Background: Raffinose, an abundant carbohydrate in plants, is degraded into galactose and sucrose by intestinal microbial enzymes.
Results: AgaSK is a protein coupling galactosidase and sucrose kinase activity. The structure of the galactosidase domain sheds light onto substrate recognition.
Conclusion: AgaSK produces sucrose-6-phosphate directly from raffinose.
Significance: Production of sucrose-6-phosphate directly from raffinose points toward a novel glycolytic pathway in bacteria.
Keywords: Bacterial Metabolism, Enzyme Mechanisms, Glycosidases, Multifunctional Enzymes, Protein Structure, Alpha-Galactosidase, Raffinose and Sucrose Degradation, Sucrose Kinase
Abstract
α-Galactosides are non-digestible carbohydrates widely distributed in plants. They are a potential source of energy in our daily food, and their assimilation by microbiota may play a role in obesity. In the intestinal tract, they are degraded by microbial glycosidases, which are often modular enzymes with catalytic domains linked to carbohydrate-binding modules. Here we introduce a bifunctional enzyme from the human intestinal bacterium Ruminococcus gnavus E1, α-galactosidase/sucrose kinase (AgaSK). Sequence analysis showed that AgaSK is composed of two domains: one closely related to α-galactosidases from glycoside hydrolase family GH36 and the other containing a nucleotide-binding motif. Its biochemical characterization showed that AgaSK is able to hydrolyze melibiose and raffinose to galactose and either glucose or sucrose, respectively, and to specifically phosphorylate sucrose on the C6 position of glucose in the presence of ATP. The production of sucrose-6-P directly from raffinose points toward a glycolytic pathway in bacteria, not described so far. The crystal structures of the galactosidase domain in the apo form and in complex with the product shed light onto the reaction and substrate recognition mechanisms and highlight an oligomeric state necessary for efficient substrate binding and suggesting a cross-talk between the galactose and kinase domains.
Introduction
Recent studies have suggested that the human intestinal microbiome plays an important role in the weight differences between human beings, namely that it could be significant in obesity (1–3). Indeed, obesity can be linked to an alteration of the ratio of Bacteroidetes and Firmicutes in the microbiome (4). Together, these bacteria represent over 90% of the bacterial phylotypes in the intestinal tract (5). This advocates a change in the fermentation pattern and a more efficient energy extraction from foodstuff operated by Firmicutes (6, 7). In particular, non-digestible carbohydrates can be metabolized by bacteria residing in the intestinal tract, and the imbalance of calorie consumption and expenditure causing obesity can be attributed to variations of the intestinal microbial ecology (8).
Among non-digestible carbohydrates, essentially originating from plant cell walls, α-galactosides are among the most abundant oligosaccharides. For example, they represent from 39 to 53 g/kg in soybean meals (9). They consist of galactose units α-(1,6) linked to a mannopyranose backbone (galactomannans) or α-(1,6) linked to glucose (melibiose) or sucrose (raffinose) or raffinose (stachyose), with sucrose and raffinose being the most abundant soluble carbohydrates found in plant cell walls (9). However, there exists no α-(1,6)-galactosidase activity in the human intestine mucosa, and α-galactosides are exclusively fermented by microbial α-(1,6)-galactosidases (EC 3.2.1.22). Studies of the bacterial metabolism of α-galactosides (10, 11) have shown that melibiose and raffinose penetrate in the cell, where they are hydrolyzed into galactose and glucose and into galactose and sucrose, respectively. Sucrose and galactose are subsequently phosphorylated by a kinase and utilized in the glycolytic pathways (12, 13).
According to the sequence-based classification of glycoside hydrolases (GHs)2 (Carbohydrate-Active Enzymes Database, CAZy, classification) (14), α-galactosidases form eukaryotic organisms are mainly grouped into families GH27 and GH36, but those from microbial sources are also found in glycoside hydrolase families GH4, GH57, GH97, and GH110. Iterative database searches have shown that families GH27 and GH36, together with families GH31 and GH66, share a common evolutionary origin with family GH13 (15). For that matter, families GH27 and GH36, together with family GH31, form glycoside hydrolase clan D (16). Crystal structures of the clinically important eukaryotic α-galactosidase and α-N-acetylgalactosaminidase have been reported by the Garman group almost a decade ago (17, 18), and a wealth of supplementary structural information for other family GH27 members has become available later on. On the basis of the high level of sequence homology between members of family GH27 and GH36, which reflects intrinsically structural homology, Comfort et al. (19) were able to infer the catalytic residues of Thermotoga maritima TmGalA, as well as to confirm their identity by elegant biochemical studies. For family GH36, only the crystal structures of T. maritima TmGalA (Protein Data Bank (PDB) 1zy9) and Lactobacillus brevis LbR11 (PDB 3mi6) have been deposited within the Protein Data Bank by Structural Genomics Consortia, and the crystal structure of Lactobacillus acidophilus NCFM LaMel36A (PDB 2xn0) has been solved very recently (20).
Family GH36 members originate from bacteria, fungi, and plants and carry out hydrolysis with a net retention of the anomeric configuration, which is why many enzymes from this family have been shown to possess transglycosylation activity (21–27). For their ability to hydrolyze α-galactosides non-digestible by humans and to synthesize diverse oligosaccharides, GH36 α-galactosidases have potential applications for the production of prebiotics. These non-digestible oligosaccharides are beneficial for human health by their ability to stimulate selectively the growth of microbiota with advantageous physiological effects on the host (28–30).
In this work, we have characterized from a functional point of view a bifunctional enzyme, hereafter called α-galactosidase/sucrose kinase (AgaSK), from Ruminococcus gnavus E1. R. gnavus is a strict anaerobic Gram-positive bacterium from the phylum Firmicutes. It belongs to the most common 57 species present in 90% of individuals (31). The enzyme AgaSK is composed of a GH36 α-galactosidase and a kinase domain and is able to release galactose from α-galactosides and to phosphorylate sucrose provided either by raffinose hydrolysis or by the environmental medium. The crystal structures of the galactosidase domain in the apo form and in complex with the product shed light onto the reaction and substrate recognition mechanisms and highlight an oligomeric state necessary for efficient substrate binding and suggesting a cross-talk between the galactose and kinase domains.
EXPERIMENTAL PROCEDURES
Cecal Content Collection and Extraction of RNA
Animal experiments were performed according to the guidelines of the French Ethics Committee. Fisher axenic rats came from the Animalerie Axénique de Micalis (ANAXEM) platform (INRA Jouy-en-Josas, France) and were reared in Trexler type isolators (La Calhène, Vélizy-Villacoublay, France). They were inoculated with ∼109 cells of the R. gnavus E1 strain (0.5 ml of fresh culture) by intragastric route. After 1 week, the animals were sacrificed, and the cecal contents were collected. Two hundred mg of fresh material were used for total RNA extraction according to the protocol described by Doré et al. (32) and cleaned up with the RNeasy mini kit (Qiagen). TURBOTM DNase (Ambion®) was used for elimination of genomic DNA as recommended by the supplier.
RT-PCR
cDNA synthesis was performed by reverse transcription (RT) of 100 ng of RNA primed with 50 ng of random primers. The reaction was carried out at 50 °C for 1 h with the SuperScriptTM III reverse transcriptase (Invitrogen), as recommended by the supplier. The enzyme was inactivated by heating for 15 min at 70 °C, and 1 μl of the mixture was used directly as a template for PCR amplifications. PCR reactions were performed in a 50-μl volume by using the Platinum® Taq DNA polymerase (Invitrogen) under the following conditions: 94 °C for 2 min; 30 cycles at 94 °C for 30 s, 50 °C for 30 s, and 72 °C for 1 min; and a final elongation step at 72 °C for 10 min. As a control, additional reactions were performed by using as a template an RT mixture without enzyme, an RT mixture without RNA, and chromosomal DNA.
Bioinformatic Analysis
Manual validation of the automatic annotation of the R. gnavus E1 genome was performed using the MaGe (Magnifying Genomes) web-based interface from Genoscope. Putative transcriptional terminators were predicted in silico using the RNAfold program. Functional predictions were based on sequence comparisons with the help of the sequence databases Profils pour l'Identification Automatique du Métabolisme (PRIAM), Clusters of Orthologous Groups of protein (COG) (www.ncbi.nlm.nih.gov/COG), and Swiss-Prot the domain/motif database InterProScan.
agaSK Amplification and Cloning into pOPIN E Expression Vector
The agaSK gene was cloned from the genomic DNA of R. gnavus E1 using DNA polymerase (PrimeSTAR HS DNA polymerase, TAKARA BIO Inc.). PCR reaction primers (forward, 5′-AGGAGATATACCATGGCAATTATATACAATCCA-3′; reverse, 5′-GTGATGGTGATGTTTCTGTTCATAAACACTTCC-3′) were used for 30 cycles of denaturation (1 min, 98 °C), annealing (1.5 min, 48 °C), and extension (3 min, 60 °C) in a Mastercycler® gradient thermocycler (Eppendorf, Hamburg, Germany). The PCR products amplified with the In-Fusion primers were combined with the expression vector pOPIN E, kindly provided by Dr. Ray Owens (Oxford University), following the manufacturer's instructions (In-FusionTM, Clontech). The recombinant vectors were checked by double-stranded DNA sequencing (Genome Express, Meylan, France). The In-Fusion system led to the production of AgaSK with a His6 tag at the C terminus.
Expression and Purification of AgaSK
Recombinant AgaSK was synthesized from Escherichia coli BL21 (DE3) grown in Luria broth, containing 50 mg·liter−1 ampicillin, for 4 h. Bacterial lysis was carried out with a cell disruptor (Constant Systems Ltd.) in the binding buffer for affinity chromatography (50 mm sodium phosphate buffer, pH 8, 40 mm imidazole) at 1.37 kbar. After a centrifugation run at 33,000 × g, the soluble fraction was loaded onto a nickel-nitrilotriacetic acid column, and the recombinant protein was eluted with 400 mm imidazole in sodium phosphate buffer, pH 8. Fractions were assayed for α-galactosidase activity, pooled, and dialyzed against McIlvaine's buffer, pH 6. The protein was purified to 95% homogeneity, with an overall yield of 20 mg per liter of culture.
α-Galactosidase Activity Measurement and Analysis of Mono- and Disaccharides
α-Galactosidase activity was assayed by monitoring the absorbance at 410 nm of p-nitrophenol released from 2 mm p-nitrophenyl-α-d-galactopyranoside (PNPG) in McIlvaine's buffer, pH 6, at 37 °C, with 9.8 nm of enzyme. One unit of α-galactosidase activity was defined as the amount of enzyme required to liberate one μmol of p-nitrophenol per minute at 37 °C. Hydrolysis products formed by recombinant AgaSK with 2 mm melibiose or raffinose in McIlvaine's buffer, pH 6, at 37 °C for 30 min were detected by thin-layer chromatography (TLC, Silica gel 60, Merck) and separated by high performance anion exchange chromatography coupled with pulsed amperometric detection (Dionex, Sunnyvale, CA). TLC plates were developed with n-propyl alcohol/water (8:2; v/v), and the sugars were detected by heating at 140 °C for 5 min after spraying with 0.1% orcinol dissolved in 5% sulfuric acid (H2SO4).
The release of products from raffinose and melibiose by the action of AgaSK was analyzed by HPLC coupled with pulsed amperometric detection equipped with a Carbo-PacPA-100 analytical column (250 × 4 mm), a GP40 gradient pump, and an AS3500 auto-sampler. The protocol used is the same as described by Cervera Tison et al. (33), except that the A buffer was composed of 5 mm NaOAc and 80 mm NaOH. To evaluate the activity of the AgaSK on α-galactosides (melibiose and raffinose), the initial slopes of progress curves were used to determine the catalytic efficiency (kcat/Km) of the reaction, following the equation of Matsui et al. (34). All assays were carried out in triplicate.
Crystallization
Crystals of the glycosidase domain of AgaSK (AgaSK-tru) were grown at room temperature by the vapor diffusion method in hanging drops consisting of equal volumes (1 + 1 μl) of full-length AgaSK solution at 7 mg/ml and crystallization solution composed of 14% (w/v) polyethylene glycol 8000 and 0.1 m Hepes, pH 7.5. Due to spontaneous in situ proteolysis, only crystals of the α-galactosidase domain could be obtained. Crystals of the complex between AgaSK-tru and galactose were obtained by soaking crystals of AgaSK-tru obtained as described above in a stabilizing solution containing 18% (w/v) polyethylene glycol 8000, 0.1 m Hepes, pH 7.5, and 20 mm galactose.
Data Collection and Structure Determination
Crystals selected for data collection were rapidly transferred into the relevant mother liquor supplemented with 30% glycerol and were flash-frozen in liquid nitrogen. X-ray data for AgaSK-tru, extending to 2.0 Å resolution, were measured at beam line Proxima1 (SOLEIL, Gif-sur-Yvette, France), and a data set at 1.45 Å resolution for AgaSK-tru and a data set at 1.35 Å resolution for AgaSK-tru in complex with galactose were subsequently measured at beam lines ID14-1 and ID29, respectively (European Synchrotron Radiation Facility (ESRF), Grenoble, France). Oscillation images of native data sets were integrated with Mosflm (35), and data for the complex of AgaSK-tru with galactose were processed with XDS (36). All scaling was performed with SCALA (37).
The structure of AgaSK-tru was determined with PHASER (38) using the structure of the α-galactosidase LbR11 from L. brevis (PDB 3mi6) as the search model. The resulting model was rebuilt in an automatic fashion with the program ARP/wARP (39) and completed manually using the program COOT (40). The resulting model was refined with REFMAC (41). For the structure of the enzyme-galactose complex, the final native model was subjected to rigid body refinement and further refined with REFMAC (41).
The final protein structures encompass residues Ala-2 to Leu-720 and, in addition to galactose in the complex structure, also contain several glycerol and polyethylene glycol molecules and a number of sodium and phosphate ions, arising form the purification, crystallization, and cryo buffers. The root mean square deviation between the apo- and sugar-bound forms is 0.07 Å for 720 Calpha atoms. The stereochemistry of the model was analyzed with MolProbity (42), and only one residue, Ser-225, was found in the disallowed regions of the Ramachandran plot. This residue is part of a loop region of the β-supersandwich domain contacting the central (β/α)8 barrel. The PISA server (43) was used for quaternary structure analysis, and figures were generated with PYMOL (44). Data collection and refinement statistics are reported in Table 1. The homology model of the kinase domain was derived with the Phyre server (45).
TABLE 1.
Data collection and refinement statistics
| AgaSK-tru | AgaSK-tru-galactose complex | |
|---|---|---|
| Data processing | ||
| Beamline | ID14–1 | ID29 |
| Space group | I222 | I222 |
| Cell dimensions, a, b, c (Å) | 105.06, 111.65, 154.89 | 105.02, 111.60, 155.29 |
| Resolution (Å)a | 36–1.45 (1.53–1.45) | 47–1.35 (1.42–1.35) |
| No. of observations | 454,881 | 1,029,088 |
| No. of unique observations | 158,241 | 198,265 |
| Rsymb | 0.077 (0.459) | 0.093 (0.470) |
| Mean I/σ(I) | 11.0 (2.2) | 11.8 (3.0) |
| Redundancy | 2.9 (2.6) | 5.2 (5.1) |
| Completeness (%) | 99.0 (80.2) | 98.6 (98.1) |
| B-factor from Wilson plot (Å2) | 10.50 | 7.99 |
| Refinement | ||
| Resolution (Å) | 36–1.45 (1.488–1.45) | 47–1.35 (1.385–1.35) |
| Rworkc (%) | 13.01 (24.0) | 12.43 (19.40) |
| Rfreed (%) | 14.96 (25.8) | 14.17 (20.09) |
| No. of reflections used in refinement | 150,207 (10,384) | 188,194 (13,810) |
| No. of free reflections | 8033 (525) | 10,028 (735) |
| r.m.s. deviationse | ||
| Bond length (Å) | 0.013 | 0.014 |
| Bond angles (°) | 1.45 | 1.01 |
| Chiral volume (Å 3) | 0.091 | 0.093 |
| Mean B-factors (Å2) | ||
| Main/side chain | 11.03/13.31 | 9.64/12.08 |
| Galactose | 8.29 | |
| Solvent/other ligands | 22.85/21.25 | 25.02/20.60 |
| r.m.s. deviations on B-factors (Å2) | ||
| Main chain | 0.534 | 0.589 |
| Side chain | 1.491 | 1.480 |
| Ramachandran plot statisticsf | ||
| % of residues in favored regions | 97.35 | 97.63 |
| % of residues in allowed regions | 99.86 | 99.86 |
| % of outliers | 0.14 | 0.14 |
a Throughout the table, the values in parentheses are for the outermost resolution shell.
b Rsym = Σhkl(Σi|Ihkl-〈Ihkl〉|)/Σhkl|〈Ihkl〉|.
c Rwork = Σhkl‖Fo| − k|Fc‖/Σhkl|Fo|.
d Rfree is calculated for randomly selected reflections excluded from refinement.
e Root mean square deviation from ideal geometry.
f Ramachandran plot statistics have been calculated with the MolProbity server.
Fluorescence Measurements
All experiments were performed at 25 ± 0.1 °C using a SAFAS flx Xenius 5117 spectrofluorometer. All spectra were corrected for buffer fluorescence. Fluorescence measurements were routinely carried out for 2 ml of AgaSK at 0.1 μm in 50 mm Hepes/KOH buffer, pH 6, containing 1.0 mm MgCl2. Increasing concentrations of ATP (from 25 to 250 μm) were added, and the emission fluorescence was scanned in the range of 310–380 nm, upon excitation at 282 nm. ATP binding was monitored by the variation of tryptophan intrinsic fluorescence of AgaSK produced by each addition of ATP. Corrections for the inner filter effect of the ligand were performed under the same conditions by using N-acetyl-tryptophanamide (Sigma). Peak integration was carried out at each ATP concentration, and the observed changes in fluorescence intensity were used for the calculation of affinity. Data collection was performed in triplicate, and the curve fitting of the data was performed by using the GraFit 4.0 software, as described by Forouhar et al. (46).
Kinase Activity Measurements
Sugar phosphorylation assays were carried out in a total volume of 10 μl. 0.44 μm AgaSK was incubated for 45 min at 37 °C with 2 mm of the tested sugar, 0.4 mm ATP, 1 mm MgCl2, and 0.05 μl of [γ-33P]ATP (0.5 μCi) in 50 mm Hepes/KOH, pH 6. The phosphorylation reaction was stopped by incubating the samples for 5 min at 80 °C and then placing them on ice. 1.5 μl of the reaction mixtures were analyzed by TLC using silica gel plates (Merck). Plates were developed at room temperature in a saturated chamber containing n-propyl alcohol and water (8:2, v/v) as a solvent system. After development, the plates were dried and exposed overnight to autoradiography. The plates were then sprayed with 0.1% orcinol dissolved in 5% H2SO4 and heated at 150 °C for 15 min for visualization of the sugar spots.
Matrix-assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry
MALDI-TOF-MS experiments were performed on an Autoflex III TOF/TOF mass spectrometer (Bruker Daltonics, Bremen, Germany), equipped with a Smartbeam laser (355 nm, 200 Hz) in positive and negative ionization mode with a reflector detection. The matrix was prepared by diluting 50 mg of 2,5-dihydroxybenzoic acid in 490 μl of a solution of H2O/acetonitrile 1:1. 10 μl of N,N-dimethylaniline (purchased from Fisher Scientific) were added to the mixture. Samples were diluted 10-fold and mixed with matrix solution of N,N-dimethylaniline/2,5-dihydroxybenzoic acid in a 1:1 ratio (v/v). 1 μl of the mixture was deposited on a polished steel MALDI target plate. Mass data were recorded using FlexControl and processed using FlexAnalysis (Bruker Daltonics). The m/z range was 180–1000.
Electrospray Ionization Quadrupole Time-of-Flight Mass Spectrometry
ESI-Q-TOF-MS experiments were performed on a Micromass Q-TOF Ultima Global mass spectrometer (Waters, Manchester, UK) in positive and negative ion mode. Samples were diluted 1000-fold in 1:1 MeOH/H2O and introduced at a flow rate of 2.5 μl/min into the electrospray ion source (the ion spray voltage was maintained at 3.1 kV). For collision-induced dissociation experiments, argon was used as the collision gas. The collision energy was optimized for each sample. The fragments were annotated according to the nomenclature of Domon and Costello (47). Mass data were recorded using MassLynx 4 (Waters). The m/z range was 150–1200.
RESULTS AND DISCUSSION
R. gnavus E1 Is Able to Metabolize Raffinose in the Intestinal Tract
R. gnavus E1 is a Gram-positive bacterium found in the human alimentary tract. To study its ability to hydrolyze non-digestible carbohydrates, and in particular α-linked galactosides, we first tested its ability to grow on melibiose and raffinose as a sole carbon and energy source. We observed that R. gnavus E1 was able to grow on these two α-galactosides, suggesting that the bacterium possesses not only a suitable sugar uptake system but also an intracellular α-galactosidase and an efficient metabolic pathway for stepwise degradation of the released monosaccharides. By activity assays using extracts of R. gnavus E1 grown on several sugars, as well as Tryptone, we confirmed the presence of a constitutively expressed enzyme, capable of hydrolyzing PNPG.
To find the enzyme(s) and the corresponding gene(s) responsible for this carbon source utilization, an analysis of the R. gnavus E1 genome was undertaken. This analysis revealed an 8.5-kb cluster, composed of six genes, potentially involved in raffinose and sucrose metabolism (supplemental Fig. 1). The first ORF, agaR, could encode a transcriptional regulator of the AraC family that is widespread among bacteria and that regulates genes having diverse functions, ranging from carbon metabolism to stress response (48). The following five genes are transcribed in the opposite direction of the first ORF. The products of agaE, agaF, and agaG could constitute part of an ABC importer specific for raffinose. Indeed, they present significant homologies with the MsmFII, MsmGII, and MsmEII proteins, the high affinity solute-binding protein, and the transmembrane domains of an ABC transporter involved in raffinose uptake in L. acidophilus NCFM (10). However, the gene encoding for the corresponding nucleotide-binding domain, necessary for an active ABC transporter (49), was not found in this cluster, but a gene with significant homology to the gene encoding this nucleotide-binding domain in L. acidophilus NCFM has been found elsewhere on the genome. The fourth gene encodes for a putative α-galactosidase, hereafter called AgaSK, and the last gene encodes for a putative sucrose phosphorylase, an enzyme that converts sucrose to fructose and glucose-1-phosphate.
To determine whether AgaSK could be involved in the α-galactosidase activity detected previously, we analyzed whether it is expressed in vivo. In particular, RT-PCR with mRNA from the cecal content obtained from germ-free mice associated with R. gnavus E1 was thus performed. A high level of agaSK transcript was detected, suggesting the presence of the enzyme in the intestinal tract (supplemental Fig. 2).
AgaSK Possesses α-Galactosidase Activity
Sequence analysis of AgaSK suggests that it does not contain a signal peptide and that it is composed of two domains (supplemental Fig. 3). The N-terminal domain encompasses residues 1–720, and a BLAST search (50) reveals homology with α-galactosidases belonging to family GH36 of the CAZy classification (14). Surprisingly, the C-terminal domain (residues 721–935) shares sequence homologies with several kinases acting on small molecules, like phosphoribulokinases, uridine kinases, or adenylyl sulfate kinases, and contains a Walker A motif ((G/A)X4GK(T/S)) (51), typical for nucleotide-binding proteins. The presence of these two domains indicates that AgaSK would not be a classical α-galactosidase but could be a bifunctional enzyme composed of two domains, each having their own enzymatic property.
To undertake a thorough biochemical characterization of AgaSK, recombinant AgaSK was heterologously expressed in E. coli BL21 (DE3). After purification, the molecular mass of the protein as determined by SDS-PAGE was about 105 kDa, consistent with its theoretical mass of 105,510 Da, deduced from the peptide sequence. The molecular mass of native AgaSK, as estimated by size-exclusion chromatography, was about 490 kDa, indicating that AgaSK is a tetramer in solution.
Then, its ability to hydrolyze carbohydrates, and in particular α-1,6 glycosidic linkages, was tested toward different molecules (Table 2). AgaSK efficiently hydrolyzed the α-galactosidic linkage in PNPG, melibiose, raffinose, and stachyose, but the activity was low toward o-nitrophenyl-β-d-galactopyranoside, p-nitrophenyl-α-d-glucopyranoside, p-nitrophenyl-α-d-xylopyranoside, locus bean, and guar gum. The release of products from raffinose and melibiose was analyzed by high performance anion exchange chromatography coupled with pulsed amperometric detection (supplemental Fig. 4). Galactose, sucrose, and glucose, but not fructose, were present in the hydrolysates of raffinose and melibiose. We then determined the enzymatic parameters of AgaSK, and we observed that the enzyme was active between pH 4.0 and 6.5 and exhibited highest activity on PNPG at pH 6.0 and 50 °C, with a specific activity of 150 ± 17 units/mg. Using PNPG as substrate, the pH profile and the kinetic parameters, Km = 1.37 ± 0.20 mm and kcat/Km = 1850 ± 24 s−1.mm−1, were similar to those of other GH36 α-galactosidases described previously (23–25, 27, 52). All these results indicate that AgaSK is an α-galactosidase specific for α-1,6 linkages.
TABLE 2.
Specific activity for hydrolysis of different natural or synthetic molecules by AgaSK
Parameters are calculated from the release of nitrophenolate from nitrophenylated substrates, and of galactose from natural substrates.
| Substrate | Specific activity |
|---|---|
| units·mg−1 | |
| PNPG | 162 ± 43 |
| Melibiose | 420 ± 8 |
| Raffinose | 231 ± 7 |
| Stachyose | 213 ± 19 |
| Locus bean gum | 21 ± 2 |
| Guar gum | 11 ± 0.6 |
| Xylose α-d-PNP | 0.96 ± 0.6 |
| Glucose α-d-PNP | 1.80 ± 4.7 |
| ONPGa | 0.70 ± 0.3 |
a ONPG, o-nitrophenyl-alpha-d-galactopyranoside.
AgaSK Is a Tetrameric Protein
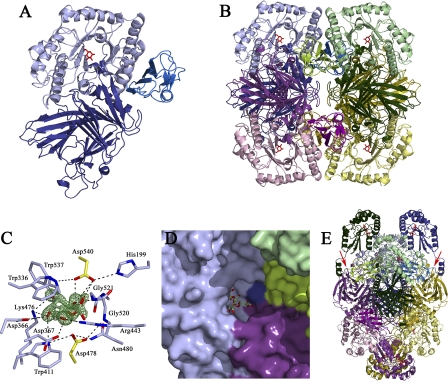
Full-length AgaSK was used for crystallization trials, but due to spontaneous in situ proteolysis, only crystals of a truncated form, AgaSK-tru, corresponding to the α-galactosidase domain, could be obtained. The structure of AgaSK-tru was solved by x-ray crystallography to 1.45 Å resolution, and the structure of a complex with galactose was obtained at 1.35 Å resolution (Table 1). The three-dimensional structure of AgaSK-tru discloses three domains: an N-terminal β-supersandwich domain (residues 2–327) followed by a canonical (β/α)8 barrel (residues 328–626) and a C-terminal β-sheet domain (residues 627–720) (Fig. 1A). AgaSK-tru forms a tetramer in the crystal structure by application of two crystallographic 2-fold axes, which is consistent with results from size-exclusion chromatography (Fig. 1B). The association of AgaSK-tru into a tetramer results in a buried surface of 21,770 Å2 as calculated by the PISA server (43), representing 26% of the total tetramer surface area. A virtually identical tetrameric assembly can be observed in the crystal structures of L. brevis α-galactosidase LbR11 and L. acidophilus NCFM LaMel36A, reflecting 35 and 46% sequence identity, respectively, with AgaSK, whereas T. maritima TmGalA (28% sequence identity with AgaSK) presents itself as a monomer.
FIGURE 1.
Structure of the α-galactosidase domain of AgaSK, tetrameric assembly, substrate-binding site, and model for full-length AgaSK. A, graphic presentation of the α-galactosidase domain of AgaSK, with the N-terminal domain in deep blue, the catalytic (β/α)8 barrel in light blue, and the C-terminal in marine blue. Galactose is shown as red sticks. B, tetrameric assembly of AgaSK-tru, with one monomer oriented and colored as in A and the other monomers shown in pink, green, and yellow hues, respectively. C, the α-galactosidase active site. Residues interacting with the product are shown as sticks, with carbons color-coded as in B. Galactose is shown as sticks with green carbons, and the catalytic residues Asp-478 and Asp-540 are shown as sticks with yellow carbons. Oxygen and nitrogen atoms are colored in red and blue, respectively. Fo − Fc electron density calculated prior to incorporation of galactose into the model and contoured at 4 σ is shown in green. D, the association of three monomers within the tetrameric assembly creates a deep tunnel for substrate binding. The substrate-binding groove is shown in surface representation, with different monomers color-coded as in B. Galactose is shown as sticks with yellow carbons, and a model of raffinose, derived from a superposition of AgaSK-tru with S. cerevisiae α-galactosidase 1, Mel1, complexed with raffinose (PDB 3lrm), is shown as sticks with green carbons. E, model for full-length AgaSK. The tetrameric assembly is color-coded as in B. The modeled C-terminal domains are colored in the same hue as the N-terminal domains of each monomer. The polypeptide chain at the junction of both domains has to bend backwards toward the (β/α)8 barrel of an adjacent subunit to avoid steric clashes. This places each nucleotide-binding domain in proximity to an α-galactosidase active site. Galactose and modeled ATP molecules are shown as red sticks. Two junctions between the galactose and the kinase domain are indicated with red arrows.
The Active Site Exemplifies Substrate Specificity
The structure of AgaSK-tru in complex with the reaction product galactose allows us to identify unambiguously the catalytic center, which is located in a shallow pocket within the (β/α)8 barrel domain (Fig. 1C). The identity of the catalytic residues can be inferred from a superposition with the crystal structure of T. maritima TmGalA, for which the catalytic nucleophile and general acid/base had been identified previously to be Asp-327 and Asp-387, respectively (19). In AgaSK-tru, Asp-478 is found at 3.0 Å from the anomeric center of the sugar and thus well positioned for nucleophilic attack. Conversely, the general acid/base Asp-540 is ideally poised to donate a hydrogen to the sugar O1 hydroxyl, found at a distance of 2.95 Å.
Asp-478 is held in place by hydrogen bonds with the side chains of Trp-411 and Asn-480 and with the main chain nitrogen of Gly-520. The protonation state of Asp-540 is modulated by interactions with the side chains of His-199 and Trp-537. The galactose, found in the standard 4C1 chair conformation, is tightly enveloped by the enzyme and establishes an extensive network of hydrogen bonds with surrounding residues, most of them conserved in family GH36. The OH-2 hydroxyl interacts with the enzyme via the side chain of the acid/base Glu-540 and the main chain nitrogen of Gly-521. The OH-3 atom hydrogen bonds to Lys-476, which in turn stabilizes the axial OH-4, together with the side chains of Trp-411 and Asp-366. The OH-6 hydroxyl contacts the side chains of Asp-367 and Arg-443. Finally, the indole ring of Trp-336 provides a stacking platform for the sugar. Some family GH36 members exhibit α-N-acetylgalactosaminidase activity, but in AgaSK-tru, the tight interactions of the substrate with the enzyme do not allow for exocyclic substitutions on the galactose moiety.
The Tetrameric Assembly Provides a Platform for Efficient Substrate Binding
Due to the tetrameric assembly of AgaSK-tru, the shallow active site pocket extends toward a deep substrate-binding tunnel formed by loop regions of the central (β/α)8 barrel and loop regions of the N- and C-terminal region of different subunits (Fig. 1D). A superposition, based on the position of galactose, of the structure of the AgaSK-tru galactose complex, with the structure of a family GH27 active site mutant of Saccharomyces cerevisiae α-galactosidase 1, Mel1, in complex with raffinose (PDB 3lrm), allows us to define structural elements possibly involved in aglycone binding. Phe-55, from the N-terminal region of a neighboring subunit, inserts tightly into a pocket situated below the active site pocket, thereby anchoring adjacent loop regions in close proximity to the putative +1 and +2 binding sites (53). Residues likely to interact with the aglycone part of raffinose are His-199, Asp-376, and Arg-443, as well as Asp-52, Arg-66, and the backbone Gly-53-Gly-54 of a neighboring subunit. Most importantly, the shallowness of the substrate-binding tunnel does not allow accommodation of substrates with glycosidic linkages other than α-1,6, confirming the strict specificity of AgaSK for α-1,6-linked galactose. Our observations let us conclude that the self-association of AgaSK into a tetramer is likely to be physiologically relevant because different subunits join near the active site to provide a platform for efficient substrate binding.
AgaSK, an α-Galactosidase with a Kinase Activity
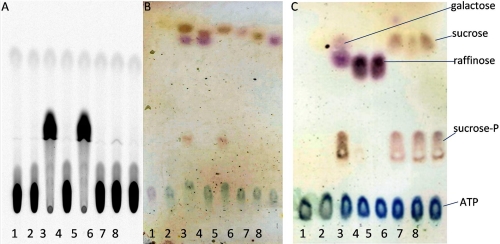
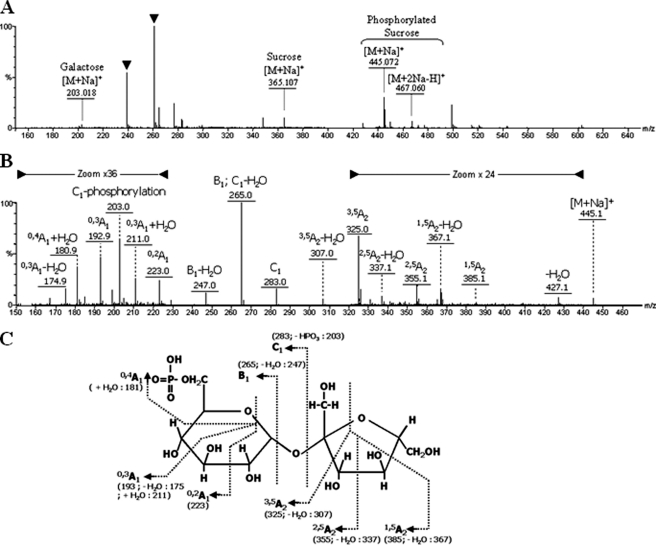
The presence of the Walker A motif in the C-terminal domain of AgaSK (supplemental Fig. 3) suggested that it was also able to bind nucleotides. We took advantage of the presence of tryptophans in the sequence of AgaSK to study nucleotide binding by intrinsic fluorescence measurements, and we showed that AgaSK binds ATP (Kdapp ∼61 ± 13 μm) (supplemental Fig. 5). We then tested its potential ability to phosphorylate several sugars, especially sugars that are substrates of the α-galactosidase domain, in the presence of ATP and [γ-33P]ATP (Fig. 2A). Raffinose and melibiose were tested (lanes 3 and 4), and a new radioactive spot was observed only when raffinose was used as substrate. Because raffinose is degraded by AgaSK into sucrose and galactose, we also tested sucrose, galactose, and the two monosaccharides composing the sucrose unit, glucose and fructose (lanes 5–8). We observed the same radioactive spot in lane 5 only, showing that sucrose was phosphorylated by AgaSK. Furthermore, to test whether AgaSK was able to phosphorylate both sugars, raffinose and sucrose, we added the α-galactosidase inhibitor d-galacto-1-deoxynojirimycin to the phosphorylation reaction mix (Fig. 2B, lanes 4, 5, 7, and 8). In the presence of the inhibitor, phosphorylated sucrose was not produced anymore when raffinose was used as substrate (lanes 4 and 5 compared with lane 3), but phosphorylated sucrose was still observed when the substrate was sucrose (lanes 6–8). We therefore conclude that in the presence of ATP, AgaSK specifically phosphorylates the sucrose unit liberated by the hydrolysis of raffinose. This specific sucrose phosphorylation by AgaSK was confirmed by MALDI-MS (Fig. 3). The control sample spectrum highlighted raffinose and adducts of ATP (supplemental Fig. 6A), whereas the spectrum of the reaction products formed by AgaSK showed two products and adducts of ADP (supplemental Fig. 6B). The structural features of phosphorylated sucrose were deduced from an MS/MS spectrum, and a set of specific related fragments was obtained, which allowed to locate the phosphate on position C6 of glucose (Fig. 3). Altogether these results show that AgaSK is also a sucrose kinase able to phosphorylate specifically sucrose on the C6 position of glucose.
FIGURE 2.
Characterization of sugar phosphorylation by AgaSK. A and B, phosphorylation of several sugars was tested in reaction mixtures containing AgaSK, ATP, MgCl2, [γ-33P]ATP (0.5 μCi), and different sugars. The radioactive spots were detected on the autoradiogram (A). The sugar spots were detected with 0.1% orcinol (B). Lanes 1, without AgaSK; lanes 2, with AgaSK; lanes 3, with raffinose; lanes 4, with melibiose; lanes 5, with sucrose; lanes 6, with glucose; lanes 7, with fructose; lanes 8, with galactose. C, phosphorylation of raffinose and sucrose was tested in reaction mixtures containing AgaSK, ATP, and MgCl2. Lane 1, without AgaSK; lane 2, with AgaSK; lane 3, with raffinose; lane 4, with raffinose and 10 μm Gal-DNJ; lane 5, with raffinose and 100 μm Gal-DNJ; lane 6, with sucrose; lane 7, with sucrose and 10 μm Gal-DNJ; lane 8, with sucrose and 100 μm Gal-DNJ. The sugar spots were detected with 0.1% orcinol.
FIGURE 3.
Positive ESI-Q-TOF-MS mass spectra and chemical structure of phosphorylated sugar. All reaction products are deduced from the MS/MS data at m/z 365.107 (sucrose monosodiated form), 445.072 (phosphorylated sucrose monosodiated form), 467.060 (phosphorylated sucrose disodiated form), and 203.018 (galactose monosodiated form). The structural features of phosphorylated sucrose are deduced from an ion m/z 445.059 MS/MS spectrum. A, full scan MS of the sample from enzymatic reaction (solvent ions are indicated by black triangles). B, MS/MS of the sodiated ion at 445,1 with corresponding annotated fragments. In two areas of the MS/MS spectrum, the peak intensity was scaled up 24 and 36 times, respectively. C, chemical structure of the phosphorylated oligosaccharide annotated with corresponding fragments.
Model of Full-length AgaSK
As all attempts to obtain crystals of full-length AgaSK failed, we used the Phyre server (45) to build a homology model for the kinase domain based on the crystal structure of putative fructose transport system kinase (YP_612366.1) from Silicibacter sp. TM1040 (PDB 3c8u). The model of the kinase domain could not be joined to the crystal structure of AgaSK-tru in an arbitrary fashion as the tetrameric assembly positions the C-terminus of the galactosidase domain from two adjacent subunits face to face (Fig. 1E). Due to these steric constraints, the kinase model had to be placed in a way such that the polypeptide chain at the galactosidase-kinase junction bends backwards toward the (β/α)8 barrel, thereby locating the active sites of the kinase domains near the galactosidase active sites of adjacent subunits within the tetramer (Fig. 1E). The proximity of the two active sites, as proposed in this model, might assure efficient cross-talk between the two activities of AgaSK, but the full-length structure of this bifunctional enzyme will be necessary to corroborate our hypothesis.
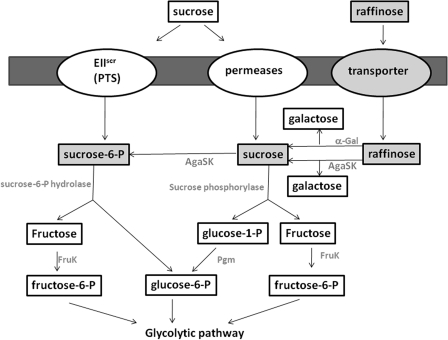
Raffinose and Sucrose Uptake and Metabolism
To date, two different pathways for sucrose utilization have been characterized in bacteria (13) (Fig. 4). The first involves a phosphoenolpyruvate-dependent phosphotransferase system (PTS), where sucrose is phosphorylated into sucrose-6-phosphate and simultaneously transported into the cell. Sucrose-6-phosphate is then cleaved by a hydrolase into glucose-6-phosphate and fructose. In the second pathway, a PTS-independent transport system allows incorporation of sucrose under unmodified form into the cell, where it is hydrolyzed by sucrose phosphorylase (54, 55). Glucose-1-phosphate is subsequently transformed by a phosphoglucomutase into glucose-6-phosphate. In both mechanisms, fructose is afterward phosphorylated by a fructokinase.
FIGURE 4.
Sucrose and raffinose pathways in R. gnavus E1 (adapted from Reid and Abratt (13)). Two pathways for sucrose transport are well characterized, the PTS-dependent sucrose system (left) and the non-PTS transport system (middle). The raffinose pathway (right) could be another possibility to provide sucrose to the bacterium.
Certain bacteria possess ABC transporters for the direct uptake of raffinose, which is subsequently cleaved by an α-galactosidase into galactose and sucrose, and the sucrose produced thereby is taken in charge by sucrose phosphorylase (54, 55). Genes coding for a putative ABC transporter for raffinose have been found in R. gnavus E1. This observation suggests that raffinose is transported within the cytoplasm of R. gnavus E1 by an ABC transporter and subsequently hydrolyzed by the α-galactosidase activity of AgaSK into galactose and sucrose. The sucrose produced thereby is instantly taken in charge by the kinase activity of AgaSK to release sucrose-6-phosphate, the substrate of sucrose-6 phosphorylase (Fig. 4). The sucrose, which escapes the kinase activity of AgaSK, is probably cleaved by sucrose phosphorylase, encoded by the gene adjacent to the AgaSK gene. These findings place AgaSK at the crossroad of the two glycolytic pathways for sucrose utilization described so far.
Concluding Remarks
In conclusion, sucrose and raffinose are the most abundant soluble carbohydrates found in plants (9), and they are probably part of the human carbohydrate energy intake. Although sucrose from diet is the substrate of the human intestinal sucrase-isomaltase, raffinose is degraded into galactose and sucrose only by intestinal microbial enzymes. Therefore, for the intestinal microbiota, a major external source of sucrose is probably raffinose, pointing out the importance of its metabolism for bacteria. In the strict anaerobic Gram-positive bacterium R. gnavus, a single enzyme, AgaSK, is able to produce sucrose-6-phosphate directly from raffinose. The two distinct enzymatic activities are related to two domains. The N-terminal domain is responsible for the α-galactosidase activity of AgaSK, whereas the C-terminal domain catalyzes the phosphorylation of the sucrose produced thereby on the C6 position of glucose. We can speculate about the interest for R. gnavus E1 to possess a bifunctional protein having two enzymatic activities within the same scaffold. We can hypothesize that it could exemplify a very efficient metabolic strategy developed by this human microbiota to metabolize raffinose, a potentially major source of carbohydrate available in the gut. Our studies contribute to a deeper understanding of the mechanisms by which Firmicutes use nutrients that are non-digestible by the host and optimize their metabolism for survival in such an extreme competitive environment as the intestinal tract.
Supplementary Material
Acknowledgments
We are indebted to S. Rabot (INRA Jouy-en-Josas, France) for providing axenic rats of the animal facility ANAXEM platform and to C. Bridonneau and P. Guillaume for skillful technical assistance. Genome sequencing was carried out by Genoscope, AP05/06 Project 27. We thank also Dr. Ray Owens from the Department for Structural Biology at the University of Oxford for the pOPIN E vector. We thank the European Synchrotron Radiation Facility and the Synchrotron Soleil for provision of beam time and assistance with data collection.
This work was supported by the CNRS, the Agence Nationale de la Recherche (ANR) contract “P-loop proteins,” and the “Ministère de l'Enseignement Supérieur.”
The atomic coordinates and structure factors (codes 2YFN and 2YFO) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The nucleotide sequences of the genes reported in this paper have been submitted to the EMBL Nucleotide Sequence Database (ENA) with accession numbers FQ790377–FQ790383.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–6.
- GH
- glycoside hydrolase
- AgaSK
- α-galactosidase/sucrose kinase
- PNPG
- p-nitrophenyl-α-d-galactopyranoside
- ABC
- ATP-binding cassette transporter
- Q-TOF
- quadrupole time-of-flight
- Gal-DNJ
- d-galacto-1-deoxynojirimycin
- PTS
- phosphotransferase system.
REFERENCES
- 1. Ley R. E., Turnbaugh P. J., Klein S., Gordon J. I. (2006) Nature 444, 1022–1023 [DOI] [PubMed] [Google Scholar]
- 2. Turnbaugh P. J., Ley R. E., Mahowald M. A., Magrini V., Mardis E. R., Gordon J. I. (2006) Nature 444, 1027–1031 [DOI] [PubMed] [Google Scholar]
- 3. Greiner T., Bäckhed F. (2011) Trends Endocrinol. Metab. 22, 117–123 [DOI] [PubMed] [Google Scholar]
- 4. Turnbaugh P. J., Hamady M., Yatsunenko T., Cantarel B. L., Duncan A., Ley R. E., Sogin M. L., Jones W. J., Roe B. A., Affourtit J. P., Egholm M., Henrissat B., Heath A. C., Knight R., Gordon J. I. (2009) Nature 457, 480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Eckburg P. B., Bik E. M., Bernstein C. N., Purdom E., Dethlefsen L., Sargent M., Gill S. R., Nelson K. E., Relman D. A. (2005) Science 308, 1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Armougom F., Henry M., Vialettes B., Raccah D., Raoult D. (2009) PLoS One 4, e7125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Venema K. (2010) Curr. Opin. Clin. Nutr. Metab. Care 13, 432–438 [DOI] [PubMed] [Google Scholar]
- 8. Neyrinck A. M., Delzenne N. M. (2010) Curr. Opin. Clin. Nutr. Metab. Care 13, 722–728 [DOI] [PubMed] [Google Scholar]
- 9. Trugo L. C., Farah A., Cabral L. (1995) Food Chem. 52, 385–387 [Google Scholar]
- 10. Barrangou R., Azcarate-Peril M. A., Duong T., Conners S. B., Kelly R. M., Klaenhammer T. R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3816–3821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hugouvieux-Cotte-Pattat N., Charaoui-Boukerzaza S. (2009) J. Bacteriol. 191, 6960–6967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Caputto R., Leloir L. R., et al. (1949) J. Biol. Chem. 179, 497–498 [PubMed] [Google Scholar]
- 13. Reid S. J., Abratt V. R. (2005) Appl. Microbiol. Biotechnol. 67, 312–321 [DOI] [PubMed] [Google Scholar]
- 14. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rigden D. J. (2002) FEBS Lett. 523, 17–22 [DOI] [PubMed] [Google Scholar]
- 16. Henrissat B., Bairoch A. (1996) Biochem. J. 316, 695–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Garman S. C., Hannick L., Zhu A., Garboczi D. N. (2002) Structure 10, 425–434 [DOI] [PubMed] [Google Scholar]
- 18. Garman S. C., Garboczi D. N. (2004) J. Mol. Biol. 337, 319–335 [DOI] [PubMed] [Google Scholar]
- 19. Comfort D. A., Bobrov K. S., Ivanen D. R., Shabalin K. A., Harris J. M., Kulminskaya A. A., Brumer H., Kelly R. M. (2007) Biochemistry 46, 3319–3330 [DOI] [PubMed] [Google Scholar]
- 20. Fredslund F., Abou Hachem M., Jonsgaard Larsen R., Gerd Sørensen P., Coutinho P. M., Lo Leggio L., Svensson B. (2011) J. Mol. Biol. 412, 466–480 [DOI] [PubMed] [Google Scholar]
- 21. Hashimoto H., Katayama C., Goto M., Okinaga T., Kitahata S. (1995) Biosci. Biotechnol. Biochem. 59, 619–623 [DOI] [PubMed] [Google Scholar]
- 22. Van Laere K. M., Hartemink R., Beldman G., Pitson S., Dijkema C., Schols H. A., Voragen A. G. (1999) Appl. Microbiol. Biotechnol. 52, 681–688 [DOI] [PubMed] [Google Scholar]
- 23. Spangenberg P., André C., Dion M., Rabiller C., Mattes R. (2000) Carbohydr. Res. 329, 65–73 [DOI] [PubMed] [Google Scholar]
- 24. Tzortzis G., Jay A. J., Baillon M. L., Gibson G. R., Rastall R. A. (2003) Appl. Microbiol. Biotechnol. 63, 286–292 [DOI] [PubMed] [Google Scholar]
- 25. Zhao H., Lu L., Xiao M., Wang Q., Lu Y., Liu C., Wang P., Kumagai H., Yamamoto K. (2008) FEMS Microbiol. Lett. 285, 278–283 [DOI] [PubMed] [Google Scholar]
- 26. Goulas T., Goulas A., Tzortzis G., Gibson G. R. (2009) Appl. Microbiol. Biotechnol. 82, 471–477 [DOI] [PubMed] [Google Scholar]
- 27. Nakai H., Baumann M. J., Petersen B. O., Westphal Y., Hachem M. A., Dilokpimol A., Duus J. Ø., Schols H. A., Svensson B. (2010) FEBS J. 277, 3538–3551 [DOI] [PubMed] [Google Scholar]
- 28. Blaut M. (2002) Eur. J. Nutr. 41, Suppl. 1, I11–I16 [DOI] [PubMed] [Google Scholar]
- 29. Tzortzis G., Goulas A. K., Baillon M. L., Gibson G. R., Rastall R. A. (2004) Appl. Microbiol. Biotechnol. 64, 106–111 [DOI] [PubMed] [Google Scholar]
- 30. Rastall R. A., Gibson G. R., Gill H. S., Guarner F., Klaenhammer T. R., Pot B., Reid G., Rowland I. R., Sanders M. E. (2005) FEMS Microbiol. Ecol. 52, 145–152 [DOI] [PubMed] [Google Scholar]
- 31. Qin J., Li R., Raes J., Arumugam M., Burgdorf K. S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T., Mende D. R., Li J., Xu J., Li S., Li D., Cao J., Wang B., Liang H., Zheng H., Xie Y., Tap J., Lepage P., Bertalan M., Batto J. M., Hansen T., Le Paslier D., Linneberg A., Nielsen H. B., Pelletier E., Renault P., Sicheritz-Ponten T., Turner K., Zhu H., Yu C., Li S., Jian M., Zhou Y., Li Y., Zhang X., Li S., Qin N., Yang H., Wang J., Brunak S., Doré J., Guarner F., Kristiansen K., Pedersen O., Parkhill J., Weissenbach J., Bork P., Ehrlich S. D., Wang J. (2010) Nature 464, 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Doré J., Sghir A., Hannequart-Gramet G., Corthier G., Pochart P. (1998) Syst. Appl. Microbiol. 21, 65–71 [DOI] [PubMed] [Google Scholar]
- 33. Cervera Tison M., André-Leroux G., Lafond M., Georis J., Juge N., Berrin J. G. (2009) Biochim. Biophys. Acta 1794, 438–445 [DOI] [PubMed] [Google Scholar]
- 34. Matsui I., Ishikawa K., Matsui E., Miyairi S., Fukui S., Honda K. (1991) J. Biochem. 109, 566–569 [DOI] [PubMed] [Google Scholar]
- 35. Leslie A. G. (1992) Joint CCP4/ESF-EACBM Newsletter, Vol. 26, pp. 27–33, Collaborative Computational Project No. 4, Daresbury Laboratory, Daresbury, Warrington, UK [Google Scholar]
- 36. Kabsch W. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Evans P. R. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 38. McCoy A. J. (2007) Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Perrakis A., Morris R., Lamzin V. S. (1999) Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 40. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 4449 [DOI] [PubMed] [Google Scholar]
- 42. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 44. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 45. Kelley L. A., Sternberg M. J. (2009) Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 46. Forouhar F., Abashidze M., Xu H., Grochowski L. L., Seetharaman J., Hussain M., Kuzin A., Chen Y., Zhou W., Xiao R., Acton T. B., Montelione G. T., Galinier A., White R. H., Tong L. (2008) J. Biol. Chem. 283, 11832–11840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Domon B., Costello C. (1988) Glycoconj. J. 5, 397–409 [Google Scholar]
- 48. Aslanidis C., Schmid K., Schmitt R. (1989) J. Bacteriol. 171, 6753–6763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Davidson A. L., Dassa E., Orelle C., Chen J. (2008) Microbiol. Mol. Biol. Rev. 72, 317–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Walker J. E., Saraste M., Runswick M. J., Gay N. J. (1982) EMBO J. 1, 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ademark P., Larsson M., Tjerneld F., Stålbrand H. (2001) Enzyme Microb. Technol. 29, 441–448 [Google Scholar]
- 53. Davies G. J., Wilson K. S., Henrissat B. (1997) Biochem. J. 321, 557–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Silverstein R., Voet J., Reed D., Abeles R. H. (1967) J. Biol. Chem. 242, 1338–1346 [PubMed] [Google Scholar]
- 55. Kitaoka M., Hayashi K. (2002) Trends Glycosci. Glycotechnol. 14, 35–50 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.