Background: PEX5 binds newly synthesized peroxisomal proteins in the cytosol and releases them in the organelle matrix.
Results: PEX5 binds monomeric catalase and releases it in the presence of PEX14.
Conclusion: PEX14 participates in the cargo release step.
Significance: Knowing how PEX5 interacts with cargo proteins and which factors disrupt this interaction are crucial for understanding this protein sorting pathway.
Keywords: Peroxisomes, Protein Sorting, Protein Targeting, Protein-Protein Interactions, Subcellular Organelles, PEX14, PEX5, Catalase
Abstract
Newly synthesized peroxisomal matrix proteins are targeted to the organelle by PEX5. PEX5 has a dual role in this process. First, it acts as a soluble receptor recognizing these proteins in the cytosol. Subsequently, at the peroxisomal docking/translocation machinery, PEX5 promotes their translocation across the organelle membrane. Despite significant advances made in recent years, several aspects of this pathway remain unclear. Two important ones regard the formation and disruption of the PEX5-cargo protein interaction in the cytosol and at the docking/translocation machinery, respectively. Here, we provide data on the interaction of PEX5 with catalase, a homotetrameric enzyme in its native state. We found that PEX5 interacts with monomeric catalase yielding a stable protein complex; no such complex was detected with tetrameric catalase. Binding of PEX5 to monomeric catalase potently inhibits its tetramerization, a property that depends on domains present in both the N- and C-terminal halves of PEX5. Interestingly, the PEX5-catalase interaction is disrupted by the N-terminal domain of PEX14, a component of the docking/translocation machinery. One or two of the seven PEX14-binding diaromatic motifs present in the N-terminal half of PEX5 are probably involved in this phenomenon. These results suggest the following: 1) catalase domain(s) involved in the interaction with PEX5 are no longer accessible upon tetramerization of the enzyme; 2) the catalase-binding interface in PEX5 is not restricted to its C-terminal peroxisomal targeting sequence type 1-binding domain and also involves PEX5 N-terminal domain(s); and 3) PEX14 participates in the cargo protein release step.
Introduction
Mammalian peroxisomal matrix proteins are synthesized on cytosolic ribosomes and post-translationally targeted to the organelle matrix by PEX5, the peroxisomal shuttle receptor (1–4). The vast majority of these proteins possess the so-called peroxisomal targeting signal type 1 (PTS1),4 a C-terminal sequence frequently ending with the tripeptide SKL or a derivative of it (5). This PTS1 interacts directly with the C-terminal half of PEX5, a domain comprising seven tetratricopeptide repeats motifs arranged into a ring-like structure (6). A minor fraction of peroxisomal matrix proteins contains instead a PTS type 2 (PTS2), a degenerated nonapeptide near their N termini (7). The PTS2-PEX5 interaction is not direct but rather is mediated by the adaptor protein PEX7 (8).
According to current models (1–4), newly synthesized peroxisomal matrix proteins interact with PEX5 while still in the cytosol. These PEX5-cargo protein complexes then dock at the peroxisomal docking/translocation machinery (DTM), a multisubunit protein complex comprising the intrinsic membrane proteins PEX13, PEX14, and the RING finger peroxins PEX2, PEX10, and PEX12 (9, 10). By a still ill-defined process, this interaction ultimately leads to the insertion of PEX5 into the DTM with the concomitant translocation of the cargo protein into the peroxisomal matrix (11–13). In at least one case, that of the PTS2-containing protein thiolase, it is also at this stage that PEX5 releases its cargo into the peroxisomal matrix (11). Interestingly, in vitro import experiments suggest that ATP hydrolysis is not needed at any of these steps, suggesting that the complete transport of a cargo protein from the cytosol into the peroxisomal matrix is driven by thermodynamically favored protein-protein interactions at the DTM (14–16). After these events, PEX5 is extracted from the DTM back into the cytosol. This involves monoubiquitination of PEX5 at a conserved cysteine residue (17–20) and the ATP-dependent extraction of the ubiquitin-PEX5 conjugate from the DTM by the mechanoenzymes PEX1 and PEX6, two members of the AAA family of ATPases (14–16). Finally, ubiquitin is removed from PEX5 probably by a combination of enzymatic and nonenzymatic processes (21, 22).
Despite all the advances made in recent years, there are still many aspects of this protein import pathway that remain unclear. A particularly important one regards the quaternary structure of the PEX5-cargo protein complex formed in the cytosol. In principle, a protein complex comprising a single PEX5 molecule and a cargo protein should be sufficient to ensure the correct targeting of that protein to the peroxisomal matrix. This is probably the case for all peroxisomal monomeric proteins (e.g. the sterol carrier protein 2 (23)), for some oligomeric enzymes in which the peroxisomal targeting signals become hidden upon oligomerization (24–27), and for natural or artificial heterodimers in which only one of the subunits contains peroxisomal targeting information (28–30). The situation for many other peroxisomal oligomeric proteins, however, is not that clear. Indeed, the observation that peroxisomes have the capacity to import some already oligomerized proteins, at least under conditions of high protein expression (28, 31–34), together with the fact that several peroxisomal oligomeric proteins may expose multiple PTS1 sequences at their surface, could suggest that these cargo proteins are transported to the organelle by more than one PEX5 molecule. Such a scenario was in fact the central premise of one hypothetical model proposed a few years ago aimed at describing the process of protein translocation across the peroxisomal membrane (35).
In an effort to understand how these proteins are sorted to the peroxisome, we started to characterize the interaction of their monomeric and oligomeric versions with PEX5. Here, we describe the results obtained with catalase, one of the most abundant peroxisomal matrix proteins and probably one of the most frequent clients of the DTM (36–38). Catalase is a heme-containing homo-tetrameric protein in its native state (four subunits of 60 kDa), with each subunit possessing a noncanonical PTS1 at its C terminus (KANL) (39–43). We selected catalase for this initial study because there are data suggesting that both its monomeric and tetrameric versions are substrates for the peroxisomal protein import machinery (27, 44–49). However, whether the peroxisomal import machinery, PEX5 in particular, displays any preference for monomeric or tetrameric catalase was unknown.
Here, we show that mammalian PEX5 binds monomeric catalase (hereafter referred to as mCat) in a much stronger manner than it binds tetrameric catalase (tCat). Actually, we were unable to detect stable PEX5-tCat complexes. Importantly, PEX5 binding to mCat blocks its tetramerization with an IC50 in the nanomolar range. The interaction of PEX5 with mCat was found to involve the PTS1-binding C-terminal half of PEX5 as well as a domain(s) present in its N-terminal half. Finally, we provide data suggesting that the PEX5-mCat interaction is disrupted by the N-terminal domain of PEX14, a central component of the DTM. The implications of these findings on the mechanism of protein translocation across the peroxisomal membrane are discussed.
EXPERIMENTAL PROCEDURES
Recombinant Proteins
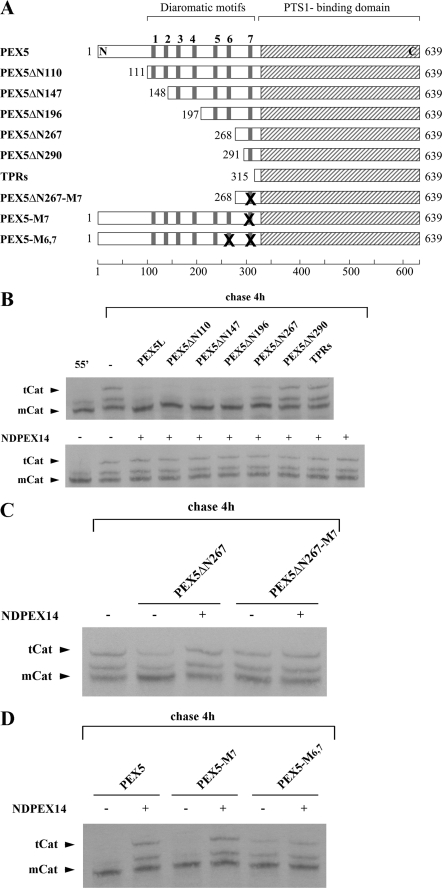
The recombinant large isoform of human PEX5 (50, 51), hereafter referred to as PEX5 for simplicity, a protein comprising the first 324 amino acid residues of PEX5 (ΔC1PEX5), a protein containing amino acid residues 315–639 of PEX5 (TPRs), PEX5 containing the missense mutation N526K (PEX5N526K), a protein comprising the first 80 amino acid residues of human PEX14 (NDPEX14), and full-length PEX19 (PEX19) were obtained as described previously (18, 52–55). The following truncated versions of human PEX5 were also produced: PEX5ΔN110 (amino acid residues 111–639 of PEX5), PEX5ΔN147 (amino acid residues 148–639 of PEX5), PEX5ΔN196 (amino acid residues 197–639 of PEX5), PEX5ΔN267 (amino acid residues 268–639 of PEX5), and PEX5ΔN290 (amino acid residues 291–639 of PEX5). The cDNAs encoding these proteins were obtained by PCR using the primers listed in supplemental Table 1 and the pQE30-PEX5 construct as template. The amplified DNA fragments were then digested with NdeI and SalI and cloned into the NdeI/SalI digested pET-28c vector (Novagene). The QuikChange® site-directed mutagenesis kit (Stratagene) was used to replace tryptophan and phenylalanine/tyrosine residues in diaromatic motifs of PEX5 by alanines (see primers in supplemental Table 1). The three proteins obtained in this way are as follows: PEX5ΔN267-M7 and PEX5-M7, proteins with a mutated 7th diaromatic motif, and PEX5-M6,7, a protein possessing both the 6th and 7th diaromatic mutated. All plasmids were sequence-verified.
The cDNA encoding mouse PEX5 was amplified from a commercially available clone (clone MMM1013-7510385, Open Biosystems) using the primers listed in supplemental Table 1, digested with NdeI and SalI, and cloned into the NdeI/SalI digested pET-28c vector. Purification of all PEX5 proteins was done as described previously (54).
Synthesis of Radiolabeled Proteins
The cDNA encoding full-length human catalase (clone IMAGE ID 5551309, Open Biosystems) was amplified by PCR using the primers listed in supplemental Table 1. This DNA was digested with XbaI and KpnI and cloned into the XbaI/KpnI-digested pGEM-4 vector (Promega), originating pGEM-4-Cat. This plasmid was used as template to produce two other plasmids, one encoding a catalase lacking its four last C-terminal amino acid residues (CatΔKANL) and the other encoding a catalase in which these four residues were replaced by ED (CatED). These plasmids were obtained using the QuikChange® site-directed mutagenesis kit (Stratagene) and the primer pairs described in supplemental Table 1. 35S-Labeled proteins were synthesized using the TnT® T7 QuickCoupled transcription/translation kit (Promega) in the presence of [35S] methionine (specific activity >1000 Ci/mmol; PerkinElmer Life Sciences) following the standard conditions of the manufacturer. Unless otherwise indicated, protein synthesis was allowed to proceed for 55 min and was then blocked with 0.5 mm of cycloheximide (final concentration). Chase incubations were done at 30 °C for the specified periods of time. Chase reactions performed in the presence of recombinant proteins typically contained 6 μl of the translation mixture in a final volume of 10 μl.
Native PAGE
Proteins were incubated in 10 μl of 50 mm Tris-HCl, pH 8.0, 2 mm DTT for 5 min at room temperature. After addition of 1 μl of 0.17% (w/v) bromphenol blue, 50% (w/v) sucrose, the samples were loaded into Tris nondenaturating discontinuous 8% polyacrylamide gels (56). The gels were run at 250 V at 4 °C for 1 h (unless indicated otherwise), blotted onto nitrocellulose membranes, stained with Ponceau S, and exposed to an x-ray film.
Size-exclusion Chromatography
35S-Labeled proteins (50 μl of in vitro transcription/translation reactions) or mixtures containing recombinant proteins and 35S-labeled proteins were diluted to 250 μl with 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA-NaOH, 1 mm DTT and injected into a Superose 12 10/300 GL column (GE Healthcare; loop volume 200 μl) running with the same buffer at 0.5 ml/min. The column was calibrated with the following globular proteins: ferritin (440 kDa), bovine serum albumin (66 kDa), and soybean trypsin inhibitor (21.5 kDa). Fractions of 500 μl were collected and subjected to trichloroacetic acid precipitation, and one-third of each sample was analyzed by SDS-PAGE. The gels were blotted onto nitrocellulose membranes, stained with Ponceau S, and exposed to an x-ray film. Soluble mouse liver peroxisomal matrix proteins were obtained by sonicating purified peroxisomes (prepared as in Ref. 57) in 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA-NaOH, 1 mm DTT, and 1:500 (v/v) mammalian protease inhibitor mixture (Sigma) and centrifuging for 30 min at 100,000 × g. Two hundred micrograms of soluble proteins, supplemented or not with 300 μg of recombinant mouse PEX5, were injected into the size-exclusion column, as above. Aliquots of 25 μl from each fraction were analyzed by SDS-PAGE/Western blotting with antibodies directed to catalase (catalog number RDI-CATALASEabr; Research Diagnostics, Inc) and L-bifunctional protein (58).
Miscellaneous
The concentration of PEX5 in rat liver cytosol (0.75 μm) was calculated from the following data: total amount of PEX5 in liver, 4 ng/μg of total peroxisomal protein; percentage of PEX5 in cytosol, 85% (59); peroxisomes, 2.5% (w/w) of total liver protein; protein content of liver, 260 mg/g (60); 1 g of liver corresponds to 0.94 ml of which 44.4% is cytosol (61).
The weighted average molecular mass of monomeric rat liver peroxisomal proteins was estimated from the densitometric analysis of a Coomassie-stained SDS gel loaded with a highly pure peroxisomal preparation (57). Peak areas were divided by the corresponding apparent molecular masses and expressed as percentage of total moles. The weighted average of these values is 49 kDa. For newly synthesized peroxisomal proteins, this value may be slightly underestimated because protein maturation processes that occur in the matrix of the organelle (e.g. the cleavage of the 75-kDa acyl-CoA oxidase into the 53- and 22-kDa subunits (62)) were not taken into account. Mole percentage for catalase (13 mol %) was calculated considering the mass percentage of the protein in rat liver peroxisomes, 15% (63), the weighted average molecular mass of rat liver peroxisomal proteins (49 kDa), the theoretical molecular mass of catalase (60 kDa), and the mass percentage of matrix proteins in total rat liver peroxisomes, 92% (57).
The amount of total peroxisomal matrix proteins in nanomoles/g of rat liver was calculated from the above referred data. A value of 122 nmol/g of liver was obtained. Because “all the major protein components of the peroxisome have the same rate of turnover” (half-life of 1.3–1.5 days (37, 38)), one can estimate the rate of total peroxisomal matrix protein synthesis (k) as 30 pmol/min/g of liver. The rate of synthesis for a particular protein is k times its mole fraction in the peroxisomal matrix. For catalase (0.13 mol fraction), this corresponds to 3.9 pmol/min/g of liver, a value similar to the one reported previously (3.87 pmol/min/g of liver (63)). The steady-state concentration of newly synthesized peroxisomal matrix proteins in the cytosol ([P]cyt) can be estimated by the following expression: [P]cyt = k × 1.443 × t½, where [P]cyt is in pmol/g of liver; k is the rate of total peroxisomal matrix protein synthesis in pmol/min/g of liver, and t½ is the cytosolic half-life of the protein in min (47). According to Lazarow and co-workers (46, 64), several peroxisomal proteins display cytosolic half-lives of about 7 min (see Fig. 5 in Ref. 64). Two outliers were noted by those authors as follows: one was catalase, a protein presenting a cytosolic half-life of 14 min; the other was urate oxidase, a protein that after 4 min of chase was already completely found in peroxisomes, an observation suggesting that its cytosolic half-life is 2 min or less. We assume that all peroxisomal matrix proteins present a similar kinetic behavior, i.e. that on average their cytosolic half-lives are 7–8 min. The total concentration of newly synthesized peroxisomal proteins in the cytosol is thus 0.73–0.83 μm, with mCat contributing with 0.19 μm.
FIGURE 5.
PEX5 diaromatic motifs involved in the NDPEX14-induced disruption of the mCat-PEX5 interaction. A, schematic representation of recombinant PEX5 proteins. The diaromatic motifs in the N-terminal half of PEX5 are numbered 1–7. Replacement of tryptophan and phenylalanine/tyrosine residues by alanines in these motifs is indicated by X. B–D, 35S-labeled catalase was synthesized in vitro for 55 min (lane 55′) and chased for 4 h in the absence (lane −) or presence of 1 μm of the indicated recombinant PEX5 proteins alone or together with NDPEX14 (30 μm). Samples were analyzed by native-PAGE/autoradiography.
RESULTS
The rabbit reticulocyte lysate-based in vitro transcription/translation system has been one of the most powerful tools for the characterization of the molecular mechanisms underlying protein sorting pathways. We reasoned that this system might also be of use to study the first step of the catalase peroxisomal import pathway, namely when and how catalase interacts with cytosolic PEX5.
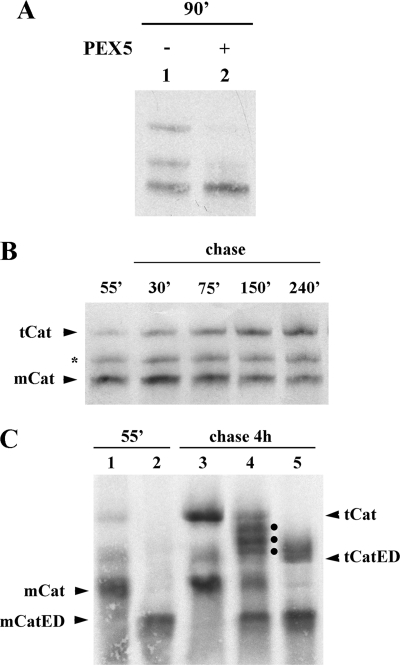
Fig. 1A shows a native-PAGE analysis of a standard in vitro transcription/translation reaction programmed with a plasmid encoding human catalase (lane 1). Three populations of 35S-labeled catalase are clearly seen in these gels. Notably, when catalase is synthesized in the presence of 1 μm human PEX5, the two slower migrating bands are no longer detected (Fig. 1A, lane 2). Apparently, some event(s) occurring in this system are blocked by PEX5.
FIGURE 1.
35S-Labeled catalase tetramerizes in vitro. A, human catalase was synthesized in vitro in a rabbit reticulocyte lysate for 90 min at 30 °C in the absence or presence of 1 μm human PEX5, as indicated, and analyzed by native-PAGE/autoradiography. B, 35S-labeled catalase was synthesized for 55 min. After adding cycloheximide, an aliquot was removed and frozen in liquid N2 (lane 55′). The remainder of the reaction was then incubated at 30 °C, and aliquots were removed and frozen at the indicated time points. The samples were subjected to native-PAGE/autoradiography. The protein bands labeled with mCat and tCat correspond to the monomeric and tetrameric forms of catalase; the band labeled with an asterisk probably represents dimeric catalase (see text for details). C, catalase and a mutant version of it possessing two acidic amino acid residues at the C terminus (CatED) were synthesized in vitro for 55 min and supplemented with cycloheximide (lanes 1 and 2, respectively). Aliquots of each reaction were then combined and incubated for 4 h at 30 °C (lane 4) or incubated individually under the same conditions (lanes 3 and 5 for catalase and CatED, respectively), and subjected to native PAGE/autoradiography. Note that this gel was run for 2.5 h to improve separation of tetramers. Longer electrophoretic runs also result in more diffuse bands. The dots in lane 4 indicate the three expected heterotetramers. mCatED and tCatED indicate the monomeric and tetrameric forms of CatED, respectively.
To understand the nature of the three catalase populations detected in these experiments and thus the inhibitory effect of PEX5, we first performed a pulse-chase analysis. In the experiment shown in Fig. 1B, catalase was synthesized for 55 min; cycloheximide was added to stop further synthesis, and the reaction was then chased for 4 h. The autoradiograph reveals that the faster migrating species (mCat; see below) is the main product after 55 min of synthesis. Its amount decreases during the chase period, with the concomitant increase of the slower migrating population (tCat). The protein band migrating between mCat and tCat (asterisk in Fig. 1B) remains fairly constant during the time course of these experiments. Its gel migration and kinetic behavior suggest that this population might be an oligomerization intermediate (probably a dimer), although further data are necessary to confirm this hypothesis.
Sedimentation analysis (data not shown) and size-exclusion chromatography (SEC; see below) of catalase before or after a 4-h chase incubation revealed that the faster migrating protein band corresponds to species displaying the hydrodynamic properties of a 60-kDa globular protein, whereas the protein in the slower migrating band behaves as a 200–250-kDa protein. Thus, the faster migrating band represents monomeric catalase, whereas the slower migrating band could be either the tetrameric enzyme or a protein complex containing catalase and some protein(s) from the in vitro protein synthesis system (e.g. a chaperone). To discriminate between these two possibilities, we adapted the strategy originally developed by Scandalios (65) to show that catalase is a tetrameric enzyme. For this purpose, we produced an acidic mutant version of catalase (CatED), which migrates faster than the normal enzyme in these gels, and we asked whether this protein is able to form heterotetramers with normal catalase upon a chase incubation of 4 h. If this is the case, then three heterotetramers containing 1, 2, or 3 molecules of the normal protein should be detected by native-PAGE; all these heterotetramers should migrate in these gels between the homotetramers of the parental molecules. If the slower migrating band corresponds to a complex containing catalase and some other protein(s), then the band pattern of the protein mixture should just correspond to the sum of the patterns obtained with each of the two catalase versions individually. The results presented in Fig. 1C indicate that the first possibility is the correct one.
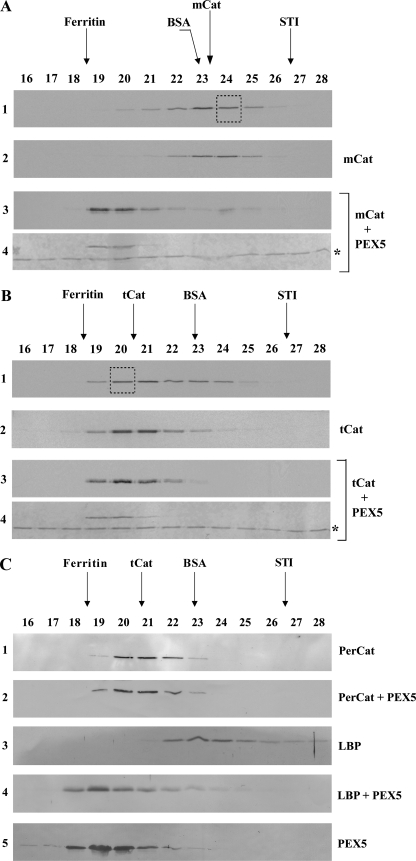
We next asked whether PEX5 can bind mCat and tCat. Because of the fact that the PEX5-catalase interaction is not preserved upon native-PAGE (see below), we used SEC, a technique in which proteins can be separated in a more physiological buffer. Translation reactions containing mCat (55 min of synthesis) and a mixture of mCat and tCat (55 min of synthesis plus 4 h of chase) were first subjected to SEC to purify mCat and tCat, respectively. Each of these proteins was then incubated with PEX5 or buffer alone and subjected to a second SEC. The results obtained with mCat are presented in Fig. 2A. In the absence of PEX5, the radiolabeled protein present in fraction 24 of the first SEC (Fig. 2A, panel 1) still elutes as a monomeric protein in the second SEC (panel 2), indicating that no tetramerization of mCat occurs during this procedure. In contrast, in the presence of PEX5, the elution volume of mCat is reduced, and the radiolabeled protein elutes now together with recombinant PEX5 (Fig. 2A, panels 3 and 4). Thus, PEX5 interacts with mCat. A different result was obtained for tCat. As shown in Fig. 2B, the elution profiles of tCat in the presence or absence of PEX5 are almost identical (Fig. 2B, panels 2 and 3) suggesting that these two proteins may interact only weakly. (Note that a major fraction of tCat co-elutes with PEX5 (Fig. 2B, compare panels 2 and 4) implying that, contrary to the situation with mCat, the two proteins are kept under near-equilibrium conditions during chromatography. This should facilitate the detection of PEX5-tCat complexes.) We also did not find evidence for the existence of a PEX5-tCat protein complex when mouse liver peroxisomal matrix proteins preincubated with either recombinant mouse PEX5 or buffer alone were subjected to SEC. Indeed, the elution volume of mouse catalase remains basically the same regardless of the presence of PEX5 (Fig. 2C, panels 1 and 2). This behavior contrasts to the one displayed by the L-bifunctional protein, a monomeric 78-kDa protein in its native state (58), which elutes much earlier in the presence of PEX5 (Fig. 2C, panels 4) than in its absence (panel 3). We still tried to detect a PEX5-tCat interaction by subjecting 35S-labeled tCat or mouse liver native catalase to ultracentrifugation through a solution containing 1 μm PEX5. Again, no evidence for a PEX5-tCat protein complex was obtained.5 Thus, if tCat interacts with PEX5, the Kd value of the interaction is larger than 1 μm. We note that interactions between PEX5 and recombinant catalase have been described before, but no quantitative binding data were reported (45, 66). Regardless of these uncertainties, it is clear from our results that PEX5 binds stronger to mCat than to tCat. Considering that tCat, unlike mCat, contains four PTS1 sequences, a property that should increase the stability of a putative complex with PEX5 because of an avidity effect, this is an unexpected finding. Apparently, some domain of the catalase polypeptide chain involved in the interaction with PEX5 is no longer accessible when the protein tetramerizes.
FIGURE 2.
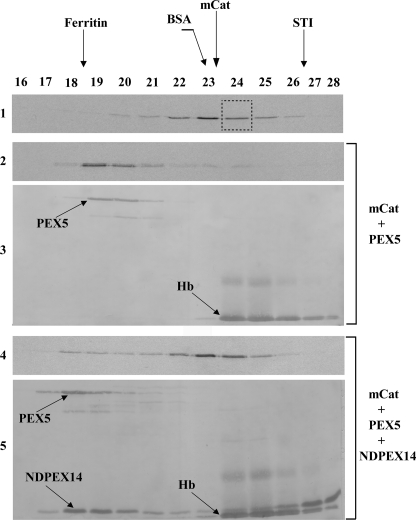
PEX5 binds monomeric catalase. A, 35S-labeled catalase was synthesized in vitro for 55 min and subjected to SEC. Radiolabeled mCat eluting in fraction 24 of this chromatography (panel 1, boxed lane) was then subjected to a second SEC either alone (panel 2) or after receiving 1 μm recombinant PEX5 (panels 3 and 4). Fractions were collected and subjected to SDS-PAGE/Western blotting. Autoradiographs (panels 1–3) and the Ponceau S-stained membrane showing PEX5 (panel 4) are presented. No recombinant PEX5 or 35S-labeled catalase were detected in the void volume of this column (fractions 14 and 15; not shown). The asterisk marks bovine serum albumin added to chromatography fractions before precipitation to control protein recoveries. B, 35S-labeled catalase, synthesized in vitro for 55 min and incubated for 4 h at 30 °C in the presence of cycloheximide, was subjected to SEC. Radiolabeled tCat eluting in fraction 20 (panel 1, boxed lane) was then subjected to a second SEC either alone (panel 2) or after receiving 1 μm recombinant PEX5 (panels 3 and 4). Fractions were processed as described above. Autoradiographs (panels 1–3) and the Ponceau S-stained membrane (panel 4) are presented. C, soluble proteins from mouse liver peroxisomes were incubated either with recombinant PEX5 or buffer alone and subjected to SEC. Fractions were subjected to SDS-PAGE/Western blotting using antibodies directed to catalase (PerCat) or L-bifunctional protein. Immunoblots (panels 1–4) and a Ponceau S-stained membrane showing PEX5 (panel 5) are presented. Note that PEX5, a monomeric 70-kDa protein in solution, displays an abnormal behavior upon SEC because a major fraction of its polypeptide chain is natively unfolded (52).
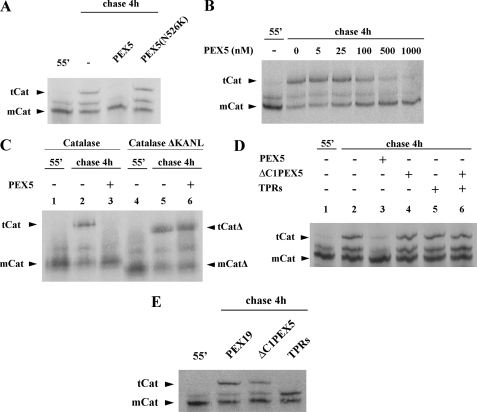
The results presented above indicate that PEX5 binds mCat blocking its tetramerization. We explored this phenomenon to further characterize the PEX5-catalase interaction. In the experiments described below, in vitro synthesized catalase was chased for 4 h in the presence of several recombinant proteins and analyzed by native-PAGE. As shown in Fig. 3A, PEX5, at 1 μm in the chase incubation, completely blocked catalase tetramerization, as expected. The inhibitory effect of PEX5 is quite strong displaying an IC50 of about 100 nm (Fig. 3B). No such effect was observed with PEX5(N526K), a mutant PEX5 molecule possessing a single missense mutation in the PTS1-binding domain that abolishes its activity (Fig. 3A) (67, 68). A similar result was obtained when a mutant version of catalase lacking the PTS1 (CatΔKANL) was used in this assay; tetramerization of this species was no longer sensitive to the inhibitory action of PEX5 (Fig. 3C, compare lane 3 with 6). Thus, inhibition of catalase tetramerization by PEX5 requires the interaction of catalase PTS1 sequence with the C-terminal PTS1-binding domain of PEX5. Interestingly, however, this domain of PEX5 alone (referred to as TPRs) does not display this capacity when tested in this assay at a 1 μm concentration (Fig. 3D, lane 5), and the same is true for a recombinant protein comprising the N-terminal half of PEX5 (ΔC1PEX5; Fig. 3D, lane 4). Likewise, a mixture of these two domains of PEX5, both at 1 μm, does not interfere with catalase tetramerization (Fig. 3D, lane 6), suggesting that these two domains of PEX5 have to reside in the same molecule (i.e. they have to be in a cis configuration) to inhibit catalase tetramerization at this concentration.
FIGURE 3.
PEX5 inhibits catalase tetramerization. A, 35S-labeled catalase was synthesized in vitro for 55 min (lane 55′) and chased for 4 h in the absence (lane −) or presence of 1 μm of the indicated recombinant proteins. B, same as in A, but using the indicated concentrations of PEX5. C, catalase (lanes 1–3) and a truncated version of it lacking the PTS1 signal (catalaseΔKANL; lanes 4–6) were synthesized for 55 min and chased in the absence (lanes 2 and 5) or presence of 1 μm PEX5 (lanes 3 and 6). D, 35S-labeled catalase was synthesized in vitro for 55 min (lane 1) and chased in the absence (lane 2) or presence of 1 μm of the indicated recombinant proteins (lanes 3–6). ΔC1PEX5 and TPRs, recombinant proteins comprising the N- and C-terminal half of PEX5, respectively. E, same as in A, but using 200 μm of the indicated recombinant proteins. Samples were analyzed by native-PAGE/autoradiography. Note that the gel shown in C was run for 2.5 h. mCat and tCat, monomeric and tetrameric versions of catalase, respectively; mCatΔ and tCatΔ, monomeric and tetrameric forms of catalaseΔKANL, respectively.
A plausible explanation for this finding is that domains present in both halves of PEX5 contribute to the interaction with mCat. We tested this hypothesis by performing additional tetramerization assays but this time using 200-fold larger concentrations of TPRs and ΔC1PEX5 in the chase incubations. PEX19, a protein involved in a different aspect of peroxisomal biogenesis (reviewed in Ref. 69), was used as a negative control. These experiments revealed that ΔC1PEX5 displays a weak but reproducible (n = 5) inhibitory effect on catalase tetramerization (Fig. 3E). A complete inhibition of catalase tetramerization is observed in the presence of 200 μm TPRs (Fig. 3E). Interestingly, there is an increase in the intensity of the band migrating between mCat and tCat in the sample chased in the presence of TPRs. This observation could suggest that TPRs does not inhibit catalase dimerization, although further data will be necessary to corroborate this interpretation. In summary, these data suggest the PEX5-mCat interaction involves domains present in both halves of PEX5.
After binding a newly synthesized peroxisomal protein in the cytosol, PEX5 docks at the DTM and promotes the translocation of its cargo across the organelle membrane. At the end of this process, DTM-embedded PEX5 has to release its cargo into the organelle matrix. Previous work in yeast suggested that PEX8, an intraperoxisomal component of the DTM, performs this task (70). However, mammals and many other organisms lack a PEX8 ortholog (71), and so the triggering mechanism for this event remains unknown. If we assume that a similar mechanism also operates in mammals, i.e. a DTM component interacts with PEX5 triggering the release of cargo into the organelle matrix, as the presently available data suggest (see Introduction), then a good candidate to perform this task is PEX14. PEX14 is an intrinsic membrane protein possessing a single putative transmembrane domain. Its C-terminal two-thirds are exposed into the cytosol, whereas its N-terminal domain is either embedded in the peroxisomal membrane or even exposed into the matrix of the organelle (72, 73). The interaction of PEX5 with the N-terminal domain of PEX14 is well documented but still poorly understood in mechanistic terms. Indeed, it is known that this domain of PEX14 (hereafter referred to as NDPEX14) interacts strongly with seven diaromatic motifs located at the N-terminal half of PEX5, some of which are indispensable for the function of PEX5 (66, 74, 75), but the reason for this complex mode of binding is unknown.
Thus, we asked whether NDPEX14 affects the PEX5-mCat interaction. For this purpose, we purified 35S-labeled mCat by SEC and incubated the radiolabeled protein with 1 μm recombinant PEX5 to generate the PEX5-mCat protein complex. This complex was then subjected to SEC either alone or after receiving 15 μm NDPEX14. As shown in Fig. 4, in the presence of NDPEX14, the elution volume of PEX5 is decreased (compare panel 3 with 5) indicating that a PEX5-NDPEX14 protein complex was formed. Importantly, under these conditions the vast majority of mCat elutes now as a monomeric protein (Fig. 4, compare panel 2 with 4). Thus, binding of NDPEX14 to PEX5 disrupts the PEX5-mCat interaction.
FIGURE 4.
N-terminal domain of PEX14 disrupts the mCat-PEX5 interaction. 35S-Labeled mCat was purified by SEC (panel 1, fraction 24), supplemented with 1 μm recombinant PEX5 and incubated for 30 min at room temperature to generate the PEX5-mCat protein complex. Half of this sample was analyzed directly by SEC (panels 2 and 3). The other half received recombinant NDPEX14 (15 μm) 30 min before chromatography (panels 4 and 5). Fractions were subjected to SDS-PAGE and blotted onto a nitrocellulose membrane. Autoradiographs (panels 1, 2, and 4) and the Ponceau S-stained membranes (panels 3 and 5) are presented. Hb, hemoglobin from the reticulocyte lysate that co-purified with mCat in the first SEC.
To better understand how NDPEX14 affects the mCat binding activity of PEX5, we produced several truncated forms of recombinant PEX5 (see Fig. 5A), and after evaluating their monodispersity and capacity to interact with NDPEX14 by native-PAGE (see supplemental Fig. S1), we tested them in the in vitro catalase tetramerization assay in the absence or presence of NDPEX14. The aim of these experiments was to identify the smallest PEX5 truncated molecule that still retains the capacity to bind mCat at low concentrations (i.e. 1 μm), as assessed by its capacity to inhibit tetramerization of the enzyme, and to determine whether the PEX14-binding diaromatic motif(s) present in this molecule is(are) involved in the disruption of the PEX5-mCat interaction. As shown in Fig. 5B (upper panel), PEX5ΔN110, PEX5ΔN147 and PEX5ΔN196, proteins lacking the first 110, 147 or 196 amino acid residues of PEX5, respectively, are as potent as full-length PEX5 in this assay. As expected from the data presented in Fig. 4, neither full-length PEX5 nor any of these truncated proteins displays an inhibitory effect on catalase tetramerization in the presence of NDPEX14 (Fig. 5B, lower panel). PEX5ΔN267, a protein containing only the 7th diaromatic motif of PEX5, still inhibits catalase tetramerization, although in a less potent manner. Again, this inhibitory effect is abolished in the presence of NDPEX14 (Fig. 5B, lower panel). In contrast, PEX5ΔN290 displays no inhibitory activity (Fig. 5B, upper panel). Taken together, these results suggest that the region between amino acid residues 197 and 290 of PEX5 is involved in the mCat-PEX5 interaction and that binding of NDPEX14 to the single diaromatic motif present in PEX5ΔN267 is sufficient to disrupt that interaction.
Unexpectedly, substitution of the tryptophan and tyrosine residues in the diaromatic motif of PEX5ΔN267 by alanines, mutations that affect its PEX14 binding activity (66, 74), results in a protein, PEX5ΔN267-M7, that no longer inhibits catalase tetramerization (Fig. 5C), suggesting that these two aromatic residues are structurally important. Interestingly, when the same mutation was introduced in full-length PEX5, the resulting protein, PEX5-M7, was found to be as potent as the normal protein in inhibiting catalase tetramerization as well as in its response to NDPEX14 (Fig. 5D). This observation suggests, on one hand, that other regions of PEX5 compensate for the alterations associated with the mutation at 7th diaromatic motif, and, on the other hand, that at least one of the remaining six diaromatic motifs present in PEX5-M7 is involved in the NDPEX14-induced disruption of the mCat-PEX5 interaction. Data suggesting that the 6th diaromatic motif of PEX5 plays a major role in the NDPEX14-induced disruption of the mCat-PEX5 interaction were obtained when recombinant PEX5-M6,7, a PEX5 protein mutated at both the 6th and 7th diaromatic motifs, was tested in the catalase tetramerization assay. As shown in Fig. 5D, PEX5-M6,7 displays an inhibitory activity in this assay, although in a less potent manner than PEX5. This finding again suggests that the structure/function of this region of PEX5 required for the interaction with mCat is not fully preserved upon mutation of the diaromatic motifs. Importantly, the inhibitory activity of PEX5-M6,7 is no longer significantly neutralized by NDPEX14. These results suggest that binding of NDPEX14 to the 6th diaromatic motif of PEX5 and probably also to the 7th motif (as inferred from the PEX5ΔN267 data) disrupts the mCat-PEX5 interaction.
DISCUSSION
The proposed mechanism for the catalase assembly pathway consists of three steps as follows: 1) apo-monomers + heme → holomonomers; 2) holomonomers → holodimers; and 3) holodimers → homotetramers (reviewed in Ref. 76). The step at which PEX5 binds and transports catalase to the peroxisome, however, has remained a controversial issue. In this work, we used an in vitro system to characterize the PEX5-catalase interaction. Our results suggest that in the presence of PEX5 step 3 no longer occurs, but whether the inhibitory effect of PEX5 is exerted at step 1 and/or 2 remains unknown. Nevertheless, it is interesting to note that the mCat species is a soluble and monodisperse protein suggesting that it already possesses a near-native conformation.
Unexpectedly, a qualitative assessment of the binding affinities of mCat and tCat for PEX5 revealed that PEX5 has a bias toward binding the former, suggesting that some domain of the catalase polypeptide chain is no longer available to interact with PEX5 when the protein tetramerizes. If we exclude catalase PTS1 from this reasoning, as the crystal structures of catalase might suggest (39, 40, 42), it is reasonable to assume that another domain of the mCat protein is involved in this phenomenon. Data supporting this possibility were obtained when we focused our attention on PEX5. Indeed, we found that the PTS1-binding domain of PEX5 is required for the mCat-PEX5 interaction, as expected, but evidence for the participation of an N-terminal domain of PEX5 in this interaction was also obtained. Each of these two PEX5 domains alone (i.e. in trans) can bind mCat inhibiting its tetramerization, but this effect is dramatically increased when these domains reside in the same molecule. Thus, they most likely bind mCat simultaneously.
The conclusion that the catalase-binding interface in PEX5 encompasses more than its PTS1-binding domain is probably also valid for other organisms. Indeed, as shown previously, yeast catalase possesses peroxisomal targeting information in two different regions of its polypeptide chain, one at the C terminus (the PTS1 sequence) and the other located in its N-terminal third (77), an observation that would be compatible with the existence of two different catalase-binding domains in yeast PEX5. Likewise, the observation that pumpkin catalase interacts with the N-terminal half of PEX5 in the yeast two-hybrid system (78), together with data on cottonseed catalase showing that its last four residues (which are conserved in pumpkin catalase) are sufficient to target a reporter protein to the peroxisome (79), could suggest that plant PEX5 possesses more than one catalase-interacting domain.
As already mentioned, binding of PEX5 to mCat inhibits its tetramerization in a potent manner. This previously unknown capacity of PEX5 evokes the properties of a family of bacterial chaperones functioning in type III secretion systems. These chaperones, some of which also contain tetratricopeptide repeats, bind proteins to be secreted in the cytoplasm preventing premature or incorrect interactions and participate in the secretion step itself (Ref. 80 and reviewed in Ref. 81). Although additional data will be necessary to reinforce the functional similarities between PEX5 and these chaperones, it is interesting to note that we have recently observed the same phenomenon when studying another oligomeric peroxisomal protein.5
Regardless of the mechanistic reasons behind the inhibitory activity of PEX5 on peroxisomal protein oligomerization, it is evident that such a property may be biologically relevant only if the amount of cytosolic PEX5 in a cell is sufficient to sequester mCat and all the other newly synthesized peroxisomal proteins that are en route to the organelle. The data available for rat liver suggest that this may well be the case. Indeed, we estimated that the cytosolic concentration of PEX5 is 0.75 μm, whereas the concentration of newly synthesized peroxisomal proteins is 0.73–0.83 μm, with mCat occupying a major fraction of this pool (0.19 μm; see under “Experimental Procedures” for details). Thus, even if we assume that newly synthesized peroxisomal proteins become available to bind PEX5 immediately after their synthesis (i.e. their folding process is not incompatible with PEX5 binding), we still reach the conclusion that there is a sufficient amount of PEX5 to bind a significant fraction of mCat.
The results presented here thus corroborate and extend the pioneering observations of Lazarow and de Duve (46, 47) suggesting that rat liver catalase arrives at the peroxisome still in its monomeric state. However, they also seem to collide with the idea that catalase is imported into the organelle only after tetramerization (49). This is not completely so. Indeed, as discussed above, the crucial factor determining whether or not catalase tetramerizes before import may well be the amount of PEX5 present in the cytosol. If a cell contains a stoichiometric excess of PEX5 over newly synthesized peroxisomal proteins, then it is likely that catalase remains monomeric and is imported as such; if not, a fraction of it will tetramerize before import. Although speculative, this possibility would explain why different results are obtained in different experimental systems (see Ref. 49 and references therein).
PEX14, a central component of the DTM, was regarded for many years as a protein involved solely in the docking of the receptor at the peroxisomal membrane (82). However, several observations have challenged this concept, and it is now clear that this protein also participates in the translocation of cargoes across the peroxisomal membrane (83, 84). The finding that the N-terminal domain of human PEX14 disrupts the PEX5-mCat interaction together with data reported earlier for Leishmania donovani PEX5 showing that its affinity for a PTS1 protein is decreased in the presence of PEX14 (85) suggest still another function for this membrane protein, a role in the release of cargoes from DTM-embedded PEX5 into the peroxisomal matrix.
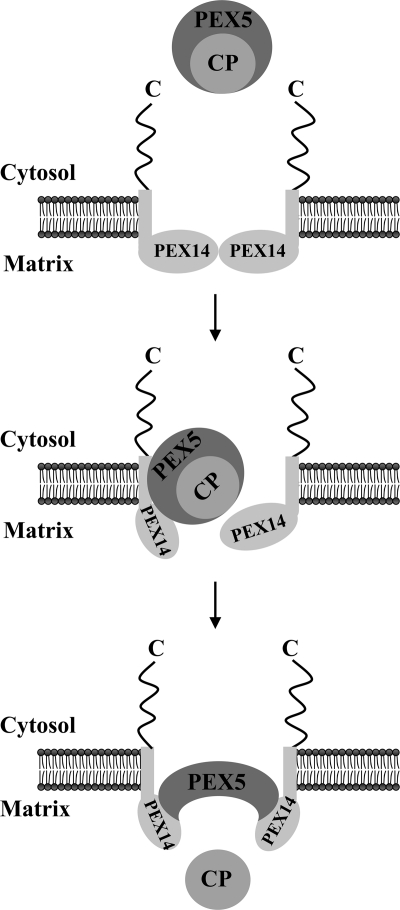
Interestingly, from the seven diaromatic motifs present in human PEX5, only one or two play a major role in the NDPEX14-induced disruption of the mCat-PEX5 interaction. This finding together with previous data showing that diaromatic motifs 2–4 of PEX5 are required for catalase import in vivo (66) suggests that the multiple interactions that are probably established between the N-terminal domain of peroxisomal PEX14 and the diaromatic motifs present in PEX5 occur in a sequential manner and may serve two different purposes. According to this hypothetical model (see Fig. 6), the first set of interactions may contribute to the docking/insertion of the PEX5-cargo protein complex into the DTM; subsequently, binding of additional PEX14 molecules to the 6th (and probably 7th) diaromatic motif(s) of PEX5 would trigger the release of the cargo protein into the peroxisomal matrix.
FIGURE 6.
Role of PEX14 in the release of cargo proteins into the peroxisomal matrix. A newly synthesized cargo protein (CP) is recognized by PEX5 in the cytosol. This protein complex then docks at and becomes inserted into the peroxisomal DTM of which only PEX14 is shown for simplicity. The DTM component(s) providing the docking site for the PEX5-cargo protein complex have not been unambiguously identified yet. As discussed elsewhere, two strong candidates for this role are PEX13 (86) and PEX14 itself (87, 88). Note that PEX13 may also participate in the cargo-release step (66). The multiple interactions of PEX5 with the N-terminal domain of the several PEX14 molecules present in the DTM ultimately trigger the release of the cargo into the peroxisomal matrix.
Supplementary Material
Acknowledgment
We thank Kalervo Hiltunen (University of Oulu, Finland) for the kind gift of the L-bifunctional antibody.
This work was supported in part by Fundação para a Ciência e Tecnologia, Portugal, and Fundo Europeu de Desenvolvimento Regional through COMPETE, Programa Operacional Factores de Competitividade in the context of QREN, Portugal, Grants PEst-C/SAU/LA0002/2011 and PTDC/BIA-BCM/64771/2006, and by the European Union VI Framework Program Grant LSHGCT-2004-512018, Peroxisomes in Health and Disease.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Table 1.
M. O. Freitas and J. E. Azevedo, unpublished results.
- PTS1
- peroxisomal targeting signal type 1
- PTS2
- peroxisomal targeting signal type 2
- DTM
- docking/translocation machinery
- mCat
- monomeric catalase
- tCat
- tetrameric catalase
- SEC
- size-exclusion chromatography
- NDPEX14
- N-terminal domain of PEX14
- TPR
- tetratricopeptide repeat.
REFERENCES
- 1. Grou C. P., Carvalho A. F., Pinto M. P., Alencastre I. S., Rodrigues T. A., Freitas M. O., Francisco T., Sá-Miranda C., Azevedo J. E. (2009) Cell. Mol. Life Sci. 66, 254–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lanyon-Hogg T., Warriner S. L., Baker A. (2010) Biol. Cell 102, 245–263 [DOI] [PubMed] [Google Scholar]
- 3. Ma C., Subramani S. (2009) IUBMB Life 61, 713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wolf J., Schliebs W., Erdmann R. (2010) FEBS J. 277, 3268–3278 [DOI] [PubMed] [Google Scholar]
- 5. Brocard C., Hartig A. (2006) Biochim. Biophys. Acta 1763, 1565–1573 [DOI] [PubMed] [Google Scholar]
- 6. Stanley W. A., Wilmanns M. (2006) Biochim. Biophys. Acta 1763, 1592–1598 [DOI] [PubMed] [Google Scholar]
- 7. Lazarow P. B. (2006) Biochim. Biophys. Acta 1763, 1599–1604 [DOI] [PubMed] [Google Scholar]
- 8. Schliebs W., Kunau W. H. (2006) Biochim. Biophys. Acta 1763, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 9. Agne B., Meindl N. M., Niederhoff K., Einwächter H., Rehling P., Sickmann A., Meyer H. E., Girzalsky W., Kunau W. H. (2003) Mol. Cell 11, 635–646 [DOI] [PubMed] [Google Scholar]
- 10. Reguenga C., Oliveira M. E., Gouveia A. M., Sá-Miranda C., Azevedo J. E. (2001) J. Biol. Chem. 276, 29935–29942 [DOI] [PubMed] [Google Scholar]
- 11. Alencastre I. S., Rodrigues T. A., Grou C. P., Fransen M., Sá-Miranda C., Azevedo J. E. (2009) J. Biol. Chem. 284, 27243–27251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gouveia A. M., Guimaraes C. P., Oliveira M. E., Reguenga C., Sa-Miranda C., Azevedo J. E. (2003) J. Biol. Chem. 278, 226–232 [DOI] [PubMed] [Google Scholar]
- 13. Gouveia A. M., Guimarães C. P., Oliveira M. E., Sá-Miranda C., Azevedo J. E. (2003) J. Biol. Chem. 278, 4389–4392 [DOI] [PubMed] [Google Scholar]
- 14. Miyata N., Fujiki Y. (2005) Mol. Cell. Biol. 25, 10822–10832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Oliveira M. E., Gouveia A. M., Pinto R. A., Sá-Miranda C., Azevedo J. E. (2003) J. Biol. Chem. 278, 39483–39488 [DOI] [PubMed] [Google Scholar]
- 16. Platta H. W., Grunau S., Rosenkranz K., Girzalsky W., Erdmann R. (2005) Nat. Cell Biol. 7, 817–822 [DOI] [PubMed] [Google Scholar]
- 17. Carvalho A. F., Pinto M. P., Grou C. P., Alencastre I. S., Fransen M., Sá-Miranda C., Azevedo J. E. (2007) J. Biol. Chem. 282, 31267–31272 [DOI] [PubMed] [Google Scholar]
- 18. Grou C. P., Carvalho A. F., Pinto M. P., Wiese S., Piechura H., Meyer H. E., Warscheid B., Sá-Miranda C., Azevedo J. E. (2008) J. Biol. Chem. 283, 14190–14197 [DOI] [PubMed] [Google Scholar]
- 19. Okumoto K., Misono S., Miyata N., Matsumoto Y., Mukai S., Fujiki Y. (2011) Traffic 12, 1067–1083 [DOI] [PubMed] [Google Scholar]
- 20. Williams C., van den Berg M., Sprenger R. R., Distel B. (2007) J. Biol. Chem. 282, 22534–22543 [DOI] [PubMed] [Google Scholar]
- 21. Debelyy M. O., Platta H. W., Saffian D., Hensel A., Thoms S., Meyer H. E., Warscheid B., Girzalsky W., Erdmann R. (2011) J. Biol. Chem. 286, 28223–28234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grou C. P., Carvalho A. F., Pinto M. P., Huybrechts S. J., Sá-Miranda C., Fransen M., Azevedo J. E. (2009) J. Biol. Chem. 284, 10504–10513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shiozawa K., Konarev P. V., Neufeld C., Wilmanns M., Svergun D. I. (2009) J. Biol. Chem. 284, 25334–25342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Faber K. N., van Dijk R., Keizer-Gunnink I., Koek A., van der Klei I. J., Veenhuis M. (2002) Biochim. Biophys. Acta 1591, 157–162 [DOI] [PubMed] [Google Scholar]
- 25. Luo B., Norris C., Bolstad E. S., Knecht D. A., Grant D. F. (2008) J. Mol. Biol. 380, 31–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stewart M. Q., Esposito R. D., Gowani J., Goodman J. M. (2001) J. Cell Sci. 114, 2863–2868 [DOI] [PubMed] [Google Scholar]
- 27. Tanaka N., Aoki K., Ishikura S., Nagano M., Imamura Y., Hara A., Nakamura K. T. (2008) Structure 16, 388–397 [DOI] [PubMed] [Google Scholar]
- 28. Elgersma Y., Vos A., van den Berg M., van Roermund C. W., van der Sluijs P., Distel B., Tabak H. F. (1996) J. Biol. Chem. 271, 26375–26382 [DOI] [PubMed] [Google Scholar]
- 29. Islinger M., Li K. W., Seitz J., Völkl A., Lüers G. H. (2009) Traffic 10, 1711–1721 [DOI] [PubMed] [Google Scholar]
- 30. Nilsen T., Slagsvold T., Skjerpen C. S., Brech A., Stenmark H., Olsnes S. (2004) J. Biol. Chem. 279, 4794–4801 [DOI] [PubMed] [Google Scholar]
- 31. Glover J. R., Andrews D. W., Rachubinski R. A. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 10541–10545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee M. S., Mullen R. T., Trelease R. N. (1997) Plant Cell 9, 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leiper J. M., Oatey P. B., Danpure C. J. (1996) J. Cell Biol. 135, 939–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McNew J. A., Goodman J. M. (1994) J. Cell Biol. 127, 1245–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gould S. J., Collins C. S. (2002) Nat. Rev. Mol. Cell Biol. 3, 382–389 [DOI] [PubMed] [Google Scholar]
- 36. Ghosh D., Berg J. M. (2010) J. Am. Chem. Soc. 132, 3973–3979 [DOI] [PubMed] [Google Scholar]
- 37. Leighton F., Coloma L., Koenig C. (1975) J. Cell Biol. 67, 281–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Poole B., Leighton F., De Duve C. (1969) J. Cell Biol. 41, 536–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maté M. J., Zamocky M., Nykyri L. M., Herzog C., Alzari P. M., Betzel C., Koller F., Fita I. (1999) J. Mol. Biol. 286, 135–149 [DOI] [PubMed] [Google Scholar]
- 40. Murthy M. R., Reid T. J., 3rd, Sicignano A., Tanaka N., Rossmann M. G. (1981) J. Mol. Biol. 152, 465–499 [DOI] [PubMed] [Google Scholar]
- 41. Purdue P. E., Castro S. M., Protopopov V., Lazarow P. B. (1996) Ann. N.Y. Acad. Sci. 804, 775–776 [DOI] [PubMed] [Google Scholar]
- 42. Putnam C. D., Arvai A. S., Bourne Y., Tainer J. A. (2000) J. Mol. Biol. 296, 295–309 [DOI] [PubMed] [Google Scholar]
- 43. Trelease R. N., Xie W., Lee M. S., Mullen R. T. (1996) Eur. J. Cell Biol. 71, 248–258 [PubMed] [Google Scholar]
- 44. Brul S., Wiemer E. A., Westerveld A., Strijland A., Wanders R. J., Schram A. W., Heymans H. S., Schutgens R. B., Van den Bosch H., Tager J. M. (1988) Biochem. Biophys. Res. Commun. 152, 1083–1089 [DOI] [PubMed] [Google Scholar]
- 45. Koepke J. I., Nakrieko K. A., Wood C. S., Boucher K. K., Terlecky L. J., Walton P. A., Terlecky S. R. (2007) Traffic 8, 1590–1600 [DOI] [PubMed] [Google Scholar]
- 46. Lazarow P. B., de Duve C. (1973) J. Cell Biol. 59, 507–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lazarow P. B., de Duve C. (1973) J. Cell Biol. 59, 491–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Middelkoop E., Strijland A., Tager J. M. (1991) FEBS Lett. 279, 79–82 [DOI] [PubMed] [Google Scholar]
- 49. Léon S., Goodman J. M., Subramani S. (2006) Biochim. Biophys. Acta 1763, 1552–1564 [DOI] [PubMed] [Google Scholar]
- 50. Braverman N., Dodt G., Gould S. J., Valle D. (1998) Hum. Mol. Genet. 7, 1195–1205 [DOI] [PubMed] [Google Scholar]
- 51. Fransen M., Brees C., Baumgart E., Vanhooren J. C., Baes M., Mannaerts G. P., Van Veldhoven P. P. (1995) J. Biol. Chem. 270, 7731–7736 [DOI] [PubMed] [Google Scholar]
- 52. Carvalho A. F., Costa-Rodrigues J., Correia I., Costa Pessoa J., Faria T. Q., Martins C. L., Fransen M., Sá-Miranda C., Azevedo J. E. (2006) J. Mol. Biol. 356, 864–875 [DOI] [PubMed] [Google Scholar]
- 53. Carvalho A. F., Grou C. P., Pinto M. P., Alencastre I. S., Costa-Rodrigues J., Fransen M., Sá-Miranda C., Azevedo J. E. (2007) Biochim. Biophys. Acta 1773, 1141–1148 [DOI] [PubMed] [Google Scholar]
- 54. Costa-Rodrigues J., Carvalho A. F., Fransen M., Hambruch E., Schliebs W., Sá-Miranda C., Azevedo J. E. (2005) J. Biol. Chem. 280, 24404–24411 [DOI] [PubMed] [Google Scholar]
- 55. Pinto M. P., Grou C. P., Alencastre I. S., Oliveira M. E., Sá-Miranda C., Fransen M., Azevedo J. E. (2006) J. Biol. Chem. 281, 34492–34502 [DOI] [PubMed] [Google Scholar]
- 56. Sullivan D. T., Carroll W. T., Kanik-Ennulat C. L., Hitti Y. S., Lovett J. A., Von Kalm L. (1985) J. Biol. Chem. 260, 4345–4350 [PubMed] [Google Scholar]
- 57. Gouveia A. M., Reguenga C., Oliveira M. E., Eckerskorn C., Sá-Miranda C., Azevedo J. E. (1999) Anal. Biochem. 274, 270–277 [DOI] [PubMed] [Google Scholar]
- 58. Palosaari P. M., Hiltunen J. K. (1990) J. Biol. Chem. 265, 2446–2449 [PubMed] [Google Scholar]
- 59. Gouveia A. M., Reguenga C., Oliveira M. E., Sa-Miranda C., Azevedo J. E. (2000) J. Biol. Chem. 275, 32444–32451 [DOI] [PubMed] [Google Scholar]
- 60. Fujiki Y., Fowler S., Shio H., Hubbard A. L., Lazarow P. B. (1982) J. Cell Biol. 93, 103–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Weibel E. R., Stäubli W., Gnägi H. R., Hess F. A. (1969) J. Cell Biol. 42, 68–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Miyazawa S., Hayashi H., Hijikata M., Ishii N., Furuta S., Kagamiyama H., Osumi T., Hashimoto T. (1987) J. Biol. Chem. 262, 8131–8137 [PubMed] [Google Scholar]
- 63. Leighton F., Poole B., Lazarow P. B., De Duve C. (1969) J. Cell Biol. 41, 521–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lazarow P. B., Robbi M., Fujiki Y., Wong L. (1982) Ann. N.Y. Acad. Sci. 386, 285–300 [DOI] [PubMed] [Google Scholar]
- 65. Scandalios J. G. (1965) Proc. Natl. Acad. Sci. U.S.A. 53, 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Otera H., Setoguchi K., Hamasaki M., Kumashiro T., Shimizu N., Fujiki Y. (2002) Mol. Cell. Biol. 22, 1639–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dodt G., Braverman N., Wong C., Moser A., Moser H. W., Watkins P., Valle D., Gould S. J. (1995) Nat. Genet. 9, 115–125 [DOI] [PubMed] [Google Scholar]
- 68. Gatto G. J., Jr., Geisbrecht B. V., Gould S. J., Berg J. M. (2000) Nat. Struct. Biol. 7, 1091–1095 [DOI] [PubMed] [Google Scholar]
- 69. Fujiki Y., Matsuzono Y., Matsuzaki T., Fransen M. (2006) Biochim. Biophys. Acta 1763, 1639–1646 [DOI] [PubMed] [Google Scholar]
- 70. Wang D., Visser N. V., Veenhuis M., van der Klei I. J. (2003) J. Biol. Chem. 278, 43340–43345 [DOI] [PubMed] [Google Scholar]
- 71. Schlüter A., Fourcade S., Ripp R., Mandel J. L., Poch O., Pujol A. (2006) Mol. Biol. Evol. 23, 838–845 [DOI] [PubMed] [Google Scholar]
- 72. Oliveira M. E., Reguenga C., Gouveia A. M., Guimarães C. P., Schliebs W., Kunau W. H., Silva M. T., Sá-Miranda C., Azevedo J. E. (2002) Biochim. Biophys. Acta 1567, 13–22 [DOI] [PubMed] [Google Scholar]
- 73. Will G. K., Soukupova M., Hong X., Erdmann K. S., Kiel J. A., Dodt G., Kunau W. H., Erdmann R. (1999) Mol. Cell. Biol. 19, 2265–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Saidowsky J., Dodt G., Kirchberg K., Wegner A., Nastainczyk W., Kunau W. H., Schliebs W. (2001) J. Biol. Chem. 276, 34524–34529 [DOI] [PubMed] [Google Scholar]
- 75. Schliebs W., Saidowsky J., Agianian B., Dodt G., Herberg F. W., Kunau W. H. (1999) J. Biol. Chem. 274, 5666–5673 [DOI] [PubMed] [Google Scholar]
- 76. Zámocký M., Koller F. (1999) Prog. Biophys. Mol. Biol. 72, 19–66 [DOI] [PubMed] [Google Scholar]
- 77. Kragler F., Langeder A., Raupachova J., Binder M., Hartig A. (1993) J. Cell Biol. 120, 665–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Oshima Y., Kamigaki A., Nakamori C., Mano S., Hayashi M., Nishimura M., Esaka M. (2008) Plant Cell Physiol. 49, 671–677 [DOI] [PubMed] [Google Scholar]
- 79. Mullen R. T., Lee M. S., Trelease R. N. (1997) Plant J. 12, 313–322 [DOI] [PubMed] [Google Scholar]
- 80. Plé S., Job V., Dessen A., Attree I. (2010) J. Bacteriol. 192, 3801–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Parsot C., Hamiaux C., Page A. L. (2003) Curr. Opin. Microbiol. 6, 7–14 [DOI] [PubMed] [Google Scholar]
- 82. Sacksteder K. A., Gould S. J. (2000) Annu. Rev. Genet. 34, 623–652 [DOI] [PubMed] [Google Scholar]
- 83. Azevedo J. E., Schliebs W. (2006) Biochim. Biophys. Acta 1763, 1574–1584 [DOI] [PubMed] [Google Scholar]
- 84. Ma C., Schumann U., Rayapuram N., Subramani S. (2009) Mol. Biol. Cell 20, 3680–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Madrid K. P., De Crescenzo G., Wang S., Jardim A. (2004) Mol. Cell. Biol. 24, 7331–7344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Williams C., Distel B. (2006) Biochim. Biophys. Acta 1763, 1585–1591 [DOI] [PubMed] [Google Scholar]
- 87. Niederhoff K., Meindl-Beinker N. M., Kerssen D., Perband U., Schäfer A., Schliebs W., Kunau W. H. (2005) J. Biol. Chem. 280, 35571–35578 [DOI] [PubMed] [Google Scholar]
- 88. Williams C., van den Berg M., Distel B. (2005) FEBS Lett. 579, 3416–3420 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.