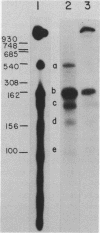
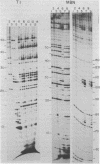
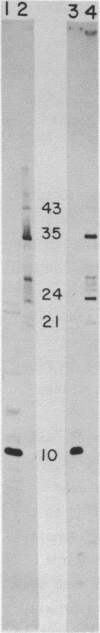
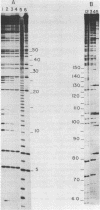
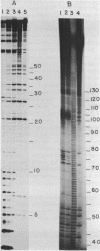
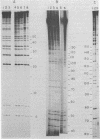
Abstract
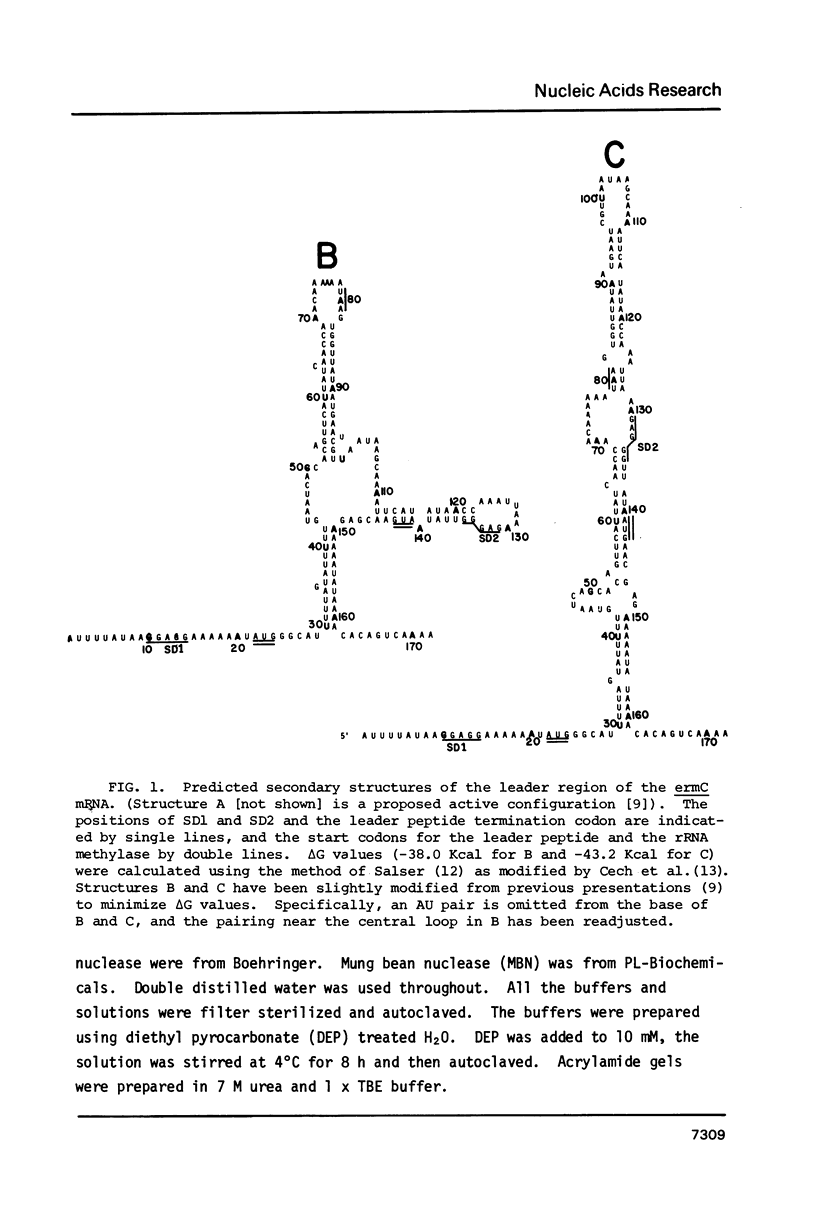
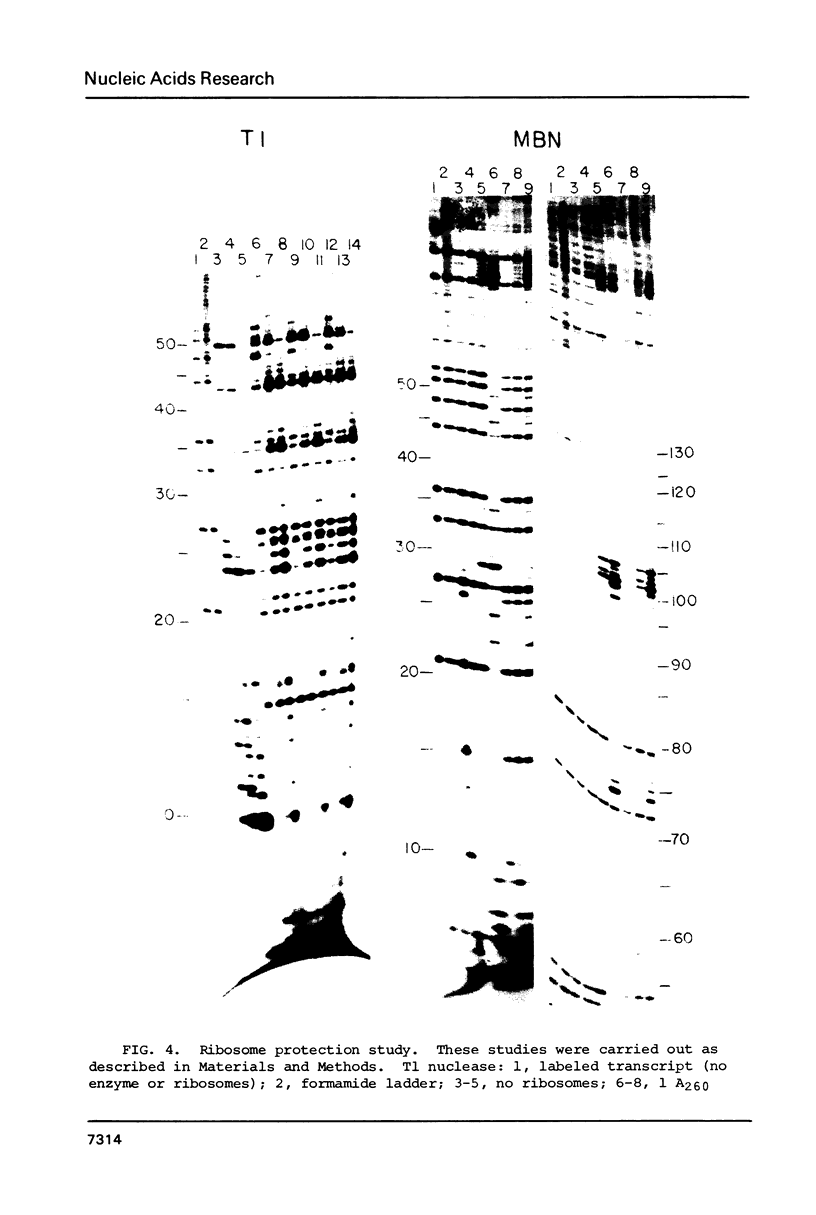
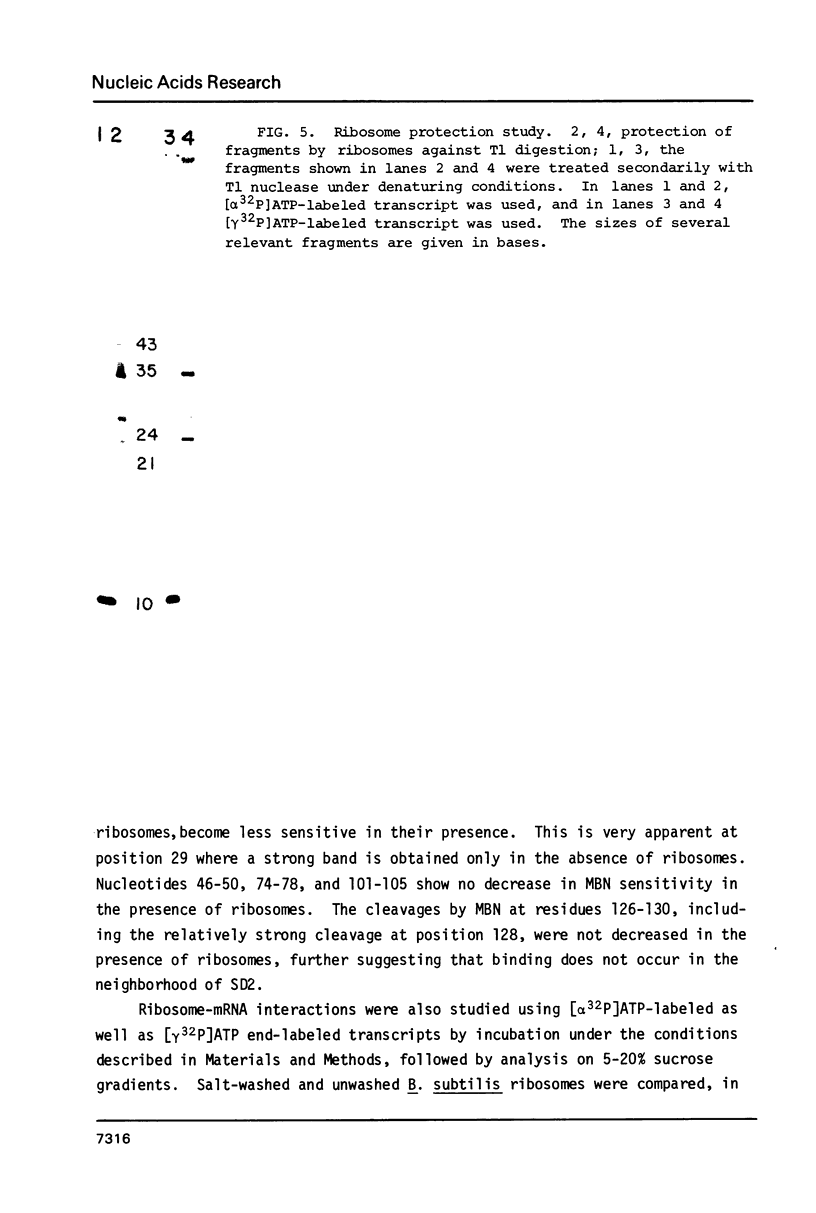
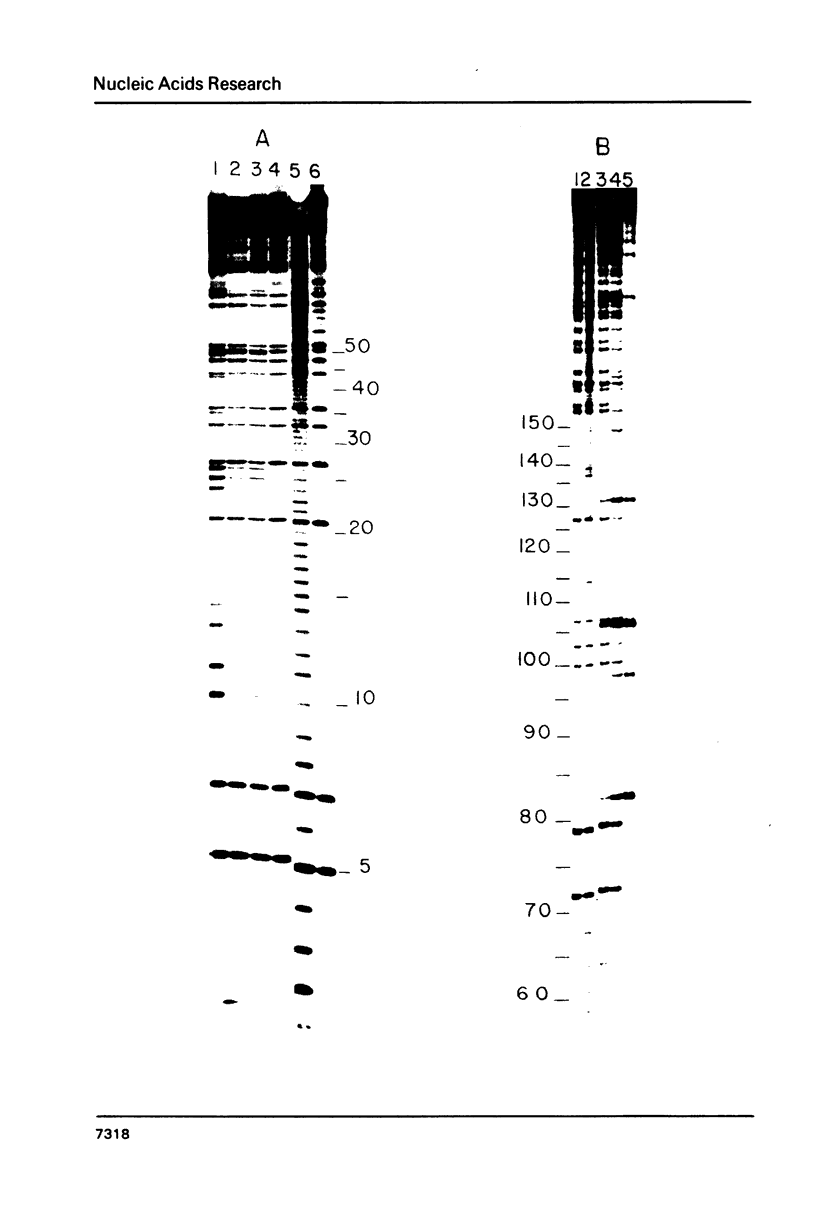
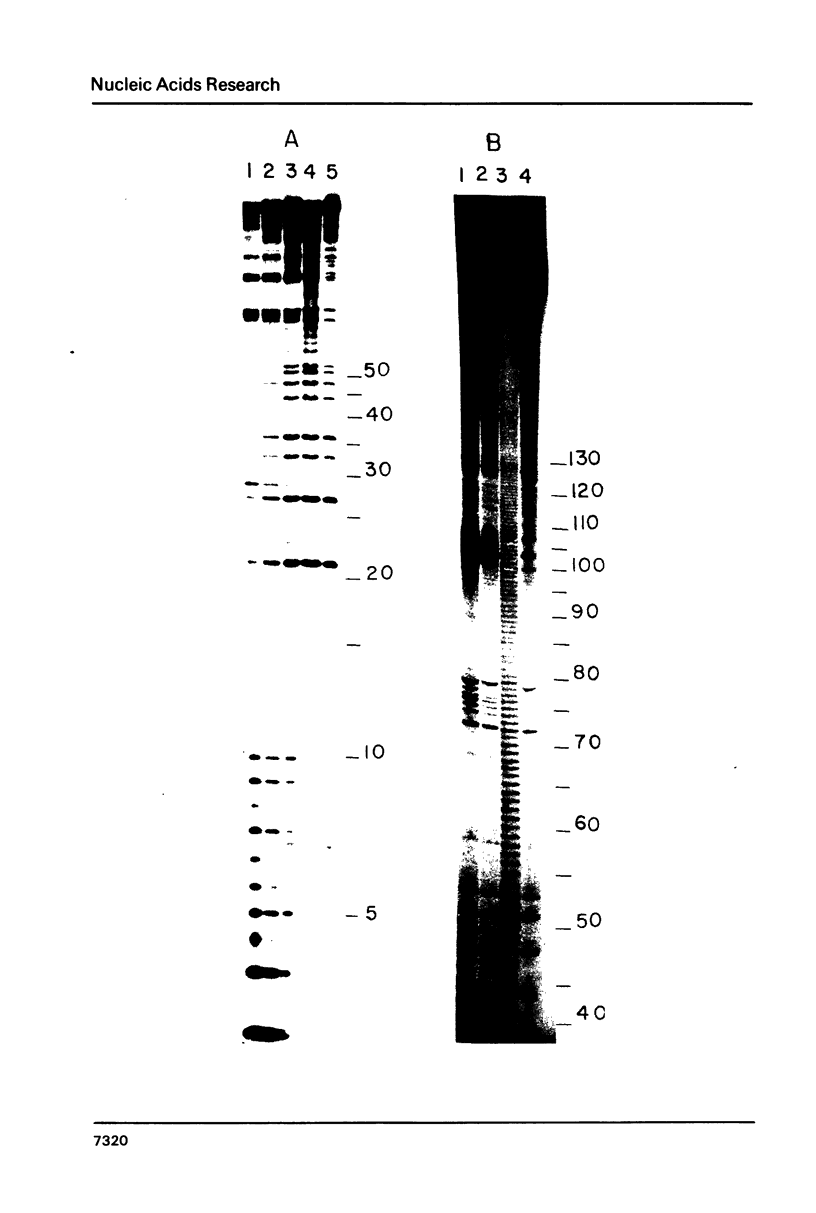
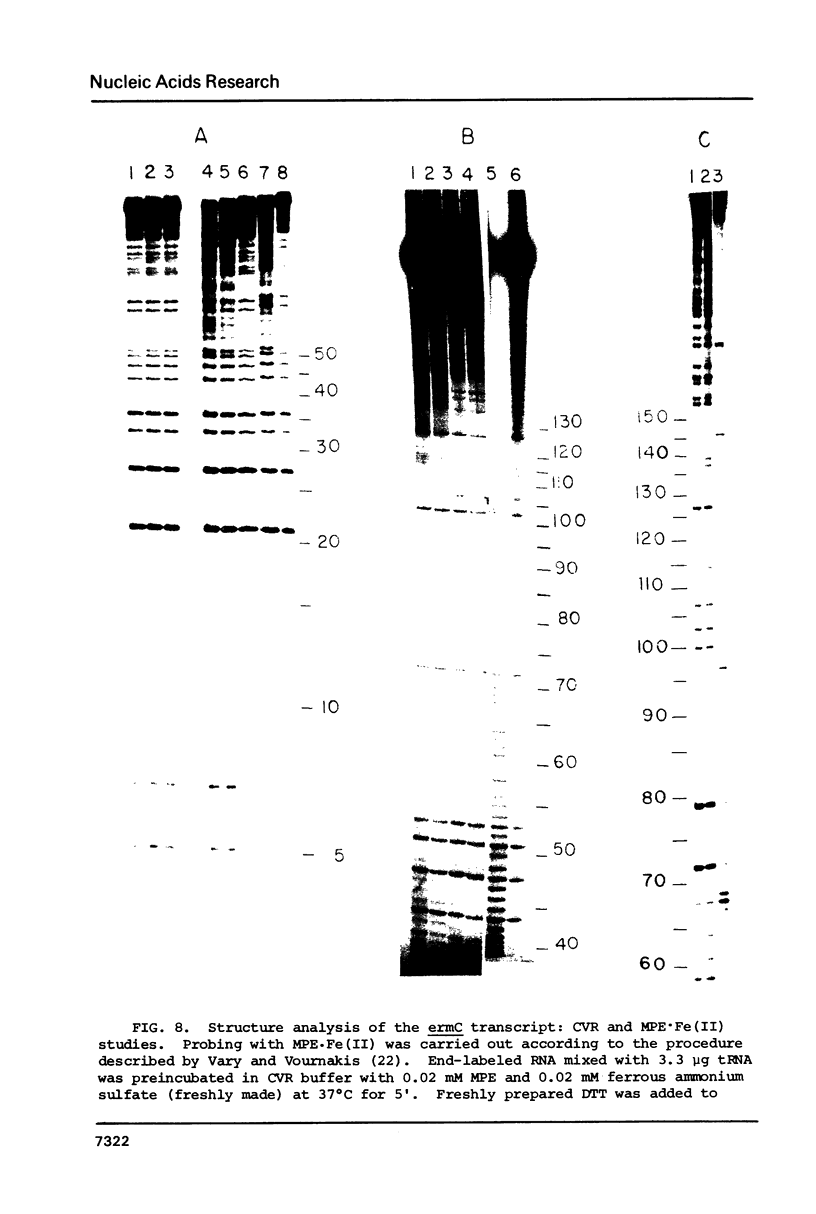
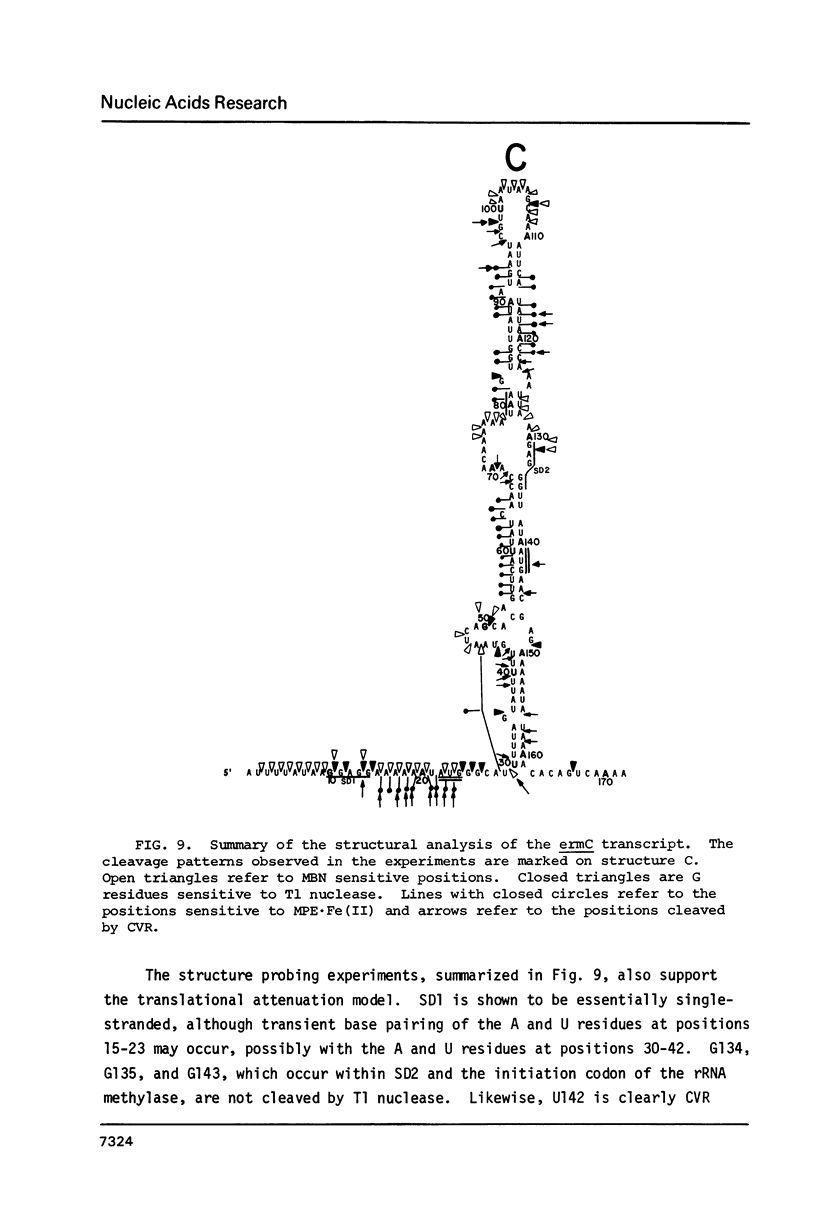
Several features of the translational attenuation model of ermC regulation were tested. This model predicts two possible secondary structures for the leader of the ermC transcript and requires that the leader contains two Shine-Dalgarno (SD) sequences. The ribosome binding site for a leader peptide (SD1) is predicted to be accessible, whereas that for the rRNA methylase protein that confers erythromycin (Em) resistance (SD2) is sequestered by base pairing. The model suggests that in the presence of inducer (Em), a ribosome stalls while translating the peptide, altering the mRNA conformation, thereby exposing SD2. The results of our ribosome binding studies demonstrate that SD1 is exposed and binds to ribosomes, whereas SD2 is unavailable. Also, the secondary structure of the 5' region of the ermC transcript was analyzed using methidium propyl-EDTA.Fe (II), T1 nuclease, and nucleases from cobra venom and mung bean sprouts as structure probes. Our results support the previously proposed model for folding of ermC mRNA, and demonstrate that SD1 is single-stranded, while SD2 and its neighboring sequences are largely base paired, consistent with the ribosome-binding results.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiba H. Autoregulation of the Escherichia coli crp gene: CRP is a transcriptional repressor for its own gene. Cell. 1983 Jan;32(1):141–149. doi: 10.1016/0092-8674(83)90504-4. [DOI] [PubMed] [Google Scholar]
- Carbon P., Ehresmann C., Ehresmann B., Ebel J. P. The sequence of Escherichia coli ribosomal 16 S RNA determined by new rapid gel methods. FEBS Lett. 1978 Oct 1;94(1):152–156. doi: 10.1016/0014-5793(78)80926-0. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Tanner N. K., Tinoco I., Jr, Weir B. R., Zuker M., Perlman P. S. Secondary structure of the Tetrahymena ribosomal RNA intervening sequence: structural homology with fungal mitochondrial intervening sequences. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3903–3907. doi: 10.1073/pnas.80.13.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond A., Dudock B. Methods of RNA sequence analysis. Methods Enzymol. 1983;100:431–453. doi: 10.1016/0076-6879(83)00072-5. [DOI] [PubMed] [Google Scholar]
- Dubnau D. Induction of ermC requires translation of the leader peptide. EMBO J. 1985 Feb;4(2):533–537. doi: 10.1002/j.1460-2075.1985.tb03661.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Contente S., Dubnau D. Characterization of Staphylococcus aureus plasmids introduced by transformation into Bacillus subtilis. J Bacteriol. 1978 Apr;134(1):318–329. doi: 10.1128/jb.134.1.318-329.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Grandi G., Hahn J., Grandi R., Dubnau D. Conformational alteration of mRNA structure and the posttranscriptional regulation of erythromycin-induced drug resistance. Nucleic Acids Res. 1980 Dec 20;8(24):6081–6097. doi: 10.1093/nar/8.24.6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha S., Szulmajster J. Effect of fusidic acid on sporulation of Bacillus subtilis. FEBS Lett. 1974 Jan 15;38(3):315–319. doi: 10.1016/0014-5793(74)80081-5. [DOI] [PubMed] [Google Scholar]
- Hahn J., Grandi G., Gryczan T. J., Dubnau D. Translational attenuation of ermC: a deletion analysis. Mol Gen Genet. 1982;186(2):204–216. doi: 10.1007/BF00331851. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Posttranscriptional modification of mRNA conformation: mechanism that regulates erythromycin-induced resistance. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7079–7083. doi: 10.1073/pnas.77.12.7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. The control region for erythromycin resistance: free energy changes related to induction and mutation to constitutive expression. Mol Gen Genet. 1981;182(2):341–348. doi: 10.1007/BF00269681. [DOI] [PubMed] [Google Scholar]
- Iordănescu S. Three distinct plasmids originating in the same Staphylococcus aureus strain. Arch Roum Pathol Exp Microbiol. 1976 Jan-Jun;35(1-2):111–118. [PubMed] [Google Scholar]
- Lai C. J., Dahlberg J. E., Weisblum B. Structure of an inducibly methylatable nucleotide sequence in 23S ribosomal ribonucleic acid from erythromycin-resistant Staphylococcus aureus. Biochemistry. 1973 Jan 30;12(3):457–460. doi: 10.1021/bi00727a015. [DOI] [PubMed] [Google Scholar]
- Lai C. J., Weisblum B. Altered methylation of ribosomal RNA in an erythromycin-resistant strain of Staphylococcus aureus. Proc Natl Acad Sci U S A. 1971 Apr;68(4):856–860. doi: 10.1073/pnas.68.4.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C. J., Weisblum B., Fahnestock S. R., Nomura M. Alteration of 23 S ribosomal RNA and erythromycin-induced resistance to lincomycin and spiramycin in Staphylococcus aureus. J Mol Biol. 1973 Feb 15;74(1):67–72. doi: 10.1016/0022-2836(73)90355-0. [DOI] [PubMed] [Google Scholar]
- Loughney K., Lund E., Dahlberg J. E. tRNA genes are found between 16S and 23S rRNA genes in Bacillus subtilis. Nucleic Acids Res. 1982 Mar 11;10(5):1607–1624. doi: 10.1093/nar/10.5.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin J. R., Murray C. L., Rabinowitz J. C. Initiation factor-independent translation of mRNAs from Gram-positive bacteria. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4912–4916. doi: 10.1073/pnas.78.8.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salser W. Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):985–1002. doi: 10.1101/sqb.1978.042.01.099. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivakumar A. G., Dubnau D. Characterization of a plasmid-specified ribosome methylase associated with macrolide resistance. Nucleic Acids Res. 1981 Jun 11;9(11):2549–2562. doi: 10.1093/nar/9.11.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivakumar A. G., Hahn J., Dubnau D. Studies on the synthesis of plasmid-coded proteins and their control in Bacillus subtilis minicells. Plasmid. 1979 Apr;2(2):279–289. doi: 10.1016/0147-619x(79)90046-5. [DOI] [PubMed] [Google Scholar]
- Shivakumar A. G., Hahn J., Grandi G., Kozlov Y., Dubnau D. Posttranscriptional regulation of an erythromycin resistance protein specified by plasmic pE194. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3903–3907. doi: 10.1073/pnas.77.7.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz J. A. Specific recognition of non-initiator regions in RNA bacteriophage messengers by ribosomes of Bacillus stearothermophilus. J Mol Biol. 1973 Jan;73(1):1–16. doi: 10.1016/0022-2836(73)90155-1. [DOI] [PubMed] [Google Scholar]
- Tamm J., Polisky B. Structural analysis of RNA molecules involved in plasmid copy number control. Nucleic Acids Res. 1983 Sep 24;11(18):6381–6397. doi: 10.1093/nar/11.18.6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vary C. P., Vournakis J. N. RNA structure analysis using methidiumpropyl-EDTA.Fe(II): a base-pair-specific RNA structure probe. Proc Natl Acad Sci U S A. 1984 Nov;81(22):6978–6982. doi: 10.1073/pnas.81.22.6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vournakis J. N., Celantano J., Finn M., Lockard R. E., Mitra T., Pavlakis G., Troutt A., van den Berg M., Wurst R. M. Sequence and structure analysis of end-labeled RNA with nucleases. Gene Amplif Anal. 1981;2:267–298. [PubMed] [Google Scholar]
- Weisblum B., Graham M. Y., Gryczan T., Dubnau D. Plasmid copy number control: isolation and characterization of high-copy-number mutants of plasmid pE194. J Bacteriol. 1979 Jan;137(1):635–643. doi: 10.1128/jb.137.1.635-643.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]