Background: The type II secretion machinery secretes large toxins across the bacterial envelope.
Results: We identified multiple interactions between secreted exoproteins and components of the machinery.
Conclusion: We propose a model for substrate recognition and transport during the secretion process.
Significance: Our data shed light on the operating mode of the type II secretion pathway and provide new potential targets for drug development.
Keywords: Chromatography, Membrane Transport, Protein Complexes, Protein Secretion, Surface Plasmon Resonance (SPR), Type II Secretion System, Exoproteins, Interactome, Pseudopilins, Secretin
Abstract
The type II secretion system enables Gram-negative bacteria to secrete exoproteins into the extracellular milieu. We performed biophysical and biochemical experiments to identify systematic interactions between Pseudomonas aeruginosa Xcp type II secretion system components and their substrates. We observed that three Xcp components, XcpPC, the secretin XcpQD, and the pseudopilus tip, directly and specifically interact with secreted exoproteins. We established that XcpPC, in addition to its interaction with the substrate, likely shields the entire periplasmic portion of the secreton. It can therefore be considered as the recruiter of the machinery. Moreover, the direct interaction observed between the substrate and the pseudopilus tip validates the piston model hypothesis, in which the pseudopilus pushes the substrate through the secretin pore during the secretion process. All together, our results allowed us to propose a model of the different consecutive steps followed by the substrate during the type II secretion process.
Introduction
Gram-negative bacteria have evolved different sophisticated secretory machines specialized for the secretion of specific categories of exoproteins (1). The type II secretion pathway is widely used by many Gram-negative bacteria for the secretion of major virulence factors in plants and animals (2). In this two-step secretion process, proteins are synthesized with a cleavable N-terminal signal sequence that enables their transport across the cytoplasmic membrane by the Sec or Tat general export machinery (3). After acquisition of their folds in the periplasm, type II secretion-dependent proteins are specifically recognized and loaded by a large macromolecular machine, the secreton, for their final release into the external milieu. The secreton spans the entire bacterial envelope and consists of at least 12 different Gsp proteins that are organized in three subcomplexes depending on their cellular localizations and mutual interactions (4).
The GspER, FS, LY, and ML proteins (see “Experimental Procedures” for nomenclature) form the inner membrane (IM)3 platform, where the GspER traffic ATPase provides energy for the system (5–8). The second subcomplex is called the pseudopilus, which is analogous to the pilus structure found in the type IV piliation system. The pseudopilus consists of five pseudopilins, all of which are subjected to maturation by the prepilin peptidase GspOA, which is also involved in the maturation of the type IV pilins. The pseudopilus is formed by the helical assembly of the major pseudopilin GspGT (9), and it has a quaternary complex of the four minor pseudopilins at its tip (10, 11). This structural similarity to the type IV piliation system suggests that the pseudopilus might also assemble on the IM platform in a pilus-like structure to push the substrate like a piston through the third subcomplex of the secreton, the secretin GspDQ (10–14).
Secretins form large, homo-multimeric pores in the outer membrane (OM). They are components of various secretion or assembly machines involved in the transport of large structures (15). Each secretin monomer has two domains: the conserved C-terminal domain, which forms the pore in the OM, and the N-terminal domain, which differs between transport systems and forms an extension of the pore cavity in the periplasm. The N-terminal domains of T2SS secretins consist of four structurally independent subdomains named N0 to N3 from the N to the C terminus. Recently, Reichow et al. (12) have reported direct interactions between GspDQ and the substrate and between GspDQ and the pseudopilus. This important finding agrees with the secreton model, in which the pseudopilus interacts with the secretin containing the substrate to push it through the pore. However, no evidence has been reported in the literature for a direct interaction between the pseudopilus and the substrate.
In different representations of the secreton, the connection between the IM platform and the OM secretin is mediated by GspCP. This component is inserted into the IM platform by its N-terminal transmembrane helix and is connected to the secretin by its periplasmic domain on the other side (16, 17).
T2SSs are prevalent among Gram-negative bacteria and are highly specific for their respective substrates. In Pseudomonas aeruginosa, two T2SSs co-exist, Xcp and Hxc, and each secretes a specific set of substrates (18).
In contrast to other secretory machines, such as the type I, type III, and type IV secretion systems, no secretion recognition signals have yet been identified for type II secretion. However, several studies have indicated the involvement of several non-adjacent regions in substrate recognition, thus suggesting the existence of a conformational secretion signal (19, 20).
Notably, no interactions between substrates and secreton components other than the secretin have been reported so far. In this work, we aimed at understanding two critical steps of the type II secretion process, the initial substrate recognition and its release after import inside the machinery. To do this, we set up a systematic protein/protein interaction study between secreted substrates and different periplasmic components of the T2SS secreton. The multiple interactions identified made it possible to propose a model of substrate recruitment and transport during the type II secretion process.
EXPERIMENTAL PROCEDURES
Nomenclature Used in This Study
Because a different nomenclature is used for non-Pseudomonas and Pseudomonas T2SSs, the alternative gene or protein nomenclature is indicated throughout this study; for example, in GspER, the “R” refers to XcpR, which is reciprocally called XcpRE. Moreover, all of the XcpPC, XcpQD, and pseudopilin variants used in this study are periplasm-soluble domains, lacking their membrane domains, and are respectively called PC, QD and TG, UH, VI, and WJ and XK for the pseudopilin variants.
Cloning, Expression, and Purification of PC, HisPC, QD-N0123, Qd-N012, and QD-N01 and the Soluble Domains of the Xcp Pseudopilins
Cloning, expression, and purification of PC, C-terminal His-tagged PC (hisPC), QD-N0123, Qd-N012, and QD-N01 followed the strategy used in Ref. 10 for TG, UH, VI, WJ, and XK cloning, expression, and purification and are described in detail in the supplemental material (Tables S1 and S2).
Exoprotein Preparation
The purified, folded, and active P. aeruginosa elastase (LasB) and lipase (LipA) used in the surface plasmon resonance (SPR) experiments were respectively purchased from Nagase ChemteX Corp., Kyoto, Japan (lot number 6528025) and obtained from Prof. Jaeger, who purified LipA as described in Stuer et al. (21). P. aeruginosa alkaline phosphatase LapA was purified from the supernatants of 1l cultures of the PAO1Δxcp strain grown under phosphate starvation as described in Ref. 18. Supernatant collected after centrifugation was concentrated and passed through a Sephadex G75 column equilibrated with 50 mm phosphate, 150 mm NaCl, pH 7. Pure LapA was recovered in fractions corresponding to its molecular mass.
Biotinylation and Immobilization of PC on Streptavidin Chip
PC was biotinylated with Sulfo-NHS-SS-Biotin (Pierce) following the protocol specified by the supplier, with minor modifications. The protein was diluted at 25 mm in 50 mm phosphate buffer (Na2HPO4/NaH2PO4), 150 mm NaCl, pH 7.5, and the reagent was added at 25 mm (molar ratio 1:1). The reaction was allowed to run for 4 h on ice. To eliminate the excess reagent, the protein solution was first filtered on an NAP-5 column (GE Healthcare) equilibrated in the same buffer as specified above and then dialyzed by a Novagen dialyzer midi (cutoff 3.5 kDa) against 2 times 400 ml of the same buffer as specified above, at 4 °C overnight.
The streptavidin chip was used in 50 mm phosphate buffer, 150 mm NaCl, pH 7. Washing with 1 m NaCl and 50 mm NaOH was carried out as specified by the supplier before fixing biotinylated PC (1.25 mm, 140 μl at 10 μl/min) at 1295 Δ response unit (RU). Non-biotinylated LipA, LasB, UH, VI, WJ, TG, and XK in 50 mm phosphate, 150 mm NaCl, pH 7 were passed over the two flow cells (10 mm, 60 μl at 10 μl/min) to test for binding. Reproducible interaction was detected with LipA and a high concentration of LasB in the presence of the inhibitor (1–10-phenanthroline (10 mm)). Binding traces were recorded for 5 concentrations of LipA, in triplicate. No regeneration was necessary as spontaneous dissociation of the analyte was observed.
Immobilization of Antibody Anti-penta-His
The CM5 sensor chip was coated with the antibody anti-penta-His (Qiagen catalog number 34660) following the procedure in Ref. 22 with minor modifications. Antibody anti-penta-His 50 μg/ml in 10 mm sodium acetate, pH 4.5, was immobilized by amine coupling (ΔRU = 7000) on Fc1 and Fc2. A solution of hisPC at 20 μm in HEPES buffer/EDTA/P20 detergent, pH 7.4, was passed over Fc2 (final ΔRU = 1300). We observed that the amount of fixed hisPC decreases slowly with time (∼50% in 3–4 h). To start with a comparable amount of bound ligand, hisPC was fixed at the same level (ΔRU = 1200–1300) before each titration cycle. Solutions of QD-N0123, QD-N012, QD-N01, TG, XK, UH, VI, WJ, LipA, LapA, and LasB in the presence of the inhibitor (1–10-phenanthroline 10 mm) and the quaternary complex UH-WJ-VI-XK (20 μm in HBS-EP, pH 7.4, 0.005% P20 detergent) were passed over the Fc2 with hisPC bound and on the control flow cell (Fc1).
Immobilization of LasB and LapA
The CM5 (carboxymethylated dextran) sensor chip was coated with LasB and LapA (80 μg/ml in 10 mm sodium acetate, pH 4 and pH 4.5, respectively, for LasB and LapA) immobilized with amine coupling (ΔRU = 800 for LasB and ΔRU = 1500 for LapA) on Fc2. Solutions of PC, QD-N0123, QD-N012, QD-N01, TG, XK, UH, VI, WJ, and the quaternary complex UH-WJ-VI-XK (20 μm in HBS-EP, pH 7.4, 0.005% P20) were passed over Fc2 with LasB or LapA bound and on the control flow cell (Fc1). In the case of LasB, reproducible signals were detected with QD-N0123, QD-N012, QD-N01, XK, UH, VI, and the quaternary complex UH-WJ-VI-XK; no evidence of interaction has been found with TG and WJ under the present experimental conditions. When QD-N0123, QD-N012, QD-N01, XK, UH, VI, and the quaternary complex UH-WJ-VI-XK were passed over Fc2 with LapA bound, no interaction was found under the present experimental conditions. Nonspecific binding was subtracted from the binding traces before calculation. Dissociation constants (Kd) were estimated based on the equilibrium level at 300 s of injection of LipA on PC and PC on LasB and at 175 s of injection of the other combinations.
Affinity Measurements
Steady-state analysis of the interactions between soluble domains of PC, QD-N0123, QD-N012, and QD-N01, pseudopilins (TG, UH, VI, WJ, and XK), and substrates (LasB, LipA, and LapA) were performed on a BIAcore X100 at 25 °C. All buffers were 0.2 μm-filtered and degassed before use.
Binding traces were recorded in duplicate for increased concentrations of analyte. In each cycle, hisPC was injected first at reproducible levels of ΔRU = 1,200–1,300, and the analyte was then injected (40 μl at 5 μl/min). Ten millimolar H3PO4 (2 μl at 5 μl/min) was used to regenerate the CM5-anti-penta-His surface. Reproducible signals were detected with QD-N0123, LipA, and LasB; no evidence of interactions was found with QD-N012, QD-N01, TG, XK, UH, VI, or WJ under the stated experimental conditions.
Nonspecific binding was subtracted from the binding traces before calculation. Dissociation constants (Kd) were estimated based on the equilibrium levels at 100 and 50 s of injection of LasB and QD-N0123, respectively. In Figs. 3 and 6–8, for all sensorgrams, the inset represents plots of equilibrium response levels (ΔRU; y axis) as a function of analyte concentration (μm; x axis), with the curve fit to a 1:1 equilibrium model for determination of KD at 50% saturation response.
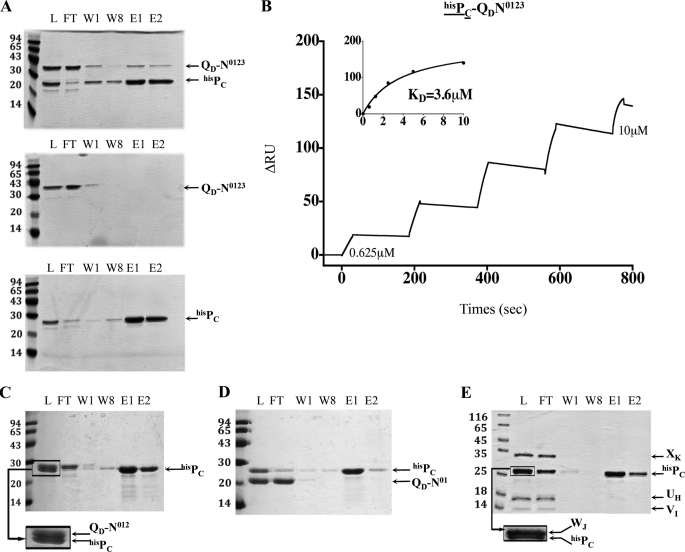
FIGURE 3.
PC/QD interaction identified by affinity chromatography and SPR experiments. A, batch co-purification of PC and QD presented in this panel were performed on affinity columns. hisPC and QD-N0123 were mixed (upper panel) or loaded individually (lower panels) on an SDS-PAGE gel. Fractions L, FT, W1, W8, E1, and E2, respectively contain loaded protein, the flow-through, the washes, and the eluates. The positions of the molecular mass markers are indicated on the left side of each gel (kDa). B, the SPR sensorgram of the positive interaction found between hisPC (bound ligand, underlined) and QD-N0123 (analyte) (see ‘Experimental Procedures‘ for details). C–E, batch co-purification experiments of hisPC and QD-N012 and hisPC and QD-N01 and hisPC and of minor pseudopilin soluble domains UH, VI, WJ, and XK, respectively, on affinity columns.
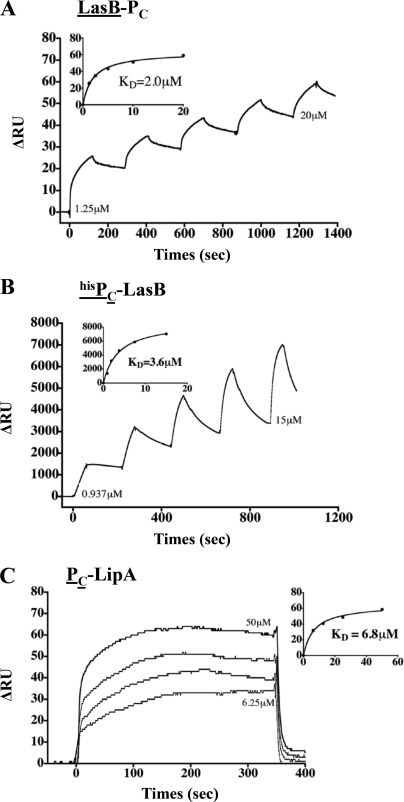
FIGURE 6.
PC/substrate interactions identified by SPR. A–C, the SPR sensorgrams of the positive interactions found between LasB and PC, hisPC and LasB, and hisPC and LipA, respectively (see ‘Experimental Procedures‘ for details). For each interaction experiment, the bound ligand is underlined.
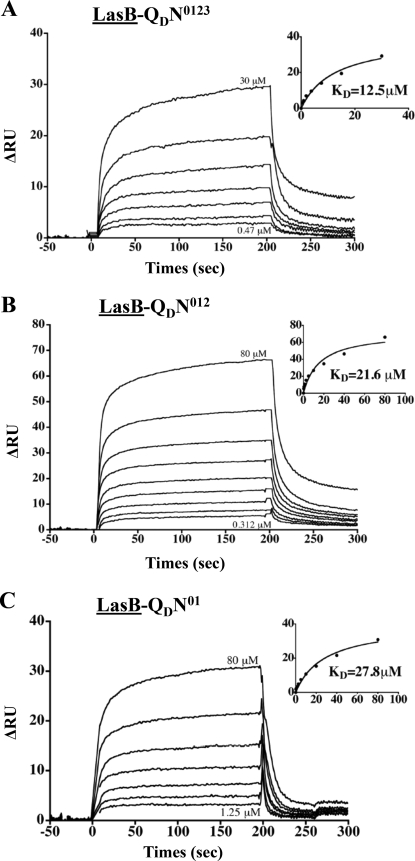
FIGURE 7.
LasB/QD interactions identified by SPR. A–C, the SPR sensorgrams of the positive interactions found between LasB (bound ligand, underlined) and QD-N0123, QD-N012, and QD-N01, respectively (see ‘Experimental Procedures‘ for details).
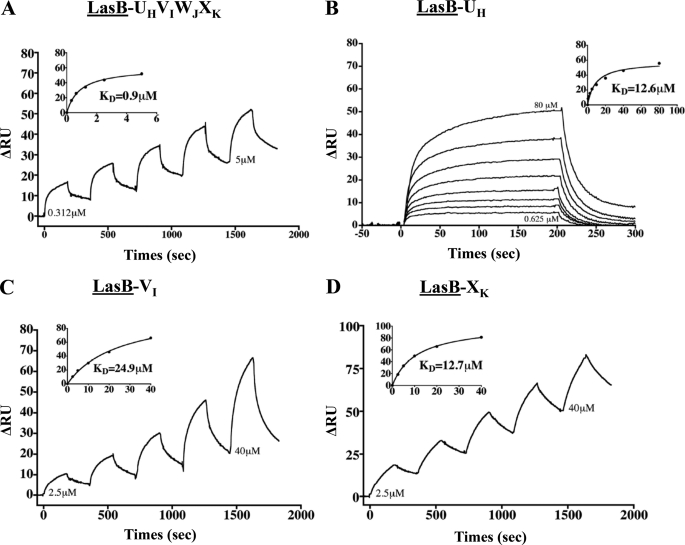
FIGURE 8.
LasB/pseudopilin interactions identified by SPR. A–D, the SPR sensorgrams of the positive interactions found between LasB (bound ligand, underlined) and the UH-VI-WJ-XK complex, UH, VI, and XK, respectively (see “Experimental Procedures” for details).
Batch Co-purification (Pulldown) of hisPC and QD-N0123, QD-N012, and QD-N01 and Quaternary Complex UH-VI-WJ-XK Periplasmic Domains
Batch co-purification experiments were performed as described in Ref. 10. The exact experimental procedure is described in the supplemental material.
Mass and Stoichiometry Calculation of XcpP-XcpQ Complex
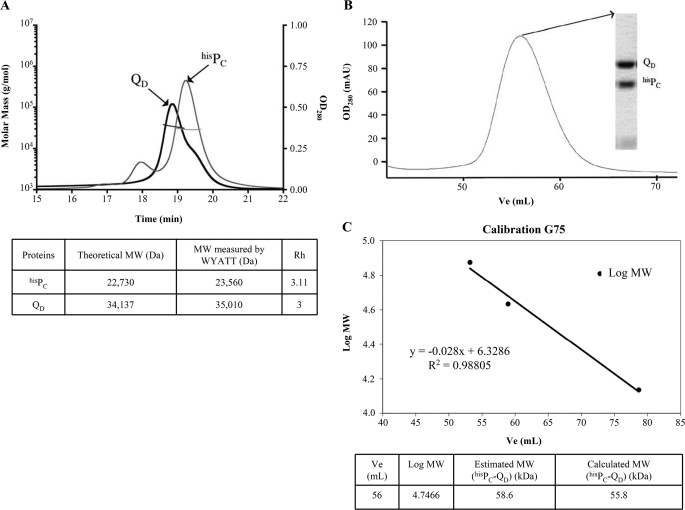
We characterized the mass of each soluble domain of hisPC and QD using the combination of UV spectrophotometry, MALS, and refractometry, coupled on-line with an analytical size-exclusion chromatography column, and we determined the hydrodynamic radii using an online QELS. UV, MALS, QELS, and refractometry measurements were achieved with a photodiode array 2996 (Waters), a miniDAWN TREOS (Wyatt Technology), a DynaPro (Wyatt technology), and an Optilab rEX (Wyatt Technology), respectively. Mass and hydrodynamic radius calculations were performed the with the ASTRA software (Wyatt Technology) using a dn/dc value of 0.185 ml/g. The column used was a 15-ml KW-804 (Shodex) with at 0.25 ml min−1 flow, on an Alliance HPLC 2695 system (Waters). The buffer was 50 mm Tris, pH 7.5, 150 mm NaCl. 50 μl (KW-804) of each protein sample at 5 mg/ml in Tris 50 mm, 150 mm NaCl, pH 7.5, were injected in each experiment. To estimate the stoichiometry of the complex, PC-QD hisPC, and QD were copurified using nickel affinity. Elution fractions were concentrated on Centricon with a 30-kDa cutoff and passed through a Sephadex 75 16/60 (GE Healthcare) equilibrated in 50 mm phosphate, 150 mm NaCl, pH 7. The Sephadex G75 was calibrated using the gel filtration LMW calibration kit (GE Healthcare). The ribonuclease A (13.7 kDa), ovalbumin (43 kDa) and conalbumin (75 kDa) were used as standard proteins to estimate the molecular mass of the PC-QD complex.
RESULTS
XcpPC Is the Peripheral Periplasmic Element of the Secreton
XcpPC is a central constituent of the Xcp T2SS. This IM protein possesses a large periplasmic domain, which directly interacts with the OM component of the machinery, the secretin XcpQD (16, 17) (Fig. 1A). To identify secretin domains as well as other secreton components that interact with the periplasmic part of XcpPC, we systematically assayed in vitro interactions between the periplasmic soluble domains of XcpPC (PC) and various periplasmic domains of secreton components. To this end, three variants of secretin soluble domains (QD) and the five pseudopilin soluble domains (TG, UH, VI, WJ, and XK) were engineered, produced, and purified in the mm range (see “Experimental Procedures”). Then, using two complementary techniques, affinity chromatography and SPR, we systematically tested in vitro protein/protein interactions using hisPC. As expected, we confirmed the interaction between PC and QD (Figs. 2 and 3, A and B). Moreover, to more precisely identify the QD subdomains involved in this interaction, we constructed and tested two other truncated versions lacking N3 (QD-N012) and the N3-N2 subdomain(s) (QD-N01), in addition to the peri-XcpQD-N0123 (QD or QD-N0123). Both SPR and pulldown experiments indicated that the N3 subdomain of QD was necessary for the interaction with PC because QD-N012 lost its capacity to bind PC (Figs. 2 and 3, C and D). This result suggests that PC covers the whole periplasmic space; at one end, XcpPC is anchored into the IM, and at the other end, it interacts with N3, the closest OM secretin periplasmic subdomain (Fig. 1A) (15). Further experiments indicate that purified PC and QD are monomeric in solution (Fig. 4A) and form a binary complex with a 1:1 ratio (Fig. 4, B and C). Based on recent cryo-electron microscopy data revealing an internal diameter of the dodecameric periplasmic secretin cavity of 55 Å (12), the binding of 12 PC inside the QD cavity is structurally not feasible; therefore, it could only take place on the external face of the secretin pore. Consequently, we propose that 12 PC subunits form a dodecameric coat around the periplasmic portion of the secreton (see model in Fig. 1A).
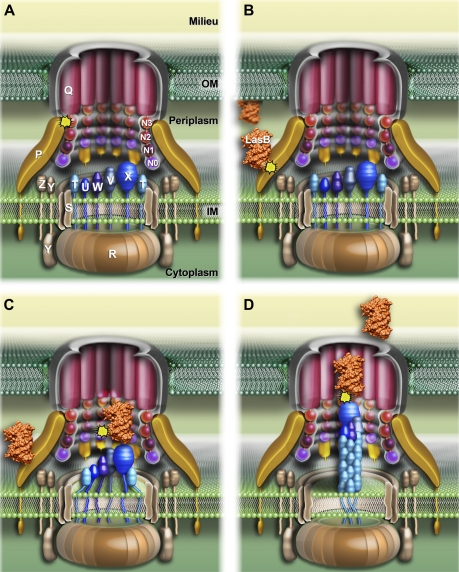
FIGURE 1.
Schematic three-dimensional model of the P. aeruginosa Xcp secreton showing substrate recognition and transport during the type II secretion process. A, the schematic representation of the P. aeruginosa Xcp secreton. All Xcp components (labeled following Xcp nomenclature only) are represented according to their cellular localization, topology, and multimerization state. B–D, the different consecutive steps followed by the substrate for its recruitment, transport, and release by the secreton during the type II secretion process (see “Discussion” for details). The interactions identified in this study are represented by yellow asterisks.
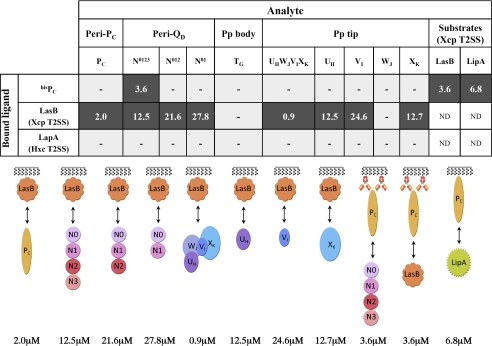
FIGURE 2.
Interaction network between the periplasmic domains of the P. aeruginosa Xcp secreton and substrates by SPR. Ligand and analytes are indicated following the code used under “Experimental Procedures.” Positive interactions are indicated by black boxes, in which the Kd values (μm) of the interactions are indicated. Gray boxes with dashes indicate that no interaction was detected. The schemes for all positive interactions are represented below the table and follow the color code used in Fig. 1. Pp, pseudopilus; ND, not determined.
FIGURE 4.
Purified PC and QD are monomeric in solution and interact in a 1:1 ratio. A, hisPC and QD-N0123 analyzed by MALS/QELS/UV/refractometry experiments. The protein masses of hisPC and QD-N0123 were measured as 23,560 and 35,010 Da, respectively. These values are close to their respective theoretical masses of 22,730 and 34,137 Da obtained by ProtParam. OD280, optical density at 280 nm. B, size-exclusion chromatography profile of the hisPC-QD-N0123 complex on a Sephadex 75 16/60 column. The inset represents an SDS-PAGE gel of the peak fraction, showing the hisPC-QD-N0123 complex. mAU, milliabsorbance units; V, elution volume. C, Sephadex 75 column calibration using gel filtration LMW standard proteins. The calculated mass of 55.8 kDa is in agreement with the estimated molecular mass of the hisPC-QD-N0123 complex by Wyatt Technology of 58.6 kDa.
Interestingly, we could not detect any interactions between PC and the pseudopilin soluble domains, either individually or as assembled in the quaternary pseudopilus tip complex, in either SPR or pulldown experiments (Figs. 2 and 3E). The absence of any interaction between PC and the pseudopilus components indicates that PC is physically separated from the pseudopilus. This result is in agreement with the secreton model, which presents the pseudopilus inside the secretin cavity (Fig. 1A) (15).
Xcp Substrates First Interact with XcpPC
Three purified P. aeruginosa T2SS substrates, the elastase LasB, the lipase LipA, and the alkaline phosphatase LapA, were used in this study (Fig. 5). LasB is the most abundant protein secreted by the P. aeruginosa Xcp T2SS. The interaction between LasB and PC was analyzed by SPR. LasB was covalently immobilized on a CM5 chip, and SPR experiments revealed a specific interaction between LasB and PC (Figs. 1B, 2, and 6A). This newly described direct interaction between a substrate and one component of the secreton was confirmed by the reciprocal SPR experiment, in which hisPC was fixed to an anti-penta-His antibody covalently linked to a CM5 chip. Purified LasB was passed over hisPC and showed a significant affinity for it (Figs. 2 and 6B). The physiological relevance of such a PC/substrate interaction during type II secretion is strengthened based on the positive interaction between PC and a second Xcp substrate, the lipase LipA (Figs. 2 and 6C). Furthermore, no interaction could be detected between PC and the alkaline phosphatase LapA, an exoprotein specifically secreted by the second P. aeruginosa T2SS system, Hxc (Fig. 2). This important result, which confirms the specificity of the interactions between PC and Xcp T2SS substrates, also validates the physiological relevance of our in vitro results. Considering that XcpPC forms the external wall of the periplasmic portion of the secreton, we propose that it is the first component of the secreton to bind the substrate, and therefore, that XcpPC is the recruiter of the system (Fig. 1B).
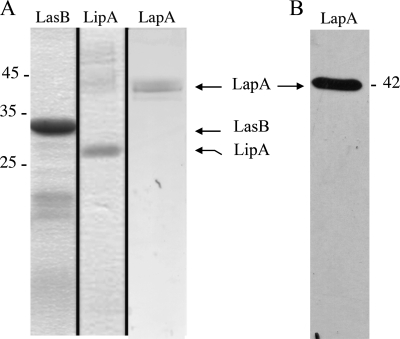
FIGURE 5.
Analysis of purified LasB, LipA, and LapA. A, samples of purified LasB, LipA, and LapA used in SPR experiments were analyzed by 15% SDS-PAGE. After electrophoresis, the gel was stained with Coomassie Blue. The positions of the molecular mass markers (kDa) are indicated by a dash. The positions of purified LasB, LipA, and LapA are indicated by arrows. B, immunodetection of purified LapA with anti-LapA antibodies.
LasB Binds the N0N1 Secretin Subdomains
Once recruited by XcpPC, the substrate is probably imported inside the machinery, where it should interact with several secreton components. One of these components is likely the partner of XcpPC: the periplasmic soluble domain of the secretin. Using LasB covalently linked to a CM5 chip, we employed SPR to analyze the interactions between substrate and the four different periplasmic subdomains of the OM secretin. We passed QD-N0123, QD-N012, and QD-N01 over LasB and found similar affinities for the three QD subdomains, indicating that the LasB-interacting domain on QD belongs to the two N0N1 subdomains (Figs. 1C, 2, and 7). This result, together with the previously identified interaction of PC with QD through the N3 subdomain, indicates that two different interaction sites exist on QD: one for PC and another for LasB (Fig. 1, A and C). Moreover and in line with the substrate specificity, no interaction was detected between secretin subdomains and the Hxc-T2SS substrate LapA (Fig. 2).
LasB Interacts with the Pseudopilus Tip
Interaction with the secretin does not explain how the substrate could be released by the machinery. It could be hypothesized that interaction with the secretin presents the substrate for expulsion by the growing pseudopilus. Using LasB immobilized on a CM5 chip, we measured its affinity for the five pseudopilins, either in a complex or individually. Our data revealed a direct interaction between the substrate and the quaternary complex of minor pseudopilins located at the tip of the pseudopilus (Figs. 1D, 2, and 8). Individual interactions between pseudopilins and LasB indicated that three of the four minor pseudopilin soluble domains, UH, VI, and XK, interacted with the substrate, whereas WJ did not show any significant binding (Figs. 2 and 8). Together with the recent SPR-identified interaction between the pseudopilus tip and the periplasmic domain of the secretin (12), our findings validate the piston model, in which the substrate, located in the secretin vestibule, is expelled out of the cell by the growing pseudopilus through the secretin pore.
Interestingly, no interaction was established between the substrate and the major pseudopilin soluble domain, TG (Fig. 2), suggesting that only the tip, and not the pseudopilus body, binds the substrate during the secretion process. This finding is in agreement with previous findings indicating that major pseudopilins could be exchanged among T2SSs without affecting their substrate specificity (23). It is noteworthy that no binding of any Xcp pseudopilins was observed when LapA was immobilized on the chip (Fig. 2).
DISCUSSION
In the actual representation of the T2SS, the OM secretin is linked to the IM platform via its interaction with GspCP, and the pseudopilus acts as a piston to push the substrate out through the secretin pore (4, 15, 24–28). This model, and more particularly the substrate recognition and transport mechanism, was very speculative because no interactions between the substrates and the components of the T2SS other than the secretin have been reported. In this study, we used SPR to identify several unprecedented specific interactions between secreted substrates and secreton components in the P. aeruginosa Xcp T2SS. For all of the substrate/secreton interactions tested, the estimated Kd values were found in the micromolar range (Fig. 2). Such weak interactions suggest a transient stay of the substrate in the secreton during the secretion process.
All together, our results allowed us to propose a sequential model for substrate recognition and secretion by the T2SS (Fig. 1). In this model, we show that XcpPC bridges the inner and outer membrane elements of the machinery and probably envelopes the whole periplasmic part of the secreton (Fig. 1A). Thanks to the multiple specific interactions found between secreted substrates and secreton components, we propose that the exported exoproteins: (i) are initially recruited in the periplasm by the XcpPC (Fig. 1B), (ii) move into the periplasmic vestibule of the secretin (Fig. 1C), and (iii) are further pushed through the pore by the pseudopilus elongating from the IM platform (Fig. 1D). Finally, (iv) the pseudopilus retracts, allowing the positioning of a new substrate in the secretin vestibule for the next secretion cycle.
Two independent studies (16, 17) have shown that GspCP interacts with GspDQ. Although none of these studies exclude the involvement of the GspDQ N3 subdomain, they clearly show that other GspDQ subdomains interact with GspCP. In our case, QD-N012 is incapable of binding PC, whereas it is capable of binding substrates. This discrepancy could be due to subtle mechanistic differences between T2SSs, reflected by the low homology among GspCP components. On the GspCP side, Korotkov et al. (16) and Login et al. (17) agreed that GspCP bound GspDQ through one specific domain, the homology region (HR) (29). Notably, the XcpPC HR is not involved in substrate binding, according to Gerard-Vincent et al. (29), who showed that an XcpPC variant harboring the HR of the Erwinia chrysanthemi OutCP homolog was still able to secrete LasB from P. aeruginosa. In the same study, it was shown that an XcpPC variant containing the Erwinia OutCP TMHR located just upstream of the HR was non-functional, thus suggesting that specific substrate recognition by GspCP might implicate the TMHR. Interestingly, the TMHR is located right after the XcpPC transmembrane domain. We therefore suggest that newly periplasmic exported substrates might enter the secreton from its periplasmic base because they successively interact with XcpPC and N0N1 secretin subdomains located in the vicinity of the inner membrane.
The specific direct interactions identified between folded exoproteins and the five Xcp components provide important information about machinery/substrate recognition. Our findings indeed suggest that the secretion signal is located on the fully active native protein and not on an intermediate state of folding nor on a dedicated periplasmic chaperone, notably the elastase propeptide and the lipase foldase (4). Our data also confirm the conformational nature of the secretion signal because there are no common linear motifs on the 13 mature Xcp secreted proteins. Future studies will try to identify the LasB and LipA motif(s) specifically involved in their recruitment by XcpPC and subsequent interactions with the secretin and the pseudopilus.
In our secretion model, stoichiometric data presented in Fig. 4 indicate that 12 molecules of XcpPC surround the periplasmic portion of the secreton through 12 PC/QD heterodimeric interactions (Fig. 1A). This organization suggests that the secreton component initially contacted by the secreted proteins is XcpPC. We found support for this hypothesis by identifying a direct interaction between XcpPC and two secreted proteins. Such a function for XcpPC in substrate recruitment and substrate specificity is in agreement with the low level of conservation among GspCP homologs in type II secretion machineries.
In our model, we propose that after recruitment by XcpPC, the substrate first enters into the secretin vestibule in a position that allows it to be subsequently pushed by the pseudopilus growing from the IM platform. Notably, the binding of substrate to the pseudopilus prior to the secretin is unlikely because substrate has been visualized inside the secretin vestibule in the absence of the pseudopilus (30).
In conclusion, we identified in this study subtle, specific, and likely transitory interactions between T2SS-secreted exoproteins and the transport machinery. Collectively, our results provide an improved understanding of two important steps of the secretion process: substrate recognition and transport by the machinery. We propose an improved model of the type II secretion process, opening new routes for academic investigation and for antimicrobial targeting utilizing organic disruptors (31).
Supplementary Material
Acknowledgments
We thank Christophe Quétard (GE Healthcare) for careful advice on SPR experiments, Sawsan Amara for assistance in lipase activity assays, Sophie Bleves, Alain Filloux, Katrina Forest, and Tâm Mignot for important and valuable discussions, and Yves-Michel Cully for preparation of Fig. 1.
This work was supported by “3D-Pilus” Young Researcher Agence Nationale de Recherches (ANR) Grant ANR-JC07-183230.
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental experimental procedures and Tables S1 and S2.
- IM
- inner membrane
- OM
- outer membrane
- T2SS
- type II secretion system
- MALS
- multiangle light scattering
- QELS
- quasi-elastic light scatter detection
- HR
- homology region
- TMHR
- transmembrane homology region
- RU
- response unit.
REFERENCES
- 1. Economou A., Christie P. J., Fernandez R. C., Palmer T., Plano G. V., Pugsley A. P. (2006) Mol. Microbiol. 62, 308–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cianciotto N. P. (2005) Trends Microbiol. 13, 581–588 [DOI] [PubMed] [Google Scholar]
- 3. Voulhoux R., Ball G., Ize B., Vasil M. L., Lazdunski A., Wu L. F., Filloux A. (2001) EMBO J. 20, 6735–6741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Filloux A. (2004) Biochim. Biophys. Acta 1694, 163–179 [DOI] [PubMed] [Google Scholar]
- 5. Patrick M., Korotkov K. V., Hol W. G., Sandkvist M. (2011) J. Biol. Chem. 286, 10378–10386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Py B., Loiseau L., Barras F. (2001) EMBO Rep. 2, 244–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen Y., Shiue S. J., Huang C. W., Chang J. L., Chien Y. L., Hu N. T., Chan N. L. (2005) J. Biol. Chem. 280, 42356–42363 [DOI] [PubMed] [Google Scholar]
- 8. Abendroth J., Bagdasarian M., Sandkvist M., Hol W. G. (2004) J. Mol. Biol. 344, 619–633 [DOI] [PubMed] [Google Scholar]
- 9. Campos M., Nilges M., Cisneros D. A., Francetic O. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 13081–13086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Douzi B., Durand E., Bernard C., Alphonse S., Cambillau C., Filloux A., Tegoni M., Voulhoux R. (2009) J. Biol. Chem. 284, 34580–34589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Korotkov K. V., Hol W. G. (2008) Nat. Struct. Mol. Biol. 15, 462–468 [DOI] [PubMed] [Google Scholar]
- 12. Reichow S. L., Korotkov K. V., Hol W. G., Gonen T. (2010) Nat. Struct. Mol. Biol. 17, 1226–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sauvonnet N., Gounon P., Pugsley A. P. (2000) J. Bacteriol. 182, 848–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hu N. T., Leu W. M., Lee M. S., Chen A., Chen S. C., Song Y. L., Chen L. Y. (2002) Biochem. J. 365, 205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Korotkov K. V., Gonen T., Hol W. G. (2011) Trends Biochem. Sci. 36, 433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Korotkov K. V., Krumm B., Bagdasarian M., Hol W. G. (2006) J. Mol. Biol. 363, 311–321 [DOI] [PubMed] [Google Scholar]
- 17. Login F. H., Fries M., Wang X., Pickersgill R. W., Shevchik V. E. (2010) Mol. Microbiol. 76, 944–955 [DOI] [PubMed] [Google Scholar]
- 18. Ball G., Durand E., Lazdunski A., Filloux A. (2002) Mol. Microbiol. 43, 475–485 [DOI] [PubMed] [Google Scholar]
- 19. Voulhoux R., Taupiac M. P., Czjzek M., Beaumelle B., Filloux A. (2000) J. Bacteriol. 182, 4051–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sauvonnet N., Pugsley A. P. (1996) Mol. Microbiol. 22, 1–7 [DOI] [PubMed] [Google Scholar]
- 21. Stuer W., Jaeger K. E., Winkler U. K. (1986) J. Bacteriol. 168, 1070–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rich R. L., Hoth L. R., Geoghegan K. F., Brown T. A., LeMotte P. K., Simons S. P., Hensley P., Myszka D. G. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8562–8567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vignon G., Köhler R., Larquet E., Giroux S., Prévost M. C., Roux P., Pugsley A. P. (2003) J. Bacteriol. 185, 3416–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Filloux A., Michel G., Bally M. (1998) FEMS Microbiol. Rev. 22, 177–198 [DOI] [PubMed] [Google Scholar]
- 25. Nunn D. (1999) Trends Cell Biol. 9, 402–408 [DOI] [PubMed] [Google Scholar]
- 26. Sandkvist M. (2001) Mol. Microbiol. 40, 271–283 [DOI] [PubMed] [Google Scholar]
- 27. Johnson T. L., Abendroth J., Hol W. G., Sandkvist M. (2006) FEMS Microbiol. Lett. 255, 175–186 [DOI] [PubMed] [Google Scholar]
- 28. Cianciotto N. P. (2009) Future Microbiol. 4, 797–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gérard-Vincent M., Robert V., Ball G., Bleves S., Michel G. P., Lazdunski A., Filloux A. (2002) Mol. Microbiol. 44, 1651–1665 [DOI] [PubMed] [Google Scholar]
- 30. Reichow S. L., Korotkov K. V., Gonen M., Sun J., Delarosa J. R., Hol W. G., Gonen T. (2011) Channels 5, 215–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shahian T., Lee G. M., Lazic A., Arnold L. A., Velusamy P., Roels C. M., Guy R. K., Craik C. S. (2009) Nat. Chem. Biol. 5, 640–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.