The article provides an overview of the development and structure of spore and pollen walls in the major plant groups and summarises progress in our understanding of the molecular genetics underpinning spore/pollen evolution and development.
Abstract
Background and aims
Many key innovations were required to enable plants to colonize terrestrial habitats successfully. One of these was the acquisition of a durable spore/pollen wall capable of withstanding the harsh desiccating and UV-B-rich environment encountered on land. The spores of ‘lower’ spore-bearing plants and the pollen of ‘higher’ seed plants are homologous. In recent years, researchers have begun to investigate the molecular genetics of pollen wall development in angiosperms (including the model organism Arabidopsis thaliana). However, research into the molecular genetics of spore wall development in more basal plants has thus far been extremely limited. This review summarizes the literature on spore/pollen wall development, including the molecular genetics associated with pollen wall development in angiosperms, in a preliminary attempt to identify possible candidate genes involved in spore wall development in more basal plants.
Presence in moss of genes involved in pollen wall development
Bioinformatic studies have suggested that genes implicated in pollen wall development in angiosperms are also present in moss and lycopsids, and may therefore be involved in spore wall development in basal plants. This suggests that the molecular genetics of spore/pollen development are highly conserved, despite the large morphological and functional differences between spores and pollen.
Future work
The use of high-throughput sequencing strategies and/or microarray experiments at an appropriate stage of ‘lower’ land plant sporogenesis will allow the identification of candidate genes likely to be involved in the development of the spore wall by way of comparison with those genes known to be involved in pollen wall development. Additionally, by conducting gene knock-out and gene swap experiments between ‘lower’ land plant species, such as the moss model species Physcomitrella patens, and the angiosperm model species arabidopsis it will be possible to test the role of these candidate genes.
Introduction
The colonization of land by plants in the Palaeozoic was a highly significant event in Earth's history, both from an evolutionary point of view and because it fundamentally changed the ecology and environment of the planet (Beerling 2007). Land plants evolved to form crucial components of all modern terrestrial ecosystems through evolutionary adaptations involving changes in anatomy, physiology and life cycle (Waters 2003; Menand et al. 2007; Cronk 2009). Key adaptations include rooting structures, conducting tissues, cuticle, stomata, and sex organs such as gametangia and spores/pollen.
Development of a durable spore wall is essential for terrestrialization as it enables the spore to withstand physical abrasion, desiccation and UV-B radiation (Wellman 2004). As part of their life cycle, sexually reproducing embryophytes manufacture either spores, or their more derived homologues pollen. The major component of the spore/pollen wall proposed to be of primary importance in enabling resistance to the conditions described above is the highly resistant biopolymer sporopollenin (Ito et al. 2007; Cronk 2009).
It seems reasonable to hypothesize that colonization of the land by plants was not possible prior to the evolution of the sporopollenin spore wall, and this adaptation is considered to be a synapomorphy of the embryophytes. Additionally, spore walls are not present in the hypothesized embryophyte antecedents, the green algae (Wellman 2004). However, the production of sporopollenin is highly likely to be pre-adaptive as it is present in a number of different algal groups such as the charophyceans, which have been proposed as the sister group to the embryophytes. In certain charophyceans, sporopollenin occurs, but is located in an inner layer of the zygote wall (Graham 1993).
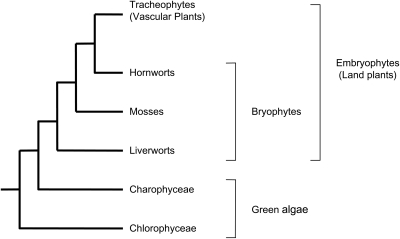
Phylogenetic studies and fossil evidence have shown that the most basal living land plants are the paraphyletic ‘bryophytes’ (Kenrick and Crane 1997; Qui et al. 2006) (Fig. 1). They comprise the liverworts, mosses and hornworts, and their phylogenetic position should allow us to further elaborate the evolutionary changes that facilitated the conquest of land by plants (Rensing et al. 2008). The moss Physcomitrella patens is the first ‘bryophyte’ genome to be sequenced. This genome, through comparisons with angiosperm genomes, is proving to be a valuable tool in experimental studies that attempt to reconstruct genome evolution during the colonization of land (Reski and Cove 2004; Quatrano et al. 2007; Rensing et al. 2008).
Fig. 1.
Phylogenetic tree for land plant evolution derived from analysis by Qui et al. (2006). The bryophytes are a paraphyletic group comprising three separate lineages. Together with the vascular plants (which include the angiosperms), bryophytes form the embryophytes, which have a sister group relationship to the green algae.
In this review, we first outline the nature of spore/pollen wall development in the major plant groups, before considering emerging understanding of the molecular genetics of pollen wall development. The latter includes identification of genes involved in sporopollenin biosynthesis and exospore formation, callose wall formation and tetrad separation. We also report results from BLAST searches of the basal land plant physcomitrella and the clubmoss Selaginella moellendorfii using genes implicated in pollen wall development in arabidopsis.
Spore and pollen wall structure and development
The spore/pollen walls of embryophytes have multiple layers and components that are laid down in a regulated manner during spore/pollen development. Layers containing the macromolecule sporopollenin are the component enabling the resistance of the spore/pollen wall to numerous environmental factors that make life on land challenging. Sporopollenin is highly resistant to physical, chemical and biological degradation procedures. Consequently, its precise chemical composition, structure and biosynthetic route have not yet been ascertained (Meuter-Gerhards et al. 1999). Traditional convention asserts that sporopollenin is a polymer of carotenoid esters (Cronk 2009). However, modern purification, degradation and analytical techniques have shown that it is comprised of polyhydroxylated unbranched aliphatic units with small quantities of oxygenated aromatic rings and phenylpropanoids (Ahlers et al. 1999; Domínguez et al. 1999).
Modes of sporopollenin deposition in spore and pollen walls
The basic mechanisms involved in the formation of the spore wall, and the deposition of sporopollenin in the exospore/exine, have been illuminated by numerous ultrastructural studies performed on extant and fossil species across the plant kingdom (Paxson-Sowders et al. 2001). Blackmore and Barnes (1987) proposed a number of sporopollenin deposition processes apparent in the spore wall. Firstly, they recognized the role of white-line-centred lamellae (WLCL) in this process. The accumulation of sporopollenin on an array of WLCL is regarded as being the most primitive method of sporopollenin deposition and has been identified in a number of algal groups and most, if not all, embryophytes (Wellman 2004). These lamellae materialize at the plasma membrane with sporopollenin polymerizing out onto either side of the white line. They accumulate in a variety of ways to form the spore/pollen wall (Blackmore and Barnes 1987; Blackmore et al. 2000; Wellman 2004).
Another mode of exospore/exine formation involves the deposition of sporopollenin from the surrounding cells of the tapetum. Transmission electron microscopy has shown that the tapetal cells possess a highly active secretory system containing lipophilic globules, which are thought to contain the precursors of sporopollenin and are deposited onto the surface either directly contributing to the exospore/exine or forming extra-exosporal layers (Piffanelli et al. 1998). Blackmore et al. (2000) suggested that a tapetal contribution to the spore wall can take place in a variety of ways, including the addition to the layers formed by the WLCL or directly onto WLCL. Studies of pollen wall formation in angiosperms highlight the role that tapetal cells play in supplying nutrients and lipid components to developing microsporocytes and microspores (Scott et al. 1991; Ariizumi et al. 2004; de Azevedo Souza et al. 2009). Interestingly, the most basal extant land plants (liverworts) lack a tapetum, which is acquired in mosses and vascular plants.
An alternative deposition process involves centripetal accumulation of sporopollenin onto previously formed layers. Blackmore et al. (2000) noted that exospore formation may be achieved by sporopollenin accumulation below a pre-existing layer, either by WLCL accumulation or by the deposition of granular or unstructured sporopollenin. A further mode of deposition is observed in seed plants where sporopollenin accumulates within a pre-patterned cell surface glycocalyx referred to as the primexine (Blackmore and Barnes 1987; Blackmore et al. 2000; Wellman 2004), which is essentially an exine precursor.
Spore wall development in bryophytes
Spore wall development has been studied in all three of the traditional bryophyte groups (reviewed in Brown and Lemmon 1988, 1990). In the majority of liverworts, immediately after meiosis, a polysaccharide wall (the spore special wall) is laid down outside the plasma membrane (Brown and Lemmon 1985). In many liverworts, this spore special wall seems to function as a primexine in which the pattern of exospore ornamentation is established (Brown and Lemmon 1993). However, in some liverworts exospore ornamentation appears to be determined by exospore precursors produced by the diploid sporocyte prior to meiosis and formation of the haploid spores (Brown et al. 1986). The exospore develops centripetally (Brown and Lemmon 1993) based on WLCL formed outside the spore cytoplasm. At completion, the entire exospore comprises sporopollenin deposited on WLCL. At maturity, the lamellate structure thus formed is clearly discernible and is highly characteristic of the liverwort exospore. Liverworts lack a tapetum and there is therefore no input from this source. The innermost layer of fibrillar intine is the final wall layer to be formed (Brown and Lemmon 1993).
Studies of spore wall development in hornworts are limited. As with liverworts, a spore special wall is formed after meiosis and functions as a primexine in which the exospore is set down. It was initially thought that the exospore formed in the absence of WLCL, but Taylor and Renzaglia have recently demonstrated their presence (W.A. Taylor, University of Wisconsin-Eau Claire, USA, pers. comm., 2011). Recent analyses of Phaeomegaceros fimbriatus have shown that the mature spore wall has a thin perine-like outer layer, but this represents the remnants of the spore mother cell wall rather than extra-exosporal material derived from a tapetum (Villarreal and Renzaglia 2006).
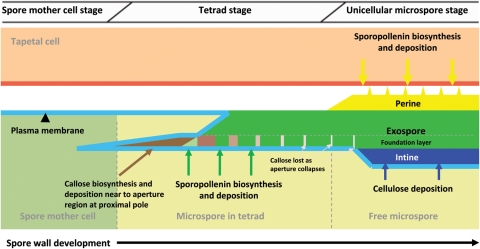
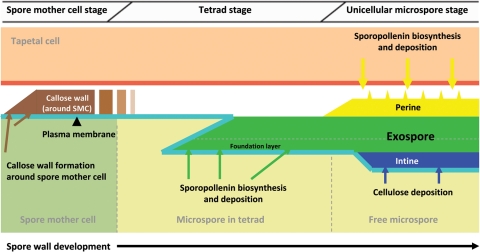
Three types of spore wall have been recognized in mosses: Bryopsida type, Andreaeidae type and Sphagnidae type (Brown and Lemmon 1990). All three of these types appear to form in the absence of a spore special wall. Bryopsida-type spore walls are homogeneous except for an inconspicuous foundation layer (Fig. 2). This foundation layer forms first via sporopollenin accumulation on WLCL. Subsequently, the homogeneous exospore layer is laid down outside the foundation layer in a centrifugal manner. This layer is probably mainly extrasporal in origin. Sometimes additional homogeneous material is also deposited inside the foundation layer. This layer is almost certainly derived from the spore. Following the accumulation of homogeneous material, the spores are coated by an additional extra-exosporal layer, referred to as the perine or perispore, which is derived from the tapetum. Finally, the intine forms.
Fig. 2.
Proposed model of spore wall development in physcomitrella. The exine foundation layer is laid down first by way of sporopollenin accumulation on WLCL. The rest of the exine layer is deposited outside the foundation layer centrifugally. Note the appearance of callose in the inner exine, which is confined to the expanded aperture region at the proximal pole.
Spore wall development in the Andreaeidae type is unique among mosses in that they have a spongy exospore that appears to form in the absence of WLCL (Brown and Lemmon 1984). By studying Andreaea rothii, Brown and Lemmon (1984) demonstrated that the exospore is instead initiated as discrete homogeneous globules within the coarsely fibrillar network of the spore mother cell. These globules accumulate and form an irregular layer with numerous interstitial spaces. The sequence of spore wall layer development is essentially the same as that of other mosses and the mature wall consists of an inner intine, a spongy exospore and an outer perine (Brown and Lemmon 1984).
Sphagnidae-type moss spore walls are more complex than those of the other mosses and consist of five layers (Brown et al. 1982). Unlike other mosses, the exospore of Sphagnidae type comprises two layers: an inner lamellate layer (A-layer) and a thick homogeneous outer layer (B-layer). In addition to the exospore, there is an intine, a unique translucent layer and the outermost perine. The A-layer is the first to form and does so by sporopollenin accumulation on WLCL, and develops evenly around the young spore immediately after meiosis. The homogeneous B-layer is deposited outside the A-layer. Overlying the exospore is a translucent layer that consists of unconsolidated exospore lamellae in a medium of unknown composition. The tapetally derived perine is deposited on top of this unique layer. The study of spore wall development in Sphagnum lescurii by Brown et al. (1982) suggests that the ontogeny of the wall layers is not strictly centripetal.
Spore wall development in pteridophytes
Spore walls have been investigated in a number of pteridophyte species representing all of the major pteridophyte groups (reviewed in Lugardon 1990; Tryon and Lugardon 1991).
Spore wall development is well understood in the homosporous lycopsid Lycopodium clavatum (Uehara and Kurita 1991). Shortly after meiosis, the plasma membrane of the sporogenous cell folds into a pattern that later becomes the reticulate spore sculpture. Small WLCL form on the plasma membrane and accumulate in a centripetal fashion, forming the greater part of the exospore. After the main lamellate part of the exospores is formed, an inner granular layer, possibly derived from the spore cytoplasm, is deposited. In some Lycopodium there are no extra-exosporal layers (Uehara and Kurita 1991) whereas in others a thin extra-exosporal layer is deposited after the completion of the exospore (Tryon and Lugardon 1991).
Spore structure and development in heterosporous lycopsids differ between microspores and megaspores. In the clubmoss selaginella, microspores possess an exospore consisting of two layers (Fig. 3). The thin inner layer is the first to develop and comprises imbricate lamellae that are formed on WLCL in a centripetal direction (Tryon and Lugardon 1991). The outer layer starts to form only once the inner layer is complete. Some selaginella species may also develop a thin perispore or a paraexospore. In the microspores of the heterosporous lycopsid Isoetes japonica, a large gap is developed between the two exospore layers (Uehara et al. 1991). The outer exospore layer is regarded as a paraexospore as it begins to form before the inner exospore, consists of similar sporopollenin, and is completed at the same time as the inner exospore.
Fig. 3.
Proposed model of spore wall development in selaginella microspores. The thin inner exine layer forms first and comprises lamellae formed centripetally on WLCL. The outer exine starts to form once the inner layer is complete. Note the presence of callose at early developmental stages around the spore mother cell.
Selaginella megaspore walls contain two layers of similar thickness (Morbelli 1995). The inner and outer layers consist of lamellae and poorly segregated components, respectively. The inner layer does not thicken during exospore development and a dense basal layer is formed by the lamellae. In contrast, the outer layer increases significantly in thickness due to self-assembly (Hemsley et al. 1994, 2000; Gabarayeva 2000). During the final stages of sporogenesis, the endospore forms between the plasma membrane and the exospore. In Isoetes, the megaspore wall is similar to that of selaginella in terms of development and structure, consisting of two layers, with the formation of the outer layer commencing prior to that of the inner layer. Substantial quantities of silica are deposited within and on top of the outer layer before the exospore is completed. Finally, the endospore is laid down between the plasma membrane and the exospore.
The exospore in homosporous ferns develops centrifugally and is once again bilayered. The inner layer acts as a substructure and consists of varying numbers of fused sheets (extensive interconnected laminae) that form by sporopollenin accumulation on WLCL. The homogeneous outer layer is considerably thicker and contains thin radial fissures and small cavities. An extra-exosporal layer (perispore) forms once the exospore is complete and is deposited from the decaying tapetum. Spore wall development in heterosporous ferns is similar to that observed in homosporous ferns, and is also similar in both microspores and megaspores.
In sphenopsids the spore walls appear to be highly derived (Lugardon 1990), and observations of Equisetum arvense have shown that four layers are present in the form of an exospore, an endospore, a middle layer and pseudoelators (Uehara and Kurita 1989). The exospores comprise inner and outer exospores. The broad and homogeneous inner exospore forms first by way of plate-like structures accumulating on the plasma membrane. The outer exospore is then formed by the deposition of granular material on the inner exospore and is similarly wide and homogeneous. Once exospore formation is complete, the middle layer forms in the gap between the exospore and the plasma membrane. The pseudoelators are the next structure to form and consist of two layers. The inner layer comprises longitudinal microfibrils during the early stages of development but eventually becomes homogeneous. The outer layer is also homogeneous and is formed by granules that are released from vesicles in the plasmodial cytoplasm. The pseudoelators are connected to the spore, by way of the middle layer, at the aperture. The endospore is the final component of the wall to form on the inside of the exospores (Taylor 1986; Uehara and Kurita 1989).
Pollen wall development in gymnosperms
Although differences in pollen wall structure and development are evident in different extant and extinct gymnosperm groups, the main ontogenetic elements appear to be homologous (summarized in Lugardon 1994; Wellman 2009). The pollen mother cell undergoes meiosis to form four haploid microspores. Subsequent development of the exine consists of a number of stages. Firstly, a callose wall forms around the pollen mother cell and subsequently extends around each of the microspores. Next, a matrix develops around each microspore upon which the fibrillar microspore surface coat and later the sexine (consisting of tectum and infratectum components) pattern is established (Zavada and Gabarayeva 1991). The microspore surface coat is deposited between the surface of the microspore and the surrounding tetrad wall prior to the formation of the wall components. This layer is regarded as being equivalent to the primexine in angiosperms. The sexine then begins to form on and within the microspore surface coat. The nexine (inner pollen exine wall) lamina is then formed below this coat; therefore, the sexine is partly developed when the nexine begins to develop. The exine as a whole appears to form in a centripetal direction from the outside inwards (Lugardon 1994). Finally, an intine is deposited on the inside of the pollen exine.
Pollen wall development in angiosperms
Pollen walls in angiosperms typically consist of an outer exine composed of sporopollenin and an inner intine composed of cellulose and pectin (Fig. 4) (Paxson-Sowders et al. 1997; Morant et al. 2007). Models of development have been proposed based on observations on numerous species, including Lilium and arabidopsis (e.g. Suzuki et al. 2008). Similar processes have been described in both these species.
Fig. 4.
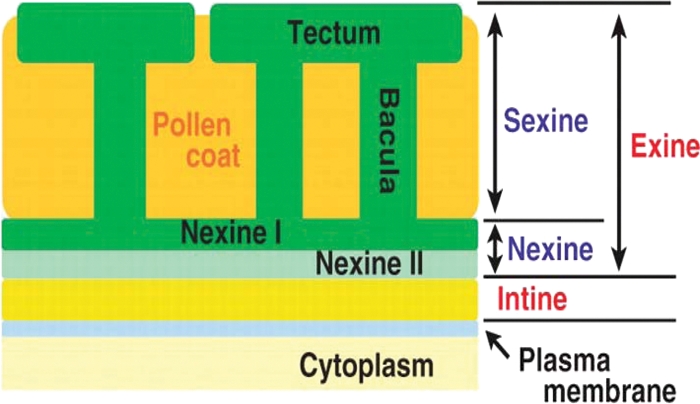
Diagram of arabidopsis pollen wall structure. The inner intine and the various components of the outer exine are indicated. Note the pollen coat (tryphine and pollenkitt) filling the cavities of the exine sculpture. Taken from Suzuki et al. (2008).
Once again, prior to meiosis, the pollen mother cell is surrounded by a callose special cell wall (Blackmore et al. 2007). Immediately after meiosis, four microspores derived from the pollen mother cell form a tetrad. A callose special wall surrounds the microspores (Blackmore et al. 2007). A cellulose primexine then forms between the plasma membrane and callose wall of each microspore. Both the callose wall and primexine are deposited at the surface of the microspore through processes mediated by the plasma membrane (Blackmore et al. 2007). A section of the primexine is then adapted to form column-like structures called the probaculae upon which sporopollenin, secreted by the microspore, will eventually accumulate and polymerize. Sporopollenin deposition and accumulation extend the probaculae, which form the baculae and the tectum (Heslop-Harrison 1963, 1968). The callose wall then degrades and the developing baculae and tectum are exposed to the fluid of the locule and receive sporopollenin secreted by the tapetum. Wall formation is complete when the nexine and intine layers are formed and the primexine recedes and disappears (Suzuki et al. 2008). The mature pollen grain is then coated by tryphine and pollenkitt, which are synthesized by the tapetum (Dickinson and Lewis 1973; Blackmore et al. 2007).
Summary
While the basic components associated with spore/pollen wall development and structure described above can be localized in different wall regions and influence wall development at different stages across the principal land plant groups, their presence in the majority or all of these groups suggests that their involvement in wall development and sporogenesis as a whole is a signature of embryophytes. The main differences concern the absence, presence and role of callose, and also the mode of sporopollenin deposition.
Callose would appear to play a major role in pollen wall development in angiosperms, where a callose wall surrounds the tetrad and serves as a template for exine development. The role of callose in wall development in other groups is less well defined. In some groups such as pteridophytes, with the exception of selaginella, callose has yet to be identified during sporogenesis and is currently thought to be absent. In some bryophytes (Hepaticopsida: Anthoceros, Geothallus, Riccia; Bryopsida: Mnium) (Waterkeyn and Bienfait 1971), callose has been identified around the spore mother cell but its link to wall development, if any, is not well understood. In this instance, it could be the case that callose is relictual as it is involved in the reproductive systems of some algal groups (Gabarayeva and Hemsley 2006).
Certain modes of sporopollenin deposition are not confined to individual plant groups and different modes have been observed on numerous occasions in a single species. For example, WLCL, while not a constant feature, have been observed in species belonging to all phylogenetic levels of land plants. However, the accumulation of sporopollenin within a pre-patterned microspore surface coat, or primexine, is seemingly confined to gymnosperms and angiosperms.
Molecular genetics of pollen wall development
In recent years there has been a surge in papers describing genes involved in pollen wall development. However, our understanding of the molecular genetics of spore/pollen development remains poor due to the complexity of the developmental process and problems in pinpointing the actual function of the genes involved. Furthermore, research has been confined to particular model angiosperms (Table 1 and Fig. 5), with little or no information on gymnosperm pollen or the spores of ‘lower’ land plants. This begs the question as to whether similar genes are involved in development of the simple walls of ‘lower’ land plant spores and the more derived pollen walls of the gymnosperms and angiosperms. However, this research is now beginning to incorporate model plant species from more primitive groups, such as the bryophytes. This extended research will enable the comparison of the molecular genetics of spore/pollen wall development in angiosperms and more primitive plants. The results from this may allow us to assess how conserved are the genes and genetic networks involved in spore/pollen wall development. We begin by reviewing what is known of the molecular genetics of pollen wall development in the angiosperms.
Table 1.
Arabidopsis genes implicated in pollen wall development.
| Role | Gene | Proposed expression |
|---|---|---|
| Sporopollenin biosynthesis and exine formation | MS2 | Sporophyte |
| YRE/WAX2/FLP1 | Sporophyte | |
| CYP703A2 | Sporophyte and microspores | |
| CYP704B1 | Sporophyte | |
| ACOS5 | Sporophyte | |
| RPG1 | Sporophyte and microspores | |
| NEF1 | Sporophyte | |
| KNS5-10 | Sporophyte | |
| KNS4 | Sporophyte | |
| ABCG26 | Sporophyte and microspores | |
| LAP5/PKSB | Sporophyte | |
| LAP6/PKSA | Sporophyte | |
| TKPR1/DRL1 | Sporophyte | |
| TKPR2/CCRL6 | Sporophyte | |
| AtMYB103/MS188 (TF) | Sporophyte | |
| MS1 (TF) | Sporophyte | |
| AtbZIP34 (TF) | Sporophyte and gametophyte | |
| Exine formation (probaculae) | DEX1 | Unknown |
| TDE1/DET2 | Unknown | |
| KNS2, 3, 12 | Sporophyte | |
| Intine formation | FLA3 | Sporophyte and microspores |
| Callose wall formation | CALS5/LAP1 | Unknown |
| KNS1, 11 | Sporophyte | |
| Tetrad separation | QRT1-3 | Sporophyte |
| A6 | Sporophyte |
TF denotes transcription factor.
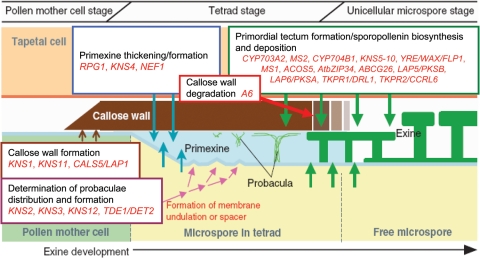
Fig. 5.
Proposed functions of genes implicated in arabidopsis pollen wall exine development. Expected time and location of gene expression are indicated. Not all arabidopsis genes described in this review are included due to a lack of information regarding the time and locality of their expression. Modified from Suzuki et al. (2008).
Arabidopsis genes implicated in sporopollenin biosynthesis and exine formation
A number of arabidopsis genes associated with the biosynthesis of exine encode proteins with sequence homology to enzymes involved in fatty acid metabolism (Dobritsa et al. 2009). Aarts et al. (1997) observed expression of the MALE STERILITY 2 (MS2) gene in the tapetum of wild-type plants at, and shortly after, the release of microspores from tetrads and noted that MS2 mutants produced pollen grains that lacked an exine layer. The exine layer had been replaced by a thin layer of unknown composition. MS2 encodes a protein with sequence similarity to long-chain fatty acyl reductases, and expression of the MS2 protein in bacteria leads to the increased synthesis of fatty alcohols (Doan et al. 2009). Taken together, these data suggest that an MS2-linked enzymatic pathway is required for the synthesis of sporopollenin (Aarts et al. 1997; Ariizumi et al. 2008; Dobritsa et al. 2009).
Another gene implicated in exine formation is YORE-YORE (YRE)/WAX2/FACELESS POLLEN1 (FLP1). Ariizumi et al. (2003) suggested that this gene encodes a transporter or catalytic enzyme that is involved in wax synthesis in stems and siliques, in the tryphine and in sporopollenin synthesis. As with MS2, the pollen exine in YRE/FLP1 mutants is poorly constructed and easily damaged, suggestive of defective sporopollenin. Expression analyses in the same study suggest that FLP1 is expressed in the tapetum, which is supported by the fact that the FLP1 mutant phenotype is sporophytically controlled (Ariizumi et al. 2003). In addition, Rowland et al. (2007) demonstrated that the ECERIFIUM 3 (CER3) gene encodes a protein of unknown function identical to YRE/WAX2/FLP1 and is therefore allelic to YRE/WAX2/FLP1.
Morant et al. (2007) showed that the arabidopsis cytochrome P450 enzyme CYP703A2 is also necessary for the synthesis of sporopollenin. The CYP703 cytochrome P450 family is specific to embryophytes and each plant species contains a single CYP703 (Morant et al. 2007). The exine layer in CYP703A2 knock-out mutants is significantly underdeveloped. Sporopollenin also appeared to be absent as the fluorescent layer around the pollen associated with the presence of phenylpropanoid units in sporopollenin was absent in CYP703A2 mutant plants (Morant et al. 2007). Morant et al. (2007) demonstrated that lauric acid and in-chain hydroxy lauric acids are present in the plant substrate and product for this enzyme. These are important building blocks in the synthesis of sporopollenin and facilitate the formation of ester and ether linkages with phenylpropanoid units. Furthermore, the same study showed that CYP703A2 is expressed in the anthers of developing arabidopsis flowers with initial expression detectable at the tetrad stage in the microspores and the tapetum (Morant et al. 2007), consistent with a role in exine formation.
Dobritsa et al. (2009) described another cytochrome P450, CYP704B1, and demonstrated that this gene is essential for exine development. CYP704B1 mutants produce pollen walls that lack a normal exine layer. The exine layer was replaced with a thin layer of material and irregular distribution of aggregates that may have been sporopollenin. The pollen walls also exhibited a characteristic striped surface, unlike the reticulate pattern displayed by the wild type, to which Dobritsa et al. (2009) designated the name zebra phenotype. It has also been shown that heterologous expression of CYP704B1 in yeast catalyses ω-hydroxylation of long-chain fatty acids, consistent with a role in sporopollenin synthesis (Dobritsa et al. 2009). Dobritsa et al. (2009) have suggested that these ω-hydroxylated fatty acids, in concert with the formation of in-chain hydroxylated lauric acids catalysed by CYP703A2, may serve as vital monomeric aliphatic building blocks in the formation of sporopollenin. Analyses of the genetic relationships between CYP704B1, CYP703A2 and MS2 (which as described above encodes a fatty acyl reductase) along with expression analyses and observation of similar zebra phenotypes in all three mutants indicate that these genes are involved in the same pathway within the sporopollenin synthesis framework and are co-expressed (Dobritsa et al. 2009). In addition, an orthologue of CYP704B1 (BnCYP704B1) has recently been identified in Brassica napus, and mutants in this gene exhibit defective exine layers (Yi et al. 2010).
Another gene reported to participate in exine formation, ACOS5, has recently been described (de Azevedo Souza et al. 2009). This encodes a fatty acyl-CoA synthetase with broad in vitro preference for the medium-chain fatty acids required in tapetal cells for sporopollenin monomer synthesis. Mutations in ACOS5 significantly compromise the development of the pollen wall, which appears to lack sporopollenin and exine. The defect in pollen formation in ACOS5 mutants coincides with the deposition of exine at the unicellular microspore stage (de Azevedo Souza et al. 2009). Additionally, after analyses of ACOS5 expression in developing anthers, de Azevedo Souza et al. (2009) proposed that it is also involved in the same biochemical pathway as the CYP703A2, CYP704B1 and MS2 genes.
The RUPTURED POLLEN GRAIN1 (RPG1) gene, which encodes a plasma membrane protein, and the NO EXINE FORMATION1 (NEF1) gene, which encodes a plastid integral membrane protein, are both required for primexine development (Ariizumi et al. 2004; Guan et al. 2008). Guan et al. (2008) revealed that exine pattern formation in RPG1 mutants is defective as sporopollenin is randomly distributed over the surface of the pollen grain. Primexine formation of microspores in RPG1 mutants is abnormal at the tetrad stage, which results in imperfect deposition of sporopollenin on the microspores (Guan et al. 2008). RPG1 plants experience microspore rupture and cytoplasmic leakage, suggesting that cell integrity had been impaired in the microspores. The same study demonstrated that RPG1 is strongly expressed in the tapetum and the microspores during male meiosis (Guan et al. 2008). Ariizumi et al. (2004) showed that NEF1 mutants exhibited similarly defective primexine and that although sporopollenin was present it was not deposited onto the plasma membrane of the microspore because of the lack of normal primexine. Ariizumi et al. (2004) tentatively suggest that NEF1 is expressed in the tapetum and is sporophytically controlled. Additionally, it was proposed that NEF1 is likely to be involved in exine formation at earlier developmental stages than other exine formation genes, such as MS2 and FLP1, since the exine is more poorly developed in NEF1 plants (Ariizumi et al. 2004).
Suzuki et al. (2008) also identified a number of genes involved in the construction of exine and pollen development in general. They managed to successfully isolate 12 KOANASHI mutants (KNS1–KNS12), which were found to be recessive and thus likely to affect pollen development sporophytically. The 12 mutants were categorized into four types. Type 3 (KNS5–KNS10) mutants displayed abnormal tectum formation on the pollen surface, and these genes therefore appear to be required either for creating primordial tectum (onto which sporopollenin is deposited) in the space between the primexine and the callose wall, or for depositing sporopollenin itself (Suzuki et al. 2008). Additionally, the type 2 mutant (KNS4) exhibits a thin exine layer mostly due to shortened baculae. It is proposed that baculae extension is closely linked to the thickening of primexine; therefore, KNS4 is likely to be a novel gene that regulates the thickening of the primexine layer (Suzuki et al. 2008).
Recently, Quilichini et al. (2010) proposed that ATP-BINDING CASSETTE G26 (ABCG26) plays a crucial role in exine formation. Abcg26-1 mutants lack an exine layer, and expression studies showed that ABCG26 is transiently and locally expressed in the tapetum post meiosis. Quilichini et al. (2010) suggest that ABCG26 transports sporopollenin precursors across the tapetum plasma membrane to the anther locule for polymerization on the surface of the developing microspores.
Other genes that have recently been associated with a defective exine include LESS ADHESIVE POLLEN 5/POLYKETIDE SYNTHASE B (LAP5/PKSB) and LESS ADHESIVE SYNTHASE 6/POLYKETIDE SYNTHASE A (LAP6/PKSA), which are also specifically and transiently expressed in the tapetum during microspore development (Kim et al. 2010). Mutant plants compromised in the expression of LAP5/PKSB and LAP6/PKSA exhibited significantly defective exine layers, and a double LAP5/PKSB LAP6/PKSA mutant appeared to completely lack an exine layer. These two genes are co-expressed with ACOS5, and recombinant LAP5/PKSB and LAP6/PKSA proteins were able to generate tri- and tetraketide alpha-pyrone compounds in vitro from a wide range of potential ACOS5-generated fatty acyl-CoA starter substrates via condensation with malonyl-CoA. These compounds would therefore appear to be required for sporopollenin biosynthesis (Kim et al. 2010). Additionally, two closely related genes, TETRAKETIDE alpha-PYRONE REDUCTASE1 (TKPR1/DRL1) and 2 (TKPR2/CCRL6), encode oxidoreductases, which have been found to be active on the tetraketide products produced by LAP5/PKSB and LAP6/PKSA. TKPR activity reduces the carbonyl function of the tetraketide alpha-pyrone compounds synthesized by LAP5/PKSB and LAP6/PKSA, and together with the activities associated with LAP5/PKSB, LAP6/PKSA and ACOS5, forms a biosynthetic pathway that ultimately produces hydroxylated alpha-pyrone compounds, potential precursors for sporopollenin (Grienenberger et al. 2010).
Arabidopsis transcription factors involved in sporopollenin and exine formation
A number of transcription factors participating in the development of exine have been described. AtMYB103/MS188 is a MYB transcription factor that is specifically expressed in the anthers and trichomes of arabidopsis (Li et al. 1999; Higginson et al. 2003). Zhang et al. (2007) have shown that AtMYB103/MS188 directly regulates the expression of the previously described exine formation gene MS2 and the callase-related A6 gene. Knock-out mutants of AtMYB103/MS188 resulted in early tapetal degeneration and abnormal microspores. Additionally, expression of the MS2 gene was not detected in the anthers of the AtMYB103/MS188 mutants (Zhang et al. 2007). The MALE STERILITY1 (MS1)/HACKLY MICROSPORE (HKM) gene, encoding a leucine zipper-like, PHD-finger motif transcription factor, is also involved in tapetum function (Ariizumi et al. 2005; Vizcay-Barrena and Wilson 2006; Ito et al. 2007; Yang et al. 2007). Phenotypic analysis of MS1 mutants by Ito et al. (2007) indicated that MS1 is required for transcriptional regulation of genes involved in primexine formation, sporopollenin synthesis and tapetum development. Lack of MS1 expression results in changes in tapetal secretion and exine structure with the appearance of autophagic vacuoles and mitochondrial swelling, suggesting that the tapetum is broken down by necrosis rather than by apoptosis as observed in the wild type (Vizcay-Barrena and Wilson 2006; Yang et al. 2007). Yang et al. (2007) further demonstrated that MS1 is expressed in the tapetal cells in a developmentally regulated manner between the late tetraspore stage and microspore release.
Another transcription factor involved in exine formation has been identified by Gibalová et al. (2009), who demonstrated that AtbZIP34 mutants exhibit defects in exine structure. The exine layer is wrinkled, and the baculae and tecta are deformed. Additionally, 50% of mutant pollen exhibited a wrinkled intine layer. Despite these abnormalities, high levels of pollen abortion or male sterility were not observed (Gibalová et al. 2009). Transcriptomic analyses revealed that expression of the proposed primexine development gene, RPG1, is significantly down-regulated in AtbZIP34 mutant pollen. Given the expression profiles of both genes, it is possible that RPG1 expression is regulated by AtbZIP34 (Gibalová et al. 2009). Analyses also suggested sporophytic and gametophytic roles for AtbZIP34 in exine and intine formation.
The observation that many of the genes described in the previous sections are predominately expressed sporophytically is somewhat at odds with the fact that exine development mostly occurs at the surface of individual microspores after meiosis. Suzuki et al. (2008) propose that this apparent contradiction may possibly be explained by many of these genes being expressed in pollen mother cells so that the relevant mRNA or proteins are inherited by the derived microspores.
Arabidopsis genes associated with probaculae formation
At present, five arabidopsis genes have been specifically associated with the formation of probaculae, which is an important component in the exine development process. The DEFECTIVE IN EXINE1 (DEX1) gene encodes a novel membrane protein that is required for anchoring sporopollenin to the surface of the microspores and is implicated in probacula formation (Paxson-Sowders et al. 1997, 2001). Sporopollenin synthesis still takes place in DEX1 mutants but primexine development is delayed and ultimately reduced, which alters membrane formation and therefore the deposition of sporopollenin. Spacers do not form in the primexine, which results in sporopollenin being randomly deposited on the plasma membrane (Paxson-Sowders et al. 2001). Additionally, sporopollenin does not appear to be anchored to the microspore and forms bulky aggregates on the developing microspore and locule walls, and the pollen wall does not form, which results in pollen degradation (Paxson-Sowders et al. 2001).
Ariizumi et al. (2008) suggested that the TRANSIENT DEFECTIVE EXINE1 (TDE1)/DE-ETIOLATED2 (DET2) gene is also involved in probacula development. Specifically, they proposed that TDE1/DET2 is involved in brassinosteroid synthesis, a hormone purported to control the rate or efficiency of the initial process of exine formation. Primexine synthesis is defective in TDE1/DET2 mutant plants which ultimately fail to produce probacula at the tetrad stage (Ariizumi et al. 2008). Additionally, globular sporopollenin is haphazardly deposited onto the microspore at the early uninucleate microspore stage (Ariizumi et al. 2008). As with DEX1 mutants, sporopollenin apparently failed to anchor to the plasma membrane of the microspore and instead aggregated on the locule wall and in the locule at the uninucleate microspore stage (Paxson-Sowders et al. 2001; Ariizumi et al. 2008). However, despite these defects, reticulate exine was clearly formed at the later stage in TDE1/DET2 mutants, which is in contrast to other mutants displaying primexine defects, such as DEX1, which always fail to produce normal exine at the later stages. This suggests that mutations in TDE1/DET2 do not result in defects at critical stages of exine development (Ariizumi et al. 2008). Expression analysis also demonstrated that brassinosteroids may be synthesized in developing microspores. The same analysis also showed that TDE1/DET2 mutations did not affect the expression of genes implicated in exine development. This suggests that brassinosteroids support exine development in a distinct pathway (Ariizumi et al. 2008).
The KNS2, 3 and 12 genes, designated type 4 genes by Suzuki et al. (2008), have also been associated with probacula formation. Type 4 mutants were shown to exhibit abnormal positioning of baculae, which were densely distributed. This suggests that the type 4 genes govern the position of probacula formation either by forming undulations on the microspore plasma membrane at the tetrad stage or by forming spacers (Suzuki et al. 2008). Additionally, Suzuki et al. (2008), using map-based cloning, were able to reveal that one of the type 4 genes, KNS2, encodes sucrose phosphate synthase, which is proposed to be potentially involved in primexine synthesis or callose wall formation, which are known to be important for the positioning of probaculae. Further studies are required to specifically determine the time and location of expression of KNS type 4 genes.
Arabidopsis genes connected to intine formation
Recently, Li et al. (2010) have proposed that the fasciclin-like arabinogalactan protein gene FLA3 is involved in the development of the intine layer by playing a role in the deposition of cellulose. The down-regulation of FLA3 via RNAi results in the appearance of a thinning intine layer and the production of ∼50% non-viable pollen grains, many of which display a wrinkled or shrunken phenotype. Expression studies showed that FLA3 is specifically expressed in pollen tubes and pollen grains, and is localized to the cell membrane (Li et al. 2010). Other arabidopsis genes have also been implicated in intine formation, including the reversibly glycosylated peptide genes, RGP1 and RGP2. Pollen grains in double-knock-out plants of RGP1 and RGP2 exhibit unusually enlarged vacuoles and a poorly defined intine layer (Drakakaki et al. 2006).
Arabidopsis genes implicated in callose wall formation
To date, three arabidopsis genes have been associated with callose wall formation. Dong et al. (2005) and Nishikawa et al. (2005) have demonstrated that the CALLOSE SYNTHASES (CALS5)/LESS ADHERENT POLLEN (LAP1) gene encodes a callose synthase essential for callose wall formation. CALS5/LAP1 mutants lack callose on the cell wall of pollen mother cells, tetrads and microspores, which ultimately results in the development of sterile pollen due to the degeneration of microspores (Dong et al. 2005). Additionally, exine structure in the mutant plants was severely deformed, affecting the baculae and tecta structure, and tryphine was haphazardly deposited as globular structures (Dong et al. 2005). This implies that the callose wall is vitally important for the formation of a properly sculpted exine (Dong et al. 2005). Expression analyses have produced varied results with regard to CALS5/LAP1 and suggest that the gene is expressed in either pollen mother cells or pollen tetrads, or possibly both cell types (Nishikawa et al. 2005).
The KNS1 and KNS11 genes constitute the type 1 genes as described and classified by Suzuki et al. (2008). Type 1 mutant plants exhibit pollen grains that display a highly collapsed exine structure in which the tecta disappear and the baculae deform into globular protrusions. Additionally, mature pollen grains of both genes were reduced in size and in number, and were distorted in shape (Suzuki et al. 2008). This phenotype closely resembled the pollen phenotype of CALS5/LAP1 mutants described above (Dong et al. 2005; Nishikawa et al. 2005; Suzuki et al. 2008). This resemblance, along with the recessive nature of the type 1 genes, suggests that KNS1 and KNS11 are expressed in pollen mother cells and are important in synthesizing or secreting callose (Suzuki et al. 2008).
Arabidopsis genes involved in tetrad separation
The QUARTET (QRT) genes have been identified as being required for pollen separation during normal pollen development (Preuss et al. 1994; Rhee and Somerville 1998; Francis et al. 2006). In wild-type arabidopsis pollen, degradation of the pollen mother cell walls takes place, which releases the individual microspores as single pollen grains (Francis et al. 2006). Mutations in any of the QRT1, QRT2 or QRT3 genes cause the outer walls of the microspores to become fused following meiosis, resulting in pollen grains being released as tetrads (Preuss et al. 1994; Rhee and Somerville 1998; Francis et al. 2006). Rhee and Somerville (1998) have demonstrated that the enzymatic removal of callose at the tetrad stage is not sufficient to release the microspores. In QRT1 and QRT2 mutants, pectic components were detectable at the time of tetrad separation, which was not the case in the wild type. This suggests that the persistence of pectin in the pollen mother cell wall is associated with tetrad separation failure (Rhee and Somerville 1998).
Pollen mother cell primary cell walls have been proposed to play a significant part in cell–cell adhesion mechanisms (Rhee and Somerville 1998). The pectins of the primary cell wall have been shown to consist mostly of homogalacturan, a polymer of β-1,4-galacturonic acid (GalUA), rhamnogalacturonan I and rhamnogalacturonan II (branching polymers of GalUA, Ara and Rha) (Brett and Waldron 1996; Tucker and Seymour 2002). As pectin is synthesized, the backbone of GalUA is in a methylesterified state that can then be demethylesterified by pectin methylesterases and cleaved by endo-polygalacturonases, which results in loosening of the cell wall (Schols and Voragen 2002; Francis et al. 2006). QRT1 and QRT2 have been proposed to encode pectin methylesterases (Francis et al. 2006). Expression analysis has shown that QRT1 is expressed shortly after meiosis is complete (Francis et al. 2006). Additionally, Rhee et al. (2003) have identified QRT3 as being an endopolygalacturonase that degrades the pectic polysaccharides of pollen mother cells. It has been demonstrated that the QRT3 gene is specifically and transiently expressed in tapetal cells during microspore release from the tetrad (Rhee et al. 2003). Immunohistochemical localization of QRT3 suggests that the protein it encodes is secreted from the tapetum during the early stages of microspore development (Rhee et al. 2003).
Genes associated with callose wall degradation have, to date, not been definitively identified. Frankel et al. (1969) and Stieglitz and Stern (1973) demonstrated that the tetrad callose wall is degraded by β-1,3-glucanase activity secreted from the tapetal cells. While a number of candidate β-1,3-glucanase-encoding genes have been identified, none has been confirmed as a callase (Hird et al. 1993). However, Hird et al. (1993) have proposed that the A6 gene may encode a component of the callase enzyme complex due to the fact that it is tapetum specific, has a strong sequence similarity to other β-1,3-glucanases, and is temporally expressed at peak levels when the plant normally expresses callase. Future identification of A6 mutant plants is needed to confirm the gene as a callase. Additionally, real-time reverse transcriptase–polymerase chain reaction analysis conducted by Zhang et al. (2007) has suggested that A6 is regulated by the AtMYB103/MS188 gene.
Summary
The genes and associated mutants described above have thus far provided clues with regard to wall development in arabidopsis, particularly with respect to exine formation and sporopollenin biosynthesis. They suggest that wall development is controlled by both the diploid sporophyte and haploid microspores, and have identified the sporophytic tapetum, in addition to the microspores themselves, as an important site for sporopollenin biosynthesis. However, large gaps in our understanding remain regarding the genetic network and biosynthetic route responsible for the formation of the pollen wall.
Flowering plant homologue genes for spore wall development are present in the moss P. patens and the lycophyte S. moellendorfii
Research into the molecular genetics of spore wall development in basal plants has thus far been extremely limited. Schuette et al. (2009), using immuno-light and immuno-electron microscopy, identified the presence of callose in the spores of physcomitrella where it was deposited in the inner exospore layer near the expanded aperture region (local expansion of the intine layer) at the proximal pole, suggesting that callose is involved in aperture expansion during wall development (Fig. 2). It is proposed that a CALS5 homologue is present in the physcomitrella genome and is involved in spore wall development (Schuette et al. 2009). However, expression studies of the CALS5 homologue, required to further address this proposition, have yet to be undertaken in physcomitrella.
In addition to CALS5, the majority of the other arabidopsis genes described in this review have been annotated and their protein sequences are available on The Arabidopsis Information Resource (TAIR) website (Website 1). We have used these protein sequences to search for homologous genes in the physcomitrella (Website 2) and selaginella genomes (Websites 3 and 4). The results suggest that homologues of all known arabidopsis pollen wall-associated genes are present in the physcomitrella genome, with the number of proposed homologues ranging from one for DEX1 and TDE1/DET2 to in excess of 50 for CYP703A2 and AtMYB103/MS188. Similar results are observed in selaginella with homologues of all but one (QRT3) of the arabidopsis pollen wall-associated genes present in its genome, ranging from two homologues in DEX1, MS1 and NEF1 to more than 50 once again for CYP703A2, AtMYB103/MS188 and the callase-related A6 gene (Table 2).
Table 2.
TBLASTN results of searches of the genomes of physcomitrella and selaginella with arabidopsis pollen wall genes. An e-value threshold of 1e−4 was used as an initial filter to determine number of homologues. Identity percentages (≥30%) and BLAST scores were then used to filter numbers further. Best match is defined as BLAST hit with the highest BLAST score.
| General function in arabidopsis | Arabidopsis gene | Gene reference | Proposed gene class | No. of proposed homologues in physcomitrella | % identity of best match in physcomitrella | BLAST score (bits) of best match in physcomitrella | No. of proposed homologues in selaginella | % identity of best match in selaginella | BLAST score (bits) of best match in selaginella |
|---|---|---|---|---|---|---|---|---|---|
| Sporopollenin biosynthesis and exine formation | MS2 | AT3G11980 | Fatty acyl reductase | 2 | 48 | 1293 | 4 | 48 | 1221 |
| YRE/WAX2/FLP1/CER3 | AT5G57800 | Aldehyde decarbonylase | 4 | 47 | 1528 | 9 | 49 | 1642 | |
| CYP703A2 | AT1G01280 | Cytochrome P450 | 31 | 46 | 1106 | >50 | 46 | 1218 | |
| CYP704B1 | AT1G69500 | Cytochrome P450 | 8 | 57 | 1612 | 27 | 60 | 1514 | |
| ACOS5 | AT1G62940 | Fatty acyl-CoA synthetase | 11 | 50 | 1378 | 26 | 51 | 1401 | |
| RPG1 | AT5G40260 | Unknown plasma membrane protein | 6 | 37 | 392 | 26 | 43 | 439 | |
| NEF1 | AT5G13390 | Unknown plastid integral membrane protein | 1 | 44 | 2487 | 2 | 41 | 2168 | |
| KNS5-10 (type 3) | – | Unknown | – | – | – | – | – | – | |
| KNS4 (type 2) | – | Unknown | – | – | – | – | – | – | |
| ABCG26 | AT3G13220 | ATP-binding cassette transporter | 3 | 46 | 1420 | 6 | 52 | 1593 | |
| LAP5/PKSB | AT4G34850 | Polyketide synthase | 21 | 57 | 1194 | 9 | 59 | 1149 | |
| LAP6/PKSA | AT1G02050 | Polyketide synthase | 16 | 50 | 944 | 9 | 47 | 932 | |
| TKPR1/DRL1 | AT4G35420 | Oxidoreductase | 7 | 51 | 868 | 32 | 52 | 850 | |
| TKPR2/CCRL6 | AT1G68540 | Oxidoreductase | 7 | 52 | 881 | 31 | 52 | 879 | |
| AtMYB103/MS188 | AT5G56110 | R2R3 MYB transcription factor | >50 | 75 | 587 | >50 | 66 | 482 | |
| MS1 | AT5G22260 | PHD-type transcription factor | 2 | 37 | 1109 | 2 | 38 | 1124 | |
| AtbZIP34 | AT2G 42380 | bZIP transcription factor | 13 | 60 | 315 | 9 | 67 | 319 | |
| Exine formation (probaculae) | DEX1 | AT3G09090 | Unknown membrane protein | 1 | 70 | 1663 | 2 | 50 | 2210 |
| TDE1/DET2 | AT2G38050 | Unknown | 1 | 40 | 455 | 4 | 43 | 495 | |
| KNS2, 3, 12 (type 4) | AT5G11110 (KNS2) | Sucrose phosphate synthase (KNS2) | 2 (KNS2) | 52 (KNS2) | 2839 (KNS2) | 4 | 53 | 2809 | |
| Intine formation | FLA3 | AT2G24450 | Fasciclin-like arabinogalactan | 0 | n/a | n/a | 5 | 33 | 221 |
| Callose wall formation | CALS5/LAP1 | AT2G13680 | Callose synthase | 12 | 64 | 6575 | 13 | 64 | 6597 |
| KNS1,11 (type 1) | – | Unknown | – | – | – | – | – | – | |
| Tetrad separation | QRT1 | AT5G55590 | Pectin methylesterase | 45 | 47 | 804 | 45 | 54 | 821 |
| QRT2 | AT3G07970 | Pectin methylesterase | 3 | 41 | 733 | 20 | 44 | 823 | |
| QRT3 | AT4G20050 | Endopolygalcturonase | 2 | 50 | 996 | 0 | n/a | n/a | |
| A6 | CAA49853 | Callase | 31 | 43 | 982 | >50 | 45 | 980 |
These results indicate that the vast majority of the pollen wall-associated genes belong to multigene families, and therefore, as with almost every developmental, signalling and metabolic context (Kafri et al. 2009), there is a high potential for genetic redundancy. Analyses of large collections of expressed sequence tag sequences have suggested that physcomitrella is a palaeopolyploid with a whole-genome duplication having occurred between 30 and 60 million years ago (Rensing et al. 2007), which may account for the presence of some of these gene duplications. While the selaginella sequence data do not indicate any ancient whole-genome duplication (Banks et al. 2011), the results here show that for most of the pollen wall-associated genes numerous copies (greater in number than in the physcomitrella genome) are also present in the selaginella genome. This suggests the occurrence of many small-scale duplication events and a greater level of gene redundancy and/or number of pseudogenes in selaginella compared with physcomitrella.
Conclusions and forward look
The presence of pollen wall-associated genes in physcomitrella and selaginella provides a strong incentive for further study in this area. Survival in a terrestrial environment has been proposed to involve the evolutionary acquisition of a number of traits, including a specialized spore wall (Wellman 2004; Cronk 2009). As described above, the morphology of moss and lycopsid spores is markedly simpler yet bears similarities to that of ‘higher’ plant pollen. Can the development of a specialized spore wall be traced to the recruitment of a few key gene products for a novel function, or did it involve a more gradual accretion of cell wall constituents in novel architectures? To what extent were the gametophyte and sporophyte involved in the deposition of the spore wall in early evolutionary history, and can this be inferred from the study of extant bryophytes and other branches of the plant evolutionary tree? The finding that homologues of angiosperm pollen cell wall-associated genes are easily identifiable in extant bryophytes and lycopsids opens the door to functional analyses of these genes. Of course, the potentially high levels of redundancy observed in physcomitrella and selaginella present challenges to a functional approach, but expression studies will help direct future research in this area and circumvent some of these difficulties. The use of high-throughput sequencing strategies and/or microarray approaches will allow researchers to identify homologues that might potentially play a role in spore wall biogenesis, and the development of techniques for gene knock-outs and gene swap experiments will allow testing of hypotheses on the conservation of gene function in the spore wall, as has already been achieved in the area of leaf, root and stomata EvoDevo studies (Harrison et al. 2005; Menand et al. 2007; Chater et al. 2011; Ruszala et al. 2011). These studies strongly suggest that true spore/pollen wall gene homologues are likely to exist in lower land plants, particularly those genes associated with wall structures that are present across embryophytes. It is also reasonable to suggest that genes associated with more specialist wall elements, such as the primexine which is only present in higher land plant groups, are less well conserved.
Although a focus in this area has been on bryophytes, such as physcomitrella (due to the availability of appropriate genetic resources), the development of novel experimental systems, such as selaginella and the liverwort Marchantia polymorpha, will allow a deeper insight into spore evolution and, more broadly, enable us to better assess whether the key mechanisms required for plant terrestrialization have been conserved over 400 million years of land plant evolution.
Sources of funding
The work was funded by the Natural Environment Research Council (UK).
Contributions by the authors
S.W. made the greatest contribution to this work and is therefore first author.
Conflicts of interest statement
None declared.
Acknowledgements
We thank Caspar Chater for advice regarding BLAST searches.
Appendix
The complete references with the full list of authors for Banks et al. (2011) and Rensing et al. (2008) are as follows:
Banks JA, Nishiyama T, Hasebe M, Bowman JL, Gribskov M, dePamphilis C, Albert VA, Aono N, Aoyama T, Ambrose BA, Ashton NW, Axtell MJ, Barker E, Barker MS, Bennetzen JL, Bonawitz ND, Chapple C, Cheng C, Correa LG, Dacre M, DeBarry J, Dreyer I, Elias M, Engstrom EM, Estelle M, Feng L, Finet C, Floyd SK, Frommer WB, Fujita T, Gramzow L, Gutensohn M, Harholt J, Hattori M, Heyl A, Hirai T, Hiwatashi Y, Ishikawa M, Iwata M, Karol KG, Koehler B, Kolukisaoglu U, Kubo M, Kurata T, Lalonde S, Li K, Li Y, Litt A, Lyons E, Manning G, Maruyama T, Michael TP, Mikami K, Miyazaki S, Morinaga S, Murata T, Mueller-Roeber B, Nelson DR, Obara M, Oguri Y, Olmstead RG, Onodera N, Petersen BL, Pils B, Prigge M, Rensing SA, Riaþo-Pachœn DM, Roberts AW, Sato Y, Scheller HV, Schulz B, Schulz C, Shakirov EV, Shibagaki N, Shinohara N, Shippen DE, Sørensen I, Sotooka R, Sugimoto N, Sugita M, Sumikawa N, Tanurdzic M, Theissen G, Ulvskov P, Wakazuki S, Weng JK, Willats WW, Wipf D, Wolf PG, Yang L, Zimmer AD, Zhu Q, Mitros T, Hellsten U, Loqué D, Otillar R, Salamov A, Schmutz J, Shapiro H, Lindquist E, Lucas S, Rokhsar D, Grigoriev IV. 2011. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332: 960–963.
Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, Tanahashi T, Sakakibara K, Fujita T, Oishi K, Shin-I T, Kuroki Y, Toyoda A, Suzuki Y, Hashimoto S, Yamaguchi K, Sugano S, Kohara Y, Fujiyama A, Anterola A, Aoki S, Ashton N, Barbazuk WB, Barker E, Bennetzen JL, Blankenship R, Cho SH, Dutcher SK, Estelle M, Fawcett JA, Gundlach H, Hanada K, Heyl A, Hicks KA, Hughes J, Lohr M, Mayer K, Melkozernov A, Murata T, Nelson DR, Pils B, Prigge M, Reiss B, Renner T, Rombauts S, Rushton PJ, Sanderfoot A, Schween G, Shiu SH, Stueber K, Theodoulou FL, Tu H, Van de Peer Y, Verrier PJ, Waters E, Wood A, Yang L, Cove D, Cuming AC, Hasebe M, Lucas S, Mishler BD, Reski R, Grigoriev IV, Quatrano RS, Boore JL. 2008. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69.
References
- Aarts MGM, Hodge R, Halantidis K, Florack D, Wilson AZ, Mulligan BJ, Stiekema WJ, Scott R, Pereira A. The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. The Plant Journal. 1997;12:615–623. doi: 10.1046/j.1365-313x.1997.00615.x. [DOI] [PubMed] [Google Scholar]
- Ahlers F, Thom I, Lambert J, Kuckuk R, Wiermann R. 1H NMR analysis of sporopollenin from Typha angustifolia. Phytochemistry. 1999;5:1095–1098. [Google Scholar]
- Ariizumi T, Hatakeyama K, Hinata K, Sato S, Kato T, Tabata S, Toriyama K. A novel male-sterile mutant of Arabidopsis thaliana, faceless pollen-1, produces pollen with a smooth surface and an acetolysis-sensitive exine. Plant Molecular Biology. 2003;53:107–116. doi: 10.1023/B:PLAN.0000009269.97773.70. [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Hatakeyama K, Hinata K, Inatsugi R, Nishida I, Sato S, Kato T, Tabata S, Toriyama K. Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. The Plant Journal. 2004;39:170–181. doi: 10.1111/j.1365-313X.2004.02118.x. [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Hatakeyama K, Hinata K, Sato S, Kato T, Tabata S, Toriyama K. The HKM gene, which is identical to the MS1 gene of Arabidopsis thaliana, is essential for primexine formation and exine pattern formation. Sexual Plant Reproduction. 2005;18:1–7. [Google Scholar]
- Ariizumi T, Kawanabe T, Hatakeyama K, Sato S, Kato T, Tabata S, Toriyama K. Ultrastructural characterization of exine development of the transient defective exine 1 mutant suggests the existence of a factor involved in constructing reticulate exine architecture from sporopollenin aggregates. Plant and Cell Physiology. 2008;49:58–67. doi: 10.1093/pcp/pcm167. [DOI] [PubMed] [Google Scholar]
- Banks JA, Nishiyama T, Hasebe M, Bowman JL, Gribskov M, De Pamphilis C, Albert VA, Aono N, Aoyama T, Ambrose BA, et al. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science. 2011;332:960–963. doi: 10.1126/science.1203810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerling DJ. Emerald planet. How plants changed Earth's history. Oxford: Oxford University Press; 2007. [Google Scholar]
- Blackmore S, Barnes SH. Embryophyte spore walls: origin, development and homologies. Cladistics. 1987;3:185–195. doi: 10.1111/j.1096-0031.1987.tb00506.x. [DOI] [PubMed] [Google Scholar]
- Blackmore S, Takahashi M, Uehara K. A preliminary phylogenetic analysis of sporogenesis in pteridophytes. In: Harley MM, Morton CM, Blackmore S, eds, editors. Pollen and spores: morphology and biology. Kew: Royal Botanic Gardens; 2000. pp. 109–124. [Google Scholar]
- Blackmore S, Wortley AH, Skvarla JJ, Rowley JR. Pollen wall development in flowering plants. New Phytologist. 2007;174:483–498. doi: 10.1111/j.1469-8137.2007.02060.x. [DOI] [PubMed] [Google Scholar]
- Brett C, Waldron K. Physiology and biochemistry of plant cell walls. London: Chapman & Hall; 1996. [Google Scholar]
- Brown RC, Lemmon BE. Spore wall development in Andreaea (Musci: Andreaeopsida) American Journal of Botany. 1984;71:412–420. [Google Scholar]
- Brown RC, Lemmon BE. Spore wall development in the liverwort, Haplomitrium hookeri. Canadian Journal of Botany. 1985;64:1174–1182. [Google Scholar]
- Brown RC, Lemmon BE. Sporogenesis in bryophytes. Advances in Bryology. 1988;3:159–223. [Google Scholar]
- Brown RC, Lemmon BE. Sporogenesis in bryophytes. In: Blackmore S, Barnes SH, eds, editors. Pollen and spores: patterns of diversification. The systematics association special. Vol. 44. Oxford: Oxford Science Publications; 1990. pp. 9–24. [Google Scholar]
- Brown RC, Lemmon BE. Spore wall development in the liverwort Fossombronia wondraczekii (Corda) Dum. Journal of the Hattori Botanical Laboratory. 1993;74:83–94. [Google Scholar]
- Brown RC, Lemmon BE, Carothers ZB. Spore wall development in Sphagnum lescurii. Canadian Journal of Botany. 1982;60:2394–2409. [Google Scholar]
- Brown RC, Lemmon BE, Renzaglia KS. Sporocytic control of spore wall pattern in liverworts. American Journal of Botany. 1986;73:593–596. [Google Scholar]
- Chater C, Kamisugi Y, Movahedi M, Fleming A, Cuming AC, Gray JE, Beerling DJ. Regulatory mechanism controlling stomatal behavior conserved across 400 million years of land plant evolution. Current Biology. 2011;12:1025–1029. doi: 10.1016/j.cub.2011.04.032. [DOI] [PubMed] [Google Scholar]
- Cronk Q. The molecular organography of plants. Oxford: Oxford University Press; 2009. [Google Scholar]
- de Azevedo Souza C, Kim SS, Koch S, Kienow l, Schneider K, Mckim SM, Haugh GW, Kombrink E, Douglas CJ. A novel fatty acyl-CoA synthase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. The Plant Cell. 2009;21:507–525. doi: 10.1105/tpc.108.062513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson HG, Lewis D. The formation of the tryphine coating the pollen grains of Raphanus, and its properties relating to the self-incompatibility system. Proceedings of the Royal Society of London. 1973;185:149–165. [Google Scholar]
- Doan TT, Carlsson AS, Hamberg M, Bülow L, Stymne S, Olsson P. Functional expression of five Arabidopsis fatty acyl-CoA reductase genes in Escherichia coli. Journal of Plant Physiology. 2009;166:787–796. doi: 10.1016/j.jplph.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Dobritsa AA, Shresta J, Morant M, Pinot F, Matsuno M, Swanson R, Møller BL, Preuss D. CYP704B1 is a long-chain fatty acid ω-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiology. 2009;151:574–589. doi: 10.1104/pp.109.144469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez E, Mercado JA, Quesada MA, Heredia A. Pollen sporopollenin: degradation and structural elucidation. Sexual Plant Reproduction. 1999;12:171–178. [Google Scholar]
- Dong X, Hong Z, Sivaramakrishnan M, Mahfouz M, Verma DP. Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant Journal. 2005;42:315–328. doi: 10.1111/j.1365-313X.2005.02379.x. [DOI] [PubMed] [Google Scholar]
- Drakakaki G, Zabotina O, Delgado I, Robert S, Keegstra K, Raikhel N. Arabidopsis reversibly glycosylated polypeptides 1 and 2 are essential for pollen development. Plant Physiology. 2006;142:1480–1492. doi: 10.1104/pp.106.086363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis KE, Lam SY, Copenhaver GP. Separation of Arabidopsis pollen tetrads is regulated by QUARTET1, a pectin methylesterase gene. Plant Physiology. 2006;142:1004–1013. doi: 10.1104/pp.106.085274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel R, Izhar S, Nitsan J. Timing of callase activity and cytoplasmic male sterility in Petunia. Biochemical Genetics. 1969;3:451–455. doi: 10.1007/BF00485605. [DOI] [PubMed] [Google Scholar]
- Gabarayeva NI. Principles and recurrent themes in sporoderm development. In: Harley MM, Morton CM, Blackmore S, eds, editors. Pollen and spores: morphology and biology. Kew: Royal Botanical Gardens,; 2000. pp. 1–16. [Google Scholar]
- Gabarayeva NI, Hemsley AR. Merging concepts: the role of self-assembly in the development of pollen wall structure. Review of Palaeobotany and Palynology. 2006;138:121–139. [Google Scholar]
- Gibalová A, Reňák D, Matczuk K, Dupl'áková N, Cháb D, Twell D, Honys D. AtbZIP34 is required for Arabidopsis pollen wall patterning and the control of several metabolic pathways in developing pollen. Plant Molecular Biology. 2009;70:581–601. doi: 10.1007/s11103-009-9493-y. [DOI] [PubMed] [Google Scholar]
- Graham LE. Origin of land plants. New York: Wiley; 1993. [Google Scholar]
- Grienenberger E, Kim SS, Lallemand B, Geoffroy P, Heintz D, Souza CD, Heitz T, Douglas CJ, Legrand M. Analysis of TETRAKETIDE alpha-PYRONE REDUCTASE function in Arabidopsis thaliana reveals a previously unknown, but conserved, biochemical pathway in sporopollenin monomer biosynthesis. The Plant Cell. 2010;22:4067–4083. doi: 10.1105/tpc.110.080036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Huang X, Zhu J, Gao J, Zhang H, Yang Z. RUPTURED POLLEN GRAIN1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis. Plant Physiology. 2008;147:852–863. doi: 10.1104/pp.108.118026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison CJ, Corley SB, Moylan EC, Alexander DL, Scotland RW, Langdale JA. Independent recruitment of a conserved developmental mechanism during leaf evolution. Nature. 2005;434:509–514. doi: 10.1038/nature03410. [DOI] [PubMed] [Google Scholar]
- Hemsley AR, Collinson ME, Kovach WL, Vincent B, Williams T. The role of self-assembly in biological systems: evidence from iridescent colloidal sporopollenin in Selaginella megaspore walls. Philosophical Transactions of the Royal Society of London, Series B. 1994;345:163–173. [Google Scholar]
- Hemsley AR, Collinson ME, Vincent B, Griffiths PC, Jenkins PD. Self-assembly of colloidal units in exine development. In: Harley MM, Morton CM, Blackmore S, eds, editors. Pollen and spores: morphology and biology. Kew: Royal Botanic Gardens,; 2000. pp. 31–44. [Google Scholar]
- Heslop-Harrison J. An ultrastructural study of pollen wall ontogeny in Silene pendula. Grana Palynologica. 1963;4:7–24. [Google Scholar]
- Heslop-Harrison J. The pollen grain wall. Science. 1968;161:230–237. doi: 10.1126/science.161.3838.230. [DOI] [PubMed] [Google Scholar]
- Higginson T, Li SF, Parish RW. AtMYB103 regulates tapetum and trichome development in Arabidopsis thaliana. Plant Journal. 2003;35:177–192. doi: 10.1046/j.1365-313x.2003.01791.x. [DOI] [PubMed] [Google Scholar]
- Hird DL, Worrall D, Hodge R, Smartt S, Paul W, Scott R. The anther-specific protein encoded by the Brassica napus and Arabidopsis thaliana A6 gene displays similarity to β-1,3-glucanase. The Plant Journal. 1993;4:1023–1033. doi: 10.1046/j.1365-313x.1993.04061023.x. [DOI] [PubMed] [Google Scholar]
- Ito T, Nagata N, Yoshiba Y, Ohme-takagi M, Ma H, Shinozaki K. Arabidopsis MALE STERILITY1 encodes a PHD-type transcription factor and regulates pollen and tapetum development. The Plant Cell. 2007;19:3549–3562. doi: 10.1105/tpc.107.054536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafri R, Springer M, Pilpel Y. Genetic redundancy: new tricks for old genes. Cell. 2009;136:389–392. doi: 10.1016/j.cell.2009.01.027. [DOI] [PubMed] [Google Scholar]
- Kenrick P, Crane PR. The origin and early diversification of land plants: a cladistic study. Washington: Smithsonian Institution Press; 1997. [Google Scholar]
- Kim SS, Grienenberger E, Lallemand B, Colpitts CC, Kim SY, Souza CD, Geoffroy P, Henitz D, Krahn D, Kaiser M, Kombrink E, Heitz T, Suh DY, Legrand M, Douglas CJ. LAP6/POLYKETIDE SYNTHASE A and LAP5/POLYKETIDE SYNTHASE B encode hydroxyalkyl alpha-pyrone synthases required for pollen development and sporopollenin biosynthesis in Arabidopsis thaliana. The Plant Cell. 2010;22:4045–4066. doi: 10.1105/tpc.110.080028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yu MA, Geng LL, Zhao J. The fasciclin-like arabinogalactan protein gene, FLA3, is involved in microspore development of Arabidopsis. The Plant Journal. 2010;64:482–497. doi: 10.1111/j.1365-313X.2010.04344.x. [DOI] [PubMed] [Google Scholar]
- Li SF, Higginson T, Parish RW. A novel MYB-related gene from Arabidopsis thaliana expressed in development anthers. Plant and Cell Physiology. 1999;40:343–347. doi: 10.1093/oxfordjournals.pcp.a029548. [DOI] [PubMed] [Google Scholar]
- Lugardon B. Pteridophyte sporogenesis: a survey of spore wall ontogeny and fine structure in a polyphyletic plant group. In: Blackmore S, Knox RB, eds, editors. Microspores: evolution and ontogeny. London: Academic Press; 1990. pp. 95–120. [Google Scholar]
- Lugardon B. Exine formation in Chamaecyparis lawsoniana (Cupressaceae) and a discussion on pteridophyte exospores and gymnosperm exine ontogeny. Review of Palaeobotany and Palynology. 1994;85:35–51. [Google Scholar]
- Menand B, Yi K, Jouannic S, Hoffman L, Ryan E, Linstead P, Schaefer DG, Dolan L. An ancient mechanism controls the development of cells with a rooting function in land plants. Science. 2007;316:1477–1480. doi: 10.1126/science.1142618. [DOI] [PubMed] [Google Scholar]
- Meuter-Gerhards A, Riegart S, Wiermann R. Studies on sporopollenin biosynthesis in Curcurbita maxima (DUCH)-II: The involvement of aliphatic metabolism. Journal of Plant Physiology. 1999;154:431–436. [Google Scholar]
- Morant M, Jørgensen K, Schaller H, Pinot F, Møller BL, Werck-Reichhart D, Bak S. CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. The Plant Cell. 2007;19:1473–1487. doi: 10.1105/tpc.106.045948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morbelli MA. Megaspore wall in Lycophyta—ultrastructure and function. Review of Palaeobotany and Palynology. 1995;85:1–12. [Google Scholar]
- Nishikawa S, Zinkl GM, Swanson RJ, Maruyama D, Preuss D. Callose (beta-1,3 glucan) is essential for Arabidopsis pollen wall patterning, but not tube growth. BMC Plant Biology. 2005;5:22. doi: 10.1186/1471-2229-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxson-Sowders DM, Owen HA, Makaroff CA. A comparative ultrastructural analysis of exine pattern development in wild-type Arabidopsis and a mutant defective in pattern formation. Protoplasma. 1997;198:53–65. [Google Scholar]
- Paxson-Sowders DM, Dodrill CH, Owen HA, Makaroff CA. DEX1, a novel plant protein, is required for exine pattern formation during pollen development in Arabidopsis. Plant Physiology. 2001;127:1739–1749. [PMC free article] [PubMed] [Google Scholar]
- Piffanelli P, Ross JHE, Murphy DJ. Biogenesis and function of the lipidic structures of pollen grains. Sexual Plant Reproduction. 1998;11:65–80. [Google Scholar]
- Preuss D, Rhee SY, Davis RW. Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science. 1994;264:1458–1460. doi: 10.1126/science.8197459. [DOI] [PubMed] [Google Scholar]
- Quatrano RS, McDaniel SF, Khandelwal A, Perroud P, Cove DJ. Physcomitrella patens: mosses enter the genomic age. Current Opinion in Plant Biology. 2007;10:182–189. doi: 10.1016/j.pbi.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Qui YL, Li LB, Wang B, Chen ZD, Knoop V, Groth-Malonek M, Dombrovska O, Lee J, Kent L, Rest J, Estabrook GF, Hendry TA, Taylor DW, Testa CM, Ambros M, Crandall-Stotler B, Duff RJ, Stech M, Frey W, Quandt D, Davis CC. The deepest divergences in land plants inferred from phylogenomic evidence. Proceedings of the National Academy of Sciences of the USA. 2006;103:15511–15516. doi: 10.1073/pnas.0603335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilichini TD, Friedmann MC, Samuels AL, Douglas CJ. ATP-binding cassette transporter G26 (ABCG26) is required for male fertility and pollen exine formation in Arabidopsis thaliana. Plant Physiology. 2010;154:678–690. doi: 10.1104/pp.110.161968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Ick J, Fawcett JA, Lang D, Zimmer A, Van De Peer Y, Reski R. An ancient genome duplication contributed to the abundance of metabolic genes in the moss Physcomitrella patens. BMC Evolutionary Biology. 2007;7:130. doi: 10.1186/1471-2148-7-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- Reski R, Cove DJ. Physcomitrella patens. Current Biology. 2004;14:261–262. doi: 10.1016/j.cub.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Rhee SY, Somerville CR. Tetrad pollen formation in quartet mutants of Arabidopsis thaliana is associated with persistence of pectic polysaccharides of the pollen mother cell. The Plant Journal. 1998;15:79–88. doi: 10.1046/j.1365-313x.1998.00183.x. [DOI] [PubMed] [Google Scholar]
- Rhee SY, Osborne E, Poindexter PD, Somerville CR. Microspore separation in the quartet 3 mutants of Arabidopsis is impaired by a defect in a developmentally regulated polygalacturonase required for pollen mother cell wall degradation. Plant Physiology. 2003;133:1170–1180. doi: 10.1104/pp.103.028266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland O, Lee R, Franke R, Schreiber L, Kunst L. The CER3 wax biosynthetic gene from Arabidopsis thaliana is allelic to WAX2/YRE/FLP1. Federation of European Biochemical Societies Letters. 2007;581:3538–3544. doi: 10.1016/j.febslet.2007.06.065. [DOI] [PubMed] [Google Scholar]
- Ruszala EM, Beerling DJ, Franks PJ, Chater C, Casson SA, Gray JE, Hetherington AM. Land plants acquired active stomatal control early in their evolutionary history. Current Biology. 2011;21:1030–1035. doi: 10.1016/j.cub.2011.04.044. [DOI] [PubMed] [Google Scholar]
- Schols HA, Voragen AGJ. The chemical structure of pectins. In: Seymour GB, Knox PG, eds, editors. Pectins and their manipulation. Boca Raton, FL: CRC Press; 2002. pp. 1–29. [Google Scholar]
- Schuette S, Wood AJ, Geisler M, Geisler-Lee J, Ligrone R, Renzaglia KS. Novel localization of callose in the spores of Physcomitrella patens and phylogenomics of the callose synthase gene family. Annals of Botany. 2009;103:749–756. doi: 10.1093/aob/mcn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RJ, Hodge R, Paul W, Draper J. The molecular biology of anther differentiation. Plant Science. 1991;80:167–191. [Google Scholar]
- Stieglitz H, Stern H. Regulation of β-1,3-glucanase activity in developing anthers of Lilium. Developmental Biology. 1973;34:169–173. doi: 10.1016/0012-1606(73)90347-3. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Masaoka K, Nishi M, Nakamura K, Ishiguro S. Identification of kaonashi mutants showing abnormal pollen exine structure in Arabidopsis thaliana. Plant and Cell Physiology. 2008;49:1465–1477. doi: 10.1093/pcp/pcn131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor WA. Ultrastructure of sphenophyllalean spores. Review of Palaeobotany and Palynology. 1986;47:105–128. [Google Scholar]
- Tryon AF, Lugardon B. Spores of the Pteridophyta. Surface, wall structure and diversity based on electron microscope studies. New York: Springer; 1991. [Google Scholar]
- Tucker GA, Seymour GB. Modification and degradation of pectins. In: Seymour GB, Knox PG, eds, editors. Pectins and their manipulation. Boca Raton, FL: CRC Press; 2002. pp. 150–173. [Google Scholar]
- Uehara K, Kurita S. An ultrastructural study of spore wall morphogenesis in Equisetum arvense. American Journal of Botany. 1989;76:939–951. [Google Scholar]
- Uehara K, Kurita S. Ultrastructural study on spore wall morphogenesis in Lycopodium clavatum (Lycopodiaceae) American Journal of Botany. 1991;78:24–36. [Google Scholar]
- Uehara K, Kurita S, Sahashi N, Ohmoto T. Ultrastructural study of microspore wall morphogenesis in Isoetes japonica (Isoetaceae) American Journal of Botany. 1991;78:1182–1190. [Google Scholar]
- Villarreal JC, Renzaglia KS. Sporophyte structure in the neotropical hornwort Phaeomegaceros fimbriatus: implications for phylogeny, taxonomy, and character evolution. International Journal of Plant Science. 2006;167:413–427. [Google Scholar]
- Vizcay-Barrena G, Wilson ZA. Altered tapetal PCD and pollen wall development in the Arabidopsis ms1 mutant. Journal of Experimental Botany. 2006;57:2709–2717. doi: 10.1093/jxb/erl032. [DOI] [PubMed] [Google Scholar]
- Waterkeyn L, Bienfait A. Morphologie et nature des parois sporocytaires chez les pteridophytes. La Cellule. 1971;69:7–23. [Google Scholar]
- Waters ER. Molecular adaptation and the origin of land plants. Molecular Phylogenetics and Evolution. 2003;29:456–463. doi: 10.1016/j.ympev.2003.07.018. [DOI] [PubMed] [Google Scholar]
- Website 1. The Arabidopsis Information Resource www.arabidopsis.org (9 May 2011)
- Website 2. Cosmoss www.cosmoss.org (9 May 2011)
- Website 3. National Center for Biotechnology Information www.ncbi.nlm.nih.gov (25 May 2011)
- Website 4. Selaginella Genome Project selaginella.genomics.purdue.edu/ (25 May 2011)
- Wellman CH. Origin, function and development of the spore wall in early land plants. In: Hemsley AR, Poole I, eds, editors. Evolution of plant physiology. Kew: Royal Botanic Gardens; 2004. pp. 43–63. [Google Scholar]
- Wellman CH. Ultrastructure of dispersed and in situ specimens of the Devonian spore Rhabdosporites langii (Eisenack) Richardson 1960: evidence for the evolutionary relationships of Progymnosperms. Palaeontology. 2009;52:139–167. [Google Scholar]
- Yang C, Vizcay-Barrena G, Conner K, Wilson ZA. MALES STERILITY1 is required for tapetal developmental and pollen wall biosynthesis. The Plant Cell. 2007;19:3530–3548. doi: 10.1105/tpc.107.054981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi B, Zeng FQ, Lei SL, Chen YN, Yao XQ, Zhu Y, Wen J, Shen JX, Ma CZ, Tu JX, Fu TD. Two duplicate CYP704B1-homologous genes BnMs1 and BnMs2 are required for pollen exine formation and tapetal development in Brassica napus. The Plant Journal. 2010;63:925–938. doi: 10.1111/j.1365-313X.2010.04289.x. [DOI] [PubMed] [Google Scholar]
- Zavada MS, Gabarayeva N. Comparative pollen wall development of Welwitschia mirabilis angiosperms. Bulletin of the Torrey Botanical Club. 1991;118:292–302. [Google Scholar]
- Zhang Z, Zhu J, Gao J, Wang C, Li H, Li H, Zhang H, Zhang S, Wang D, Wang Q, Huang H, Xia H, Yang Z. Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis. The Plant Journal. 2007;52:528–538. doi: 10.1111/j.1365-313X.2007.03254.x. [DOI] [PubMed] [Google Scholar]