein. In fact, the error rate is about one in 1000–100,000 (see Gallant and Foley 1980). How is this level of accuracy achieved?
Figure 8.
Codon–anticodon interactions. Left panel: a cognate codon–anticodon interaction. These is strict Watson-Crick base pairing between the first two bases in the mRNA codon and the corresponding anticodon bases, but wobble pairing is acceptable in the third position. Middle panel: a near-cognate codon–anticodon interaction. The distinction between this interaction and a cognate interaction is that a wobble pairing occurs in the first or second position. Right panel: a noncognate interaction. In a noncognate interaction no Watson-Crick or wobble pairs form between the codon and the anticodon.
Two kinds of models have been advanced to explain the high fidelity of protein synthesis. Some have suggested that the ribosome might interact with the mRNA-tRNA complexes that form on its surface in such a way as to favor cognate complexes over near-cognate complexes energetically by enough to increase the fidelity of translation by 100–1000-fold. Others have proposed that the accuracy of translation is explained by proofreading. In order for the translation system to proofread, its duty cycle would have to be organized so that the free energy difference between cognate mRNA-tRNA pairings and near-cognate mRNA-tRNA pairings could be exploited (at least) twice every time the ribosome selects an aminoacyl tRNA from the cytoplasmic pool.
An historical comment is in order here. At the time proofreading mechanisms were first proposed for the translation system, it was believed that the free energy difference between cognate and near cognate pairings could explain a fidelity of 1 in 100, and thus if one iteration yielded a fidelity of 1 in 100, two iterations that were sequential and independent would yield 1 in 10,000, which is about what is needed. As already noted, more recent thermodynamic studies suggest that the free energy “penalty” associated with noncognate pairings is much smaller than first thought, perhaps so small that the error rates predicted from energy differences could be as high as 1 in 10 (!) (Xia et al. 1998). Thus by itself, proofreading would probably not be enough.
Our understanding of translational fidelity has been revolutionized by the small subunit crystal structures that have appeared since 2000. We now know that interactions between the small subunit and codon–anticodon complexes do indeed contribute substantially to the free energy advantage of cognate interactions, but that kinetic proof reading also contributes to fidelity.
14. THE tRNA BINDING SITES IN THE SMALL RIBOSOMAL SUBUNIT
Remarkably, the location of the A-, P-, and E sites on the small subunit, and much of what we now know about how they interact with tRNAs was revealed by a structure of the Tth small subunit obtained from crystals that contained no tRNA (Wimberly et al. 2000). The reason this structure is informative is that the Tth small subunit has a protruding “spur” (helix 6 of 16S rRNA), the conformation of which is essentially the same as that of the anticodon stem-loop of a tRNA. Fortuitously, in the crystals studied, the spur of each subunit is inserted into the P site of a neighbor, where its “anticodon” sequence makes a noncognate interaction with a three-base sequence at the 5′ end of the host subunit's 16S rRNA, which happens to fold back into the P-site so that it can act like a mRNA (Carter et al. 2000). The accuracy with which this crystal packing interaction mimics P-site tRNA binding could be shown by superimposing the small subunit structure in question on a lower resolution structure of the Tth 70S ribosome that did have tRNAs and mRNA bound (Cate et al. 1999). In addition to validating the P-site interactions seen in the tRNA-free small subunit crystals in question, this superposition identified the small subunit's A and E sites in enough detail so that inferences could be drawn about the way tRNAs interact with those sites also.
Compositional arguments like the one that proves that peptide bond formation is RNA-catalyzed cannot be used to show that small subunit function is RNA-driven. Although the 3 tRNA binding sites in the 30S ribosomal subunit all contain 16S rRNA sequences, none of them is protein-free. However, there is a suggestive compositional gradient. Protein is least abundant in the A site, which is arguably the most important site on the small subunit because it is where decoding occurs; only a short segment of protein S12 interacts with tRNAs bound there. By contrast, tRNA-protein interactions dominate in the small subunit's E site, the site that appears the least important of the three in the sense that one could imagine a ribosome that lacks an E site, but not one that lacks an A- or P site (Carter et al. 2000). This compositional gradient is even more pronounced in the large ribosomal subunit; its E site includes protein (Schmeing et al. 2003), but its A- and P-sites are protein-free (Nissen et al. 2000).
Only in the case of the P site have experiments been performed that directly test the functional importance of its protein components (Hoang et al. 2004). The P site of the small subunit includes sequences from the C-terminal tails of proteins S9 and S13, and cells that have S9 and S13 genes that lack their normal C-terminal tail sequences are viable, but grow slowly. Thus the proteins in the P site optimize it, but do not give it functional properties it would otherwise lack. Until similar studies are performed on the A site, we will have to rely on purely structural evidence to tell us what RNA and protein contribute to its function.
15. THE ROLE OF rRNA IN mRNA DECODING
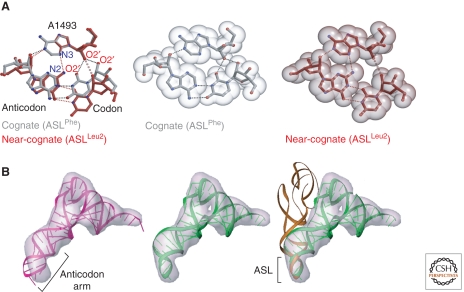
Much of what is now known about the way the A site operates comes from structures of the Tth small subunit with mRNAs and anticodon stem/loops bound in the A site (see Ogle and Ramakrishan, 2005). The first such structures showed that when cognate tRNA-mRNA complexes bind to the A site, three 16S rRNA bases, which genetic and biochemical experiments had earlier showed are vital for decoding (A1492, A1493 and G530), change conformation (Ogle et al. 2001). In the absence of A-site ligands, A1492 and A1493 are buried in the interior of helix 44, the secondary structure element of which they are a part, and G530 is in the syn conformation. When cognate tRNA-mRNA complexes bind to the A site, the three adopt positions that permit them to interact with the minor groove of the three-base-pair helix formed by mRNA codons and tRNA anticodons. A1493 swings out of the interior of helix 44, and makes a type I A-minor interaction (Nissen et al. 2001) with the base pair that includes the first base of the codon. The three hydrogen bonds so created stabilize the interaction between the small subunit and the A-site complex, and the reason this interaction is so significant for fidelity is that any Watson-Crick pair between the first base of a mRNA codon and a tRNA anticodon can interact with A1493 in exactly the same way, but non Watson-Crick base pairs cannot. Thus the A1493 interaction strongly favors cognate pairing over noncognate, or near-cognate pairing in the A site (Fig. 9A).
Figure 9.
A-site tRNA interactions in the small ribosomal subunit. (A) interactions in the first position. Left Panel: The structure formed by a Watson-Crick pair in the first position with the ribosome compared with that formed when the base pair in the first position is a wobble base pair. In both cases A1493 of 16S rRNA inserts into the minor groove side of the base pair. More hydrogen bonds form and the fit is tighter if the pair is Watson-Crick than it does if it is a wobble pair. Middle Panel: A space-filling model of a cognate pair/A1493 interaction. Right Panel: A space-filling model of a wobble pair/A1493 interaction. (Reproduced with permission from Ogle et al. 2002 [© Elsevier].) (B). The conformation of aminoacyl tRNAs bound to the 70S ribosome in the pre-accommodation state. Left panel: Failure of the crystal structure of free tRNA to fit the cryoelectron microscopic electron density for tRNA in images of the 70S ribosome with aminoacyl tFNA together with EF-Tu bound in the pre-accommodation state. The tRNA crystal structure is shown in purple and the electron density in gray. Middle panel: Model obtained for tRNA in the pre-accommodated state by marrying the anticodon stem structure of a tRNA in the accommodated state to the crystal structure of a tRNA fit into the rest of tRNA electron density. The new model is green, and its anticodon stem is bent. Right Panel: Comparison of the orientation of a accommodated tRNA with a pre-accommodated tRNA The accommodated tRNA is brown and the pre-accommodated tRNA is green. (Reproduced with permission from Valle et al. (2003) [© Elsevier].)
The minor groove face of the base pair formed by the second base of a codon and the second base of a tRNA anticodon is also “monitored.” In this instance, G530 changes its conformation from syn to anti, and in that conformation, it interacts both with A1492, which like A1493 has swung out of helix 44, and the middle anticodon base in the helix. In addition to interacting with G530, A1492 makes a type II A-minor interaction with the middle codon base in the same base pair. Again the resulting 4 base complex will accommodate any codon–anticodon base pair that is a Watson-Crick base pair, but no non-Watson-Crick base pairs. Thus this interaction too favors cognate pairing over non-, or near-cognate pairings energetically.
The small subunit interacts with the third base pair in a codon–anticodon helix in a much less specific manner. It does not “measure” the width of the minor groove side of that pair the way it measures the width of the minor groove side of the other two base pairs. Thus not surprisingly sequence differences between codons in the third, or wobble, position are much less critical for coding than sequence differences in the first two, as has long been known. Non-Watson-Crick pairings are often acceptable in that position; GU pairings, for example, are almost as good as GC pairings.
The inferences drawn from the cognate A-site codon–anticodon structures have been tested, and extended using a series of small subunit structures in which the A site contains codon–anticodon complexes that have a non-cognate GU instead of a cognate GC at either the first or at the second codon position (Ogle et al. 2002; Ogle et al. 2003). Interestingly, the codon–anticodon interaction is so poor when there is a GU mismatch in the first position that the anticodon stem loop component of the complexes formed could not be visualized crystallographically unless the antibiotic paromomycin was included in the crystal to stabilize it. Paromomycin binds to helix 44 of 16S rRNA, filling space normally occupied by bases 1492 and 1493, and forcing them to adopt a conformation close to that seen when there is a cognate codon–anticodon pair in the A site. Given this extra “help,” codon–anticodon complexes with GUs in the first position will bind in the A site, but the A minor-like interaction A1493 makes with them is imperfect. Fewer hydrogen bonds form, and the fit of the minor groove side of A1492 into the minor groove side of the GU pair is suboptimal. Worse, some hydrogen bond donors are left entirely without acceptors in these complexes, which imposes a large energetic cost. However, it is clear that the energetic penalty paid when there are first positions mismatches is reduced in the presence of the drug, and it should thus it makes sense that translational fidelity falls when cells are exposed to it.
GU mismatches in the second position are also problematic structurally. The electron density in the vicinity of the middle base pair of the codon–anticodon helix suggests that the small subunit is not able to find an energetically satisfactory way to accommodate GU mismatches. There appear to be two structures present in the crystal, which may be in equilibrium with each other. Sometimes the mRNA base occupies the position it would occupy if it were part of cognate base pair, and A1492 makes the expected type II A-minor interaction with it, but when this is so, the anticodon with which it is supposed to pair is positioned so that it cannot interact with G530 properly, which remains in the syn conformation. At other times, positions are reversed. The anticodon base occupies its cognate position, and interacts normally with an anti G530, but its codon mate is out of location, and does not interact properly with A1492. In either case, the full energetic value of the ribosome interactions that accompany cognate pairing is not realized.
The local conformational changes that occur when the A site of the small subunit interacts with cognate codon–anticodon complexes are accompanied by global changes in the over-all conformation of the small subunit that appear to be critical for fidelity (see the following) (Ogle et al. 2002). The three tRNA binding sites of the small ribosomal subunit are located at the interface between the subunit's head domain and its “shoulder” and “body” domains. When cognate interactions occur in the A site, the head rotates toward the large ribosomal subunit, clamping down on its tRNA binding sites, and the shoulder rotates in toward those sites also. These conformational changes do not occur when paromomycin binds to the subunit, nor when near-cognate codon–anticodon complexes occupy the A site. However, they do occur when near-cognate codon–anticodon complexes form in the presence of the drug. These observations indicate that these conformational changes are functionally significant. It is likely they contribute to the still poorly understood process that causes the rate of GTP hydrolysis by EF-Tu to accelerate when cognate interactions between mRNA and tRNA occur in the A site.
The decoding puzzle has one last piece that we have not yet considered. In all the codon–anticodon/small subunit complex structures examined at high resolution, the anticodon stem-loop in the A site is in its accommodated position, which is to say that when a 70S ribosome has an intact tRNA bound to its 30S A site in that orientation, its CCA end will occupy the A site in the large subunit's peptidyl transferase center. This is not the orientation of aminoacyl tRNAs on the ribosome when they first bind to it. As mentioned earlier, the tRNA-containing entity delivered to the ribosome is a ternary complex containing EF-Tu·GTP as well as aminoacyl tRNA, and while the anticodon loop of a ribosome-bound ternary complex occupies the A site, the rest of the molecule is in an entirely different orientation; their CCA ends are far from the peptidyl transferase center (Valle et al. 2002; Stark et al. 2002; Valle et al. 2003). tRNA reorientation, i.e., accommodation, occurs only after the GTP bound to the ternary complex has hydrolyzed, and EF-Tu · GDP has left the ribosome (Stark et al. 2002). Does the picture just provided for A-site interactions with codon and anticodons apply to the pre-accommodation state, or is the interaction completely different?
Amazingly, recent electron microscopic findings as well as a recent crystal structure suggest that as far as the anticodon stem-loop bound to the small subunit A site is concerned, the pre-accommodation state is the same as the accommodated state. It appears that the anticodon stems of tRNAs bound to ribosomes in the pre-accommodation state are bent so that their anticodon loops will fit into the A site in the orientation seen after accommodation (Valle et al. 2003; Li et al. 2008; Schmeing et al. 2009) (Fig. 9B). Accommodation appears to be driven by a springlike straightening out of the anticodon stem that becomes possible once a pre-accommodated tRNA has been released from its interactions with EF-Tu. In any case, it appears that tRNAs are bound to the ribosome only through the interactions they make with A-site components during accommodation. If that interaction is weak, as it will be if the codon–anticodon interaction is near-cognate, instead of cognate, the likelihood that accommodation will result in the dissociation of the tRNA from the ribosome will be high.
Recent kinetic studies have provided important insights into how the system just described actually works (Gromadski and Rodnina 2004; Blanchard et al. 2004). The binding of ternary complexes to the ribosome is a two-step process, the first of which is nonspecific. After a ternary complex has become engaged with the ribosome, a second, codon recognition step occurs that does depend on interactions. Kinetic data show that if the system were to reach equilibrium at that stage, which it normally does not, cognate interactions would be favored over near-cognate interactions by a factor of 300–400, which is almost enough by itself to account for the fidelity of translation. The high potential discrimination of this step unquestionably reflects the interactions 16S rRNA makes with complexes in the A site that are discussed earlier. However when the system operates normally, it does not come to equilibrium at this stage. The rates of subsequent steps are too fast, and hence it does not discriminate effectively between cognate and near-cognate interactions at this point. In fact, the fidelity of the tRNA selection phase of protein synthesis is determined mainly by the next step in the process, the activation of the GTPase of EF-Tu. The rate of activation is ∼600 times faster if a cognate interaction has occurred in the A site of the 30S subunit than it is if the interaction in the A site is near-cognate. (All the steps that follow activation, including accommodation, are fast compared with cognate activation.) Furthermore, the rate of near-cognate activation is so slow compared with the rate of dissociation of near-cognate complexes from the ribosome that the probability of rejection of near-cognate complexes is very high at this step. Although the system discriminates quite effectively between cognate and near-cognate interactions during tRNA selection, the overall fidelity of the protein synthesizing system is higher than that process allows. It is the accommodation step that provides the additional fidelity required, and it does so because the probability of dissociation of near-cognate complexes during accommodation is higher than that of cognate complexes. Thus the system does proofread. It appears that the cognate/near-cognate discrimination provided by the tRNA selection phase we just discussed is about 60-fold, whereas that provided by the accommodation step is about 15-fold.
16. STRUCTURAL BASIS OF ELONGATION FACTOR FUNCTIONS
Although a detailed discussion of the crystal structures of 70S ribosomes with elongation factors bound is beyond the scope of this review, the factors being, after all “merely” proteins, a brief summary of the major conclusions derived from them seems in order. In the last year crystal structures have appeared of the Tth 70S ribosomes with three elongation factors bound: EF-P, EF-Tu and EF-G (Blaha et al. 2009; Schmeing et al. 2009; Gao et al. 2009). The least well known factor is EF-P, which stimulates the formation of the first peptide bond (Aoki et al. 1997; Glick and Ganoza 1975). The structure of this complex shows that it binds adjacent to a tRNAfmet in the P site, on its E-site side (Blaha et al. 2009). It appears to stabilize that placement of fmet-tRNAfmet, preventing both its back translocation and forward translocation. The structure of aminoacyl-tRNA-EF-Tu bound to the 70S ribosome with tRNAs in the P and E sites extends the resolution of previous cryo-EM studies to atomic resolution. The bend in the RNA of the anticodon stem and the interactions of the ribosome GTPase center with EF-Tu can be seen in this structure (Schmeing et al. 2009). The structure reported for the complex with EF-G is similarly revealing (Gao et al. 2009).
17. CONCLUDING COMMENTS
Many important aspects of ribosome function remain to be explained in molecular terms, but recent progress gives one hope that the crystal structures needed to address them may be obtained before long. Although this review has emphasized crystallographic findings, it is important to remember that information provided by genetics, biochemistry, and enzymology has been just as important to recent progress.
Finally, the long-standing hypothesis that the first ribosome-like object, which probably appeared on this planet around 3.5 billion years ago, was made entirely of RNA seems more likely than ever. Given what we now know about ribosome structure and function, it is easy to imagine an all-RNA particle having similar capabilities. That said, it would be foolish to assume that the biological world in which that first ribosome appeared was one in which RNA was the only polymer of consequence. Why would an RNA structure evolve that makes polypeptides if polypeptides did not already exist that would confer a selective advantage on the proto-organisms capable of synthesizing them?
ACKNOWLEDGMENTS
We thank Joachim Frank, Venki Ramakrishnan, Jeffrey Hansen, and Martin Schmeing for providing figures. The work was supported by a grant from the National Institutes of Health (GM022778) to P.B.M. and T.A.S.
Footnotes
Editors: John F. Atkins, Raymond F. Gesteland, and Thomas R. Cech
Additional Perspectives on RNA Worlds available at www.cshperspectives.org
REFERENCES
- Aoki H, Dekany K, Adams SL, Ganoza MC 1997. The gene encoding elongation factor protein P is essential for viability and it required for protein synthesis. J Biol Chem 272: 32254–32259 [DOI] [PubMed] [Google Scholar]
- Ban N, Freeborn B, Nissen P, Penczek P, Grassucci RA, Sweet R, Frank J, Moore PB, Steitz TA 1998. A 9 Å resolution X-ray crystallographic map of the large ribosomal subunit. Cell 93: 1105–1115 [DOI] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA 2000. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289: 905–920 [DOI] [PubMed] [Google Scholar]
- Bennett WS, Steitz TA 1976. Glucose-induced conformational change in yeast hexokinase. Proc Natl Acad Sci USA 75: 4848–4852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk V, Cate JHD 2007. Insights into protein biosynthesis from structures of bacterial ribosomes. Curr Opin Struct Biol 17: 302–309 [DOI] [PubMed] [Google Scholar]
- Blaha G, Stanley RE, Steitz TA 2009. Formation of the first peptide bond. Science 325: 966–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaha G, Gurel G, Schroeder SJ, Moore PB, Steitz TA 2008. Mutations outside the anisomycin-binding site can make ribosomes drug-resistant. J Mol Biol 379: 505–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard SC, Gonzalez RL Jr, Kim HD, Chu S, Puglisi JD 2004. tRNA selection and kinetic proofreading in translatioin. Nature Struct Mol Biol 11: 1008–1014 [DOI] [PubMed] [Google Scholar]
- Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407: 340–348 [DOI] [PubMed] [Google Scholar]
- Caskey CT, Beaudet AL, Scolnick EM, Rosman M 1971. Hydrolysis of fMet-tRNA by peptidyl transferase. Proc Natl Acad Sci USA 68: 3163–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate JH, Yusupov MM, Yusupova GZ, Earnest TN, Noller HF 1999. X-ray crystal structures of 70S ribosome functional complexes. Science 285: 2095–2104 [DOI] [PubMed] [Google Scholar]
- Crick FHC 1968. The origin of the genetic code. J Mol Biol 38: 367–379 [DOI] [PubMed] [Google Scholar]
- Dorner S, Panuschka F, Schmid W, Barta A 2003. Mononucleoditde derivatives as ribosomal P-site substrates reveal an important conribution of the 2′-OH activity. Nucl Acids Res 31: 6536–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant J, Foley D 1980. On the causes and prevention of mistranslation. In Ribosomes, (ed. Chambliss G., Craven G.R., Davies J., Davis K., Kahan L., Nomura M.), pp. 615–638 Baltimore: University Park Press [Google Scholar]
- Gao Y.-G, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V 2009. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 326: 694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Sengupta J, Valle M, Korostelev A, Eswar N, Stagg SM, Van Roey P, Agrawal RK, Harvey SC, Sali A et al. 2003. Study of the structural dynamics of the E. coli 70S ribosome using real-space refinement. Cell 113: 789–801 [DOI] [PubMed] [Google Scholar]
- Gavrilova LP, Kostiashkina OE, Kotelianshy VE, Ruthkevitch HM, Spirin AS 1976. Factor-free (“unenzymatic”) and factor-dependent systems of translocation of polyuridylicacial by Escherichia coli ribosomes. J Mol Biol 101: 537–552 [DOI] [PubMed] [Google Scholar]
- Gindulyte A, Bashan A, Agmon I, Massa L, Yonath A, Karle J 2006. The transition state for the formation of the peptide bond. Proc Natl Acad Sci USA 103: 13327–13332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BR, Ganoza MC 1975. Identification of a soluble protein that stimulates peptide bond formation. Proc Natl Acad Sci USA 72: 4257–4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R, Noller HF 1997. Ribosomes and translation. Annu Rev Biochem 66: 679–716 [DOI] [PubMed] [Google Scholar]
- Gromadski KB, Rodnina M 2004. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Molec Cell 13: 191–200 [DOI] [PubMed] [Google Scholar]
- Gurel G, Blaha G, More PB, Steitz TA 2009. U2504 determined the species specificity of the A-site cleft antibiotics. The structures of tiamulin, homoherringtonine and bruceantin bound to the ribosome. J Mol Biol 389: 146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JL, Moore PB, Steitz TA 2003. Structure of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J Mol Biol 330: 1061–1075 [DOI] [PubMed] [Google Scholar]
- Hansen JL, Schmeing TM, Moore PB, Steitz TA 2002. Structural insights into peptide bond formation. Proc Natl Acad Sci USA 99: 11670–11675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Francheschi F, Yonath A 2001. High resolution structure of the large ribosomal subunit from a mesophilic bacterium. Cell 107: 679–688 [DOI] [PubMed] [Google Scholar]
- Hoang L, Frederick K, Noller HF 2004. Creating ribosomes with an all-RNA 30S subunit P site. Proc Nat Acad Sci USA 101: 12439–12433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippolito JA, Kanyo ZF, Wang D, Franeschi FJ, Moore PB, Steitz TA, Duffy EM 2008. Crystal structure of the oxazolidinone antibiotic linezolid bound to the 50S ribosomal subunit. J Med Chem 51: 3353–3356 [DOI] [PubMed] [Google Scholar]
- Kim DF, Green R 1999. Base-pairing between 23S rRNA and tRNA in the ribosomal A site. Molec Cell 4: 859–864 [DOI] [PubMed] [Google Scholar]
- Klein DJ, Moore PB, Steitz TA 2004. The roles of ribosomal proteins in the structure, assembly and evolution of the large ribosomal subunit. J Mol Biol 340: 141–177 [DOI] [PubMed] [Google Scholar]
- Korostelev A, Laurberg M, Noller HF 2009. Multistart simulated annealing refinement of the crystal structure of the 70S ribosome. Proc Natl Acad Sci USA 106: 18195–18200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev A, Asahara H, Lancaster L, Laurberg M, Hirschi A, Zhu J, Trakhanov S, Scott WG, Noller HF 2008. Crystal structure of a translation termination complex formed with RF2. Proc Natl Acad Sci USA 105: 19684–19689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev A, Trakhanov S, Laurberg M, Noller HF 2006. Crystal structure of a 70S ribosome-tRNA complex reveals functional rearrangements. Cell 126: 1065–1077 [DOI] [PubMed] [Google Scholar]
- Koshland DE, in The Enzymes (eds. Boyer P.D., Lardy H., Myrback K.) 305–346 Academic Press, New York, 1959 [Google Scholar]
- Lauerberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF 2008. Structural basis for translation termination of the 70S ribosome. Nature 454: 852–857 [DOI] [PubMed] [Google Scholar]
- Li W, Agirrezabala X, Lei JL, Bouakaz L, Brunelle JL, Ortiz-Meoz RF, Green R, Sanyal S, Ehrenberg M, Frank J 2008. Recognition of aminoacyl-tRNA: a common molecular mechanism revealed by cryo-EM. EMBO J 27: 3322–3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monro RE, Staehlin T, Celma ML, Vazquez D 1969. The peptidyl transferase activity of ribosomes. Cold Spring Harbor Symp Quant Biol 34: 357–366 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Ito K 2004. Intraribosomal regulation of expression and fate of proteins. ChemBioChem 5: 48–51 [DOI] [PubMed] [Google Scholar]
- Nierhaus KH, Schulze H, Cooperman BS 1980. Molecular mechanisms of the peptidyl transferase center. Biochem Internat 1: 185–192 [Google Scholar]
- Nissen P, Ban N, Hansen J, Moore PB, Steitz TA 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289: 920–930 [DOI] [PubMed] [Google Scholar]
- Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA 2001. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc Natl Acad Sci USA USA 98: 4899–4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller HF 1991. Ribosomal RNA and translation. Annu Rev Biochem 60: 191–227 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Ramakrishnan V 2005. Structural insights into translational fidelity. Annu Rev Biochem 74: 129–177 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Carter AP, Ramakrishnan V 2003. Insights into the decoding mechanism from recent ribosome structures. Trends in Biochem Sci 28: 259–266 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM, Tarry MJ, Carter AP, Ramakrishnan V 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292: 897–902 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Murphy FVI, Tarry MJ, Ramakrishnan V 2002. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111: 721–732 [DOI] [PubMed] [Google Scholar]
- Page MI, Jencks WP 1971. Entropic contributions to rate acceleration in enzymatic and intramolecular reactions and the chelate effect. Proc Natl Acad Sci USA 68: 1678–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnell KM, Seila AC, Strobel SA 2002. Evidence against stabilization of the transitionstate oxyanion by a pKa-perturbed RNA base in the peptidyl transferase center. Proc Natl Acad Sci USA 99: 11658–11663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka S 1974. Assay for nonenzymatic and enzymatic translocation with Escherichia coli ribosomes. Meth Enzymol 30: 462–470 [DOI] [PubMed] [Google Scholar]
- Pioletti M, Schlunzen F, Harms J, Zarivach R, Glumann M, Avila H, Bashan A, Bartels H, Auerbach T, Jacobi C et al. 2001. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J 20: 1829–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaha RR, Green R, Noller HF 1995. A base pair between tRNA and 23S rRNA in the peptidyl transferase centre of the ribosome. Nature 377: 309–314 [DOI] [PubMed] [Google Scholar]
- Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartles H, Agmon I, Franceschi F et al. 2000. Structure of functionally activated small ribosomal subunit at 3.3 A resolution. Cell 102: 615–623 [DOI] [PubMed] [Google Scholar]
- Schleunzen F, Zarivach R, Harms J, Bashan A, Tocilj A, Albrecht R, Yonath A, Franceschi F 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413: 814–821 [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Moore PB, Steitz TA 2003. Structure of deacylated tRNA mimics bound to the E site of the large ribosomal subunit. RNA 9: 1345–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeing TM, Huang KS, Strobel SA, Steitz TA 2005b. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl tRNA. Nature 438: 520–524 [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Kitchen D, Scaringe SA, Strobel SA, Steitz TA 2005a. Structural insights into the roles of water and the 2′ hydroxyl of the P-site tRNA in the peptidyl transferase reaction. Molec Cell 20: 437–448 [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Voorhees RM, Kelley AC, Goa Y-G, Murphy FV IV, Weir JR, Ramakrishnan V 2009. The crystal structure of the ribosome bound to EF-Tu and aminoacyl tRNA. Science 326: 688–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjuro A, Holton JH, Doudna-Cate JH 2005. Structure of the bacterial ribosome at 3.5 Å resolution. Science 310: 827–834 [DOI] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FV IV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V 2006. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 3313: 1935–1942 [DOI] [PubMed] [Google Scholar]
- Sievers A, Beringer M, Rodnina MV, Wolfenden R 2004. The ribosome as an entropy trap. Proc Natl Acad Sci USA 101: 7897–7901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonovic M, Steitz TA 2008. Cross-crystal averaging reveals that the structure of the peptidyl transferasemcneter is the same in the 70S ribosome and the 50S subunit. Proc Natl Acad Sci USA 105: 500–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southworth DR, Brunelle JL, Green R 2002. EFT-independent translation of the mRNA-tRNA complexes promoted by modification of the ribosomes with thiol-specific reagents. J Mol Biol 324: 611–623 [DOI] [PubMed] [Google Scholar]
- Stark H, Rodnina MV, Wieden H.-J., Zemlin F, Wintermeyer W, van Heel M 2002. Ribosome interactions of aminoacyl-tRNA and elongation factor Tu in the codon-recognition complex. Nature Struc Biol 9: 849–854 [DOI] [PubMed] [Google Scholar]
- Tenson T, Lovmar M, Ehrenberg M 2003. The mechanism of action macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J Mol Biol 330: 1005–1014 [DOI] [PubMed] [Google Scholar]
- Tu D, Blaha G, Moore PB, Steitz TA 2005. Structures of MLSbK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell 121: 257–270 [DOI] [PubMed] [Google Scholar]
- Valle M, Sengupta J, Swami NK, Grassucci R, Burkhardt N, Nierhaus KH, Agrawal RK, Frank J 2002. Cryo-EM reveals an active role for aminoacyl tRNA in the accommodation process. EMBO J 21: 3557–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Zavialov AV, Li W, Stagg SM, Seugupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J 2003. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nature Struc Biol 10: 899–906 [DOI] [PubMed] [Google Scholar]
- Voss NR, Gerstein M, Stetiz TA, Moore PB 2006. The geometry of the ribosomal polypeptide exit tunnel. J Mol Biol 360: 893–906 [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V 2009. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nature Struc Biol 16: 528–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD 1963. Involvement of RNA in the synthesis of proteins. Science 140: 17–26 [DOI] [PubMed] [Google Scholar]
- Weinger JS, Parnell KM, Dorner S, Green R, Strobel SA 2004. Substrate-assisted catalysis of peptide bond formation by the ribosome. Nature Struc Biol 11: 1101–1106 [DOI] [PubMed] [Google Scholar]
- Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, Ramakrishnan V 2008. Science 322: 953–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DN, Harms JM, Nierhaus KH, Schlunzen F, Fucini P 2005. Species-specific antibiotic-ribosome interactions: implications for drug development. Biol Chem 386: 1239–1252 [DOI] [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WM, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V 2000. Structure of the 30S ribosomal subunit. Nature 407: 327–339 [DOI] [PubMed] [Google Scholar]
- Wittmann-Liebold B 1986. Ribosomal proteins: their structure and evolution. In Structure, Function, and Genetics of Ribosomes (ed. Hardesty B., Kramer G.), pp. 326–361 New York: Springer-Verlag [Google Scholar]
- Wool IG 1986. Studies of the structure of eukaryotic (mammalian) ribosomes. In Structure, Function, and Genetics of Ribosomes (ed. Hardesty B., Kramer G.), pp. 391–411 New York: Springer-Verlag [Google Scholar]
- Wool IG, Chan Y.-L., Gluck A 1995. Structure and evolution of mammalian ribosomal proteins. Biochem Cell Biol 73: 933–947 [DOI] [PubMed] [Google Scholar]
- Xia T, SantaLucia J Jr, Burkard ME, Kierzek R, Schroeder SJ, Jiao C, Cox C, Turner DH 1998. Thermodynamic paramenters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry 37: 14719–14735 [DOI] [PubMed] [Google Scholar]
- Yim G, Wang HH, Davies J 2007. Antibiotics as signaling molecules. Phil Transact Roy Soc B – Biol Sci 362:1195–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonath A, Bashan A 2004. Ribosome crystallography: mutation, peptide bond formation, and amino acid polymerization are hampered by antibiotics. Annu Rev Microbiol 58: 233–251 [DOI] [PubMed] [Google Scholar]
- Youngman EM, Brunelle JL, Kochaniak AB, Green R 2004. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell 117: 589–599 [DOI] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JHD, Noller HF 2001. Crystal structure of the ribosome at 5.5 A resolution. Science 292: 883–896 [DOI] [PubMed] [Google Scholar]