Background: The roles of PP5 in normal biology are poorly understood.
Results: To help evaluate the biological actions of PP5, a Cre/loxP-conditional mouse line was generated.
Conclusion: In response to UV light, PP5 regulates the phosphorylation of Chk1 at Ser-345.
Significance: Understanding the biological roles for phosphatases is critical for understanding the role of reversible phosphorylation in the control of signaling networks.
Keywords: Mouse, Phosphatase, Phosphatidylserine, PP2A, Protein Phosphatase, Serine/Threonine Protein Phosphatase
Abstract
PP5 is a ubiquitously expressed Ser/Thr protein phosphatase. High levels of PP5 have been observed in human cancers, and constitutive PP5 overexpression aids tumor progression in mouse models of tumor development. However, PP5 is highly conserved among species, and the roles of PP5 in normal tissues are not clear. Here, to help evaluate the biological actions of PP5, a Cre/loxP-conditional mouse line was generated. In marked contrast to the early embryonic lethality associated with the genetic disruption of other PPP family phosphatases (e.g. PP2A and PP4), intercrosses with mouse lines that ubiquitously express Cre recombinase starting early in development (e.g. MeuCre40 and ACTB-Cre) produced viable and fertile PP5-deficient mice. Phenotypic differences caused by the total disruption of PP5 were minor, suggesting that small molecule inhibitors of PP5 will not have widespread systemic toxicity. Examination of roles for PP5 in fibroblasts generated from PP5-deficient embryos (PP5−/− mouse embryonic fibroblasts) confirmed some known roles and identified new actions for PP5. PP5−/− mouse embryonic fibroblasts demonstrated increased sensitivity to UV light, hydroxyurea, and camptothecin, which are known activators of ATR (ataxia-telangiectasia and Rad3-related) kinase. Further study revealed a previously unrecognized role for PP5 downstream of ATR activation in a UV light-induced response. The genetic disruption of PP5 is associated with enhanced and prolonged phosphorylation of a single serine (Ser-345) on Chk1, increased phosphorylation of the p53 tumor suppressor protein (p53) at serine 18, and increased p53 protein levels. A comparable role for PP5 in the regulation of Chk1 phosphorylation was also observed in human cells.
Introduction
The genome of all living cells is attacked relentlessly by both reactive by-products of normal metabolism and environmental mutagens. Accordingly, all cells have evolved elaborate mechanisms to maintain genomic fidelity. One, the DNA damage response, is a signal transduction network that is dependent upon reversible phosphorylation and functions to regulate cell cycle transition, DNA replication, DNA repair, and apoptosis (for review see Refs. 1 and 2). In response to genomic damage caused by reactive oxygen species, UV light, chemicals, or ionizing radiation, three PI3K-related protein kinases, ATM (ataxia-telangiectasia mutated), ATR (ataxia-telangiectasia and Rad3-related), and DNA-PKcs (DNA-dependent proteins kinase catalytic subunit) become activated (3–6). Via elaborate mechanisms, these kinases phosphorylate proteins that help initiate and coordinate: 1) the recruitment of repair proteins to damaged DNA, 2) the suppression of cell cycle progression while repairs are in progress, and 3) the induction of apoptosis when damage cannot be adequately repaired. Other Ser/Thr kinases (e.g. ASK1 (apoptosis signaling kinase 1), Chk1 (checkpoint kinase Chk1), and JNK) together with transcription factors (notably the p53 tumor suppressor protein) help coordinate cell cycle arrest and progression into apoptosis (6). To allow a dynamic response in which cells that have made sufficient repairs can resume DNA replication and continue cell cycle progression, Ser/Thr protein phosphatases are needed to counter the kinases that initiate and maintain the DNA damage response. Currently, the roles of individual protein phosphatases are not well understood.
PP5 is an okadaic acid/cantharidin/microcystin-sensitive member of the PPP family of Ser/Thr phosphatases, which also includes PP1, PP2A, PP4, and PP6 (7–9). Currently the roles played by PP5 in biology and disease are not clear. PP5 is expressed ubiquitously in normal tissues, and elevated PP5 protein levels are associated with the development of breast, liver, and possibly other forms of cancer (10–15). In mouse models of tumor progression, constitutive PP5 overexpression aids tumor growth but does not induce spontaneous tumor formation (12–14). At the molecular level, PP5 is found in multiprotein complexes. In cells that have not encountered stress, PP5 is found in complexes containing heat shock protein 90 (Hsp90), heat shock protein 70 (Hsp70), stress-induced phosphoprotein 1 (STIP1), and cell division cycle 37 homolog (Cdc37) (16–18). In response to various forms of genomic stress, PP5 forms complexes with several stress-activated protein kinases, including DNA-PKcs, ATM, ATR, and ASK1 (19–25). When PP5 levels are suppressed with siRNA or antisense oligonucleotides, several stress-responsive proteins (e.g. ASK1, Cdc37, DNA-PKcs, GRs, p53, and Raf1) display enhanced phosphorylation (16, 17, 19–21, 24–27). Phosphorylation activates ASK1, triggering a response leading to apoptosis, and PP5 appears to negatively regulate ASK1 (19–21, 25, 27). Following exposure to ionizing radiation or UV light, PP5 contributes to ATM-mediated (23, 28, 29) or ATR-mediated (22) checkpoint regulation, respectively. Together, these observations suggest that PP5 may play an underappreciated role in the regulation of signaling networks that allow a cell to respond appropriately to genomic damage.
The high conservation of PP5 among species (mouse, rabbit, bovine, and human PP5 protein share >98% identity) suggests that the actions of PP5 are conserved in mammals. Here, to help identify specific actions of PP5, we generated a PP5-Cre/loxP-conditional mouse line. When animals expressing the Cre/loxP-conditional allele of PP5 were crossed with the MeuCre40 line, viable PP5−/− offspring were produced. Mating with a second mouse strain (ACTB-Cre), in which the Cre recombinase is under the human β-actin gene promoter and expressed in all cells of the embryo by the blastocyst stage, also produced viable PP5-deficient mice. After fully backcrossing (F10-F12) the PP5−/− mice on the C57BL/6J background, studies conducted with PP5−/− mice and embryonic fibroblasts generated from PP5−/− mice (PP5−/− MEFs)3 reveal insight into the roles of PP5 in cellular responses to damage produced by UV light.
EXPERIMENTAL PROCEDURES
Targeting Construct
The murine PP5 gene (Ppp5c) contains 13 exons within a 23-kb region of chromosome 7. To generate the targeting vector, a mouse 129/SvJ genomic bacterial artificial chromosome library was obtained from Incyte Genomics and screened for the presence of both exon 1 and exon 2 (located ∼5 kb downstream of exon 1) using Southern analysis. 32P-Labeled DNA probes were generated using PCR. First a 242-base pair probe targeting exon 2 was created using synthetic primers: 5′-GTCAGGCCATCGAGTTGAACC-3′ and 5′-CTCGTAGTCACGCAGGGCAGC-3′. Three plasmids containing exon 2 (27821, 27822, and 27823) were then screened for the presence of exon 1 using a PCR-generated 32P-labeled probe produced with synthetic primers: 5′-GCTTTGCGGCATGGCGATGGC-3′ and 5′-GATCGCGTTCTCGTAGTCC-3′. Analysis of mouse genetic data predicted that digestion with BamHI would produce a ∼10-kb DNA fragment containing exon 1 flanked on both sides with >4 kb, which was produced by digestion of 27822 with BamHI. The targeting vector (see Fig. 1A) containing three loxP sites, neomycin (Neo)/TK genes, and enhanced GFP was then constructed using conventional cloning techniques (30). The final construct was sequenced in its entirety to verify integrity and correct orientation.
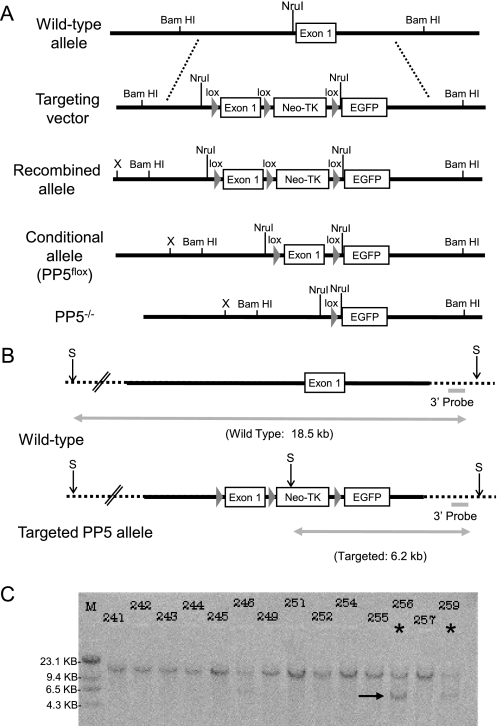
FIGURE 1.
Southern analysis of ES cells indicating homologous recombination. A, schematic representation of the wild-type PP5 mouse allele, targeting vector, recombined allele, conditional allele (PP5flox), and the final PP5 knock-out (PP5−/−) allele generated by Cre recombinase. loxP sites are indicated by triangles (lox). The relative positions of sequences recognized by restriction endonucleases used in analysis are indicated above the lines. B, diagram showing the position of sites recognized by SpeI (S) and the expected size of DNA produced by SpeI digestion of genomic DNA from ES cells containing wild-type or the targeted PP5 alleles. C, autoradiogram showing the hybridization of a 32P-labeled 3′ external probe to genomic DNA from ES cells following digestion with SpeI. For Southern analysis, probes that target a region of the PP5 gene outside the targeting vector (indicted by dashed line) were used to ensure the identification of clones in which homologous recombination had occurred. An asterisk designates an example of a positive clone (i.e. clone 256), and an arrow indicates the 6.2-kb band produced by SpeI digestion of genomic DNA in which homologous recombination has occurred.
Embryonic Stem Cells
The targeting plasmid was digested with SacII, and uptake of linear DNA by 129/X1A embryonic stem (ES) cells derived from 129/SvJ blastocysts (Yale Animal Genomics) was facilitated by electroporation (Bio-Rad Gene Pulser II at 230 V and 500 mF). Following electroporation, ES cells were plated on neo-transgenic mouse embryo fibroblasts inactivated by γ-irradiation and grown in ES cell medium (DMEM, 15% FBS, 1000 units/ml leukemia inhibitory factor, 2 mm glutamine, 0.001% β-mercaptoethanol, and 0.1 mm nonessential amino acids) containing 300 μg/ml G418 for selection. Individual G418-resistant clones were selected and cultured in 24-well plates coated with 0.1% gelatin. Southern analysis in combination with SpeI digestion was used to identify clones containing the PP5 targeted allele as outlined in Fig. 1A. Clones with the correct orientation were detected with a 3′ external probe generated via PCR using the primer pair: 5′-CCTGCTGTAGTATCCTGCTGTCC-3′ and 5′-GGTGCCATCCCTCCACGTCCAC-3′ (see Fig. 1C). Conformation of the correct orientation was also obtained by the generation of a 6.5-kb band following digestion with 6.5 DrdI, which can be detected with both an external 5′ probe and a PCR-generated probe to exon (supplemental Fig. S1).
Chimeric Mice and Mouse Genotyping
A correctly targeted clone (clone 256) was chosen and injected into C57BL/6J blastocysts (Yale Animal Genomics). Chimeric male offspring were mated with C57BL/6J female mice (Jackson Laboratories) and transmission of the conditional allele (PP5flox) was determined using PCR with the primer pair (P1, 5′-CATCCGGAACGACACTTGTGCGGCAG-3′; and P2, 5′-CCAGTGAGTGGCTTCTGC TTGGTATCC-3′ (see Fig. 2). Genotype determinations were performed on genomic DNA prepared by digestion of either tail or embryonic tissues. Mouse tail DNA was isolated using the Qiagen DNeasy blood and tissue kit (Qiagen). The amplification parameters were as follows: 94 °C for 3 min, 94 °C for 45 s, 60 °C for 45 s, and 72 °C for 1 min 40 s for 30 cycles with a final extension of 72 °C for 2.5 min. The resulting PCR product was separated by electrophoresis on a 1% agarose gel and stained with ethidium bromide. The 1.6-kb band indicates the presence of the floxed (PP5flox) allele; a 1.3-kb product represents the PP5 knock-out (PP5−/−) allele.
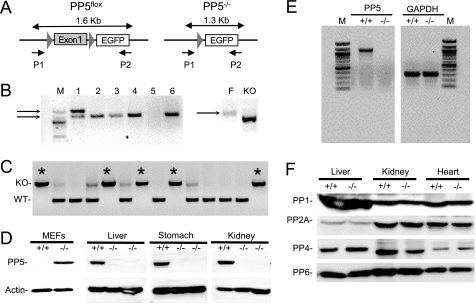
FIGURE 2.
Generation of PP5flox and PP5−/− mice. A, schematic representation of PP5flox (left panel) and PP5−/− alleles (right panel). PCR using primers targeting a region 5′of exon 1 (P1) and enhanced GFP (P2) produce a 1.6-kb product with DNA from mice containing Exon 1. A 1.3-kb product is produced when exon 1 is removed. DNA recognized by P2 is not present in wild-type mice. B, left panel, PCR analysis of litter 160 illustrating the detection of heterozygous (PP5flox/−; lane 1) and PP5−/− (lanes 2–4 and 6) animals. Right panel, PCR products produced from DNA of homozygous PP5flox (F) and PP5−/− (KO) mice. C, representative PCR products used for genotyping, showing PP5+/+ (WT; single lower band), heterozygous (two bands), and PP5−/− mice (KO; single upper band; *) produced using a sense primer complementary to the PP5 promoter and an antisense primer complementary to a region in intron one (see “Experimental Procedures”). D, Western analysis of MEFs and the indicated whole tissue homogenates produced from wild-type (+/+) and PP5 knock-out (−/−) mice. E, ethidium bromide staining of agarose gels to detect PCR products separated by electrophoresis. Under identical conditions, RNA from wild-type (+/+) and PP5 knock-out (−/−) mice was used to produce cDNA, which was then used as template for PCR-mediated amplification using primers that amplify PP5 (5′-GCTTTGCGGCATGGCGATGGC-3′ and 5′-CAGCACTTTGCCATTGATAC-3′). GAPDH was amplified as a control (5′-GCCCATCACCATCTTCCAG-3′ and 5′-TGAGCCCTTCCACAATGCC-3′). F, Western blots showing the levels of PP1, PP2A, PP4, and PP6 in whole organ homogenates obtained from wild-type (+/+) and PP5 knock-out (−/−) mice.
Mice
C57BL/6J mice were purchased from Jackson Laboratories. The MeuCre40 transgenic line (31) was kindly provided by the laboratory of Professor Martin Holzenberger (Paris, France). The β-actin-Cre (ACTB-Cre) transgenic line (32) was obtained from Yale Animal Genomics. All of the animal procedures were preapproved by the institutional animal care and use committee of the University of South Alabama.
In Vivo Selection Cassette Removal
Removal of the Neo-TK selection cassette was achieved selectively in vivo using the MeuCre40 line (supplemental Figs. S2 and S3) as described previously (31).
MEF Generation and Culture
MEFs were isolated from embryonic day 13.5 embryos from PP5+/− intercrosses using conventional methods. Embryo tissues were mechanically disrupted in 100 ml of 0.25% trypsin and placed into individual 6-cm dishes in RPMI medium with 10% FBS. After a few days, PP5−/− MEFs failed to thrive in conventional MEF medium. Subsequent attempts from PP5−/− intercrosses revealed that PP5−/− MEFs grew well when plated on gelatin-coated (0.1%) plates and grown in DMEM-F12 medium supplemented with 5% horse serum (Invitrogen), 30% conditioned medium (filtered medium from the previous passage), insulin (10 μg/ml), EGF (10 μg/ml), hydrocortisone (500 ng/ml), and cholera toxin (100 ng/ml), following adenovirus-mediated transformation with SV40 large T-antigen.
Cell Viability Assays
Littermate PP5−/− and PP5+/+ MEFs (passages 3–6) were transformed with SV40 large T-antigen and plated at 0.2 × 105 cells/well onto 12-well (2 cm in diameter) dishes. After the cells attached and grew to ∼50% confluence, they were treated with UV light (irradiated with 0, 6, 12, 18, 24, or 36 J/m2 of shortwave UV from a 23-watt, shortwave UV light source (254 nm)), camptothecin, or hydroxyurea, at the concentrations indicated. After 24 h, cell viability was measured using ViaLight Plus Cell proliferation assays (Lonza Inc., Rockland, ME) or CellTiter-Glo Luminescence cell viability assay (Promega Corporation, Madison, WI) according to methods provided by the manufacturer.
Generation of cDNA and PCR Detection of PP5
RNA was isolated from equal amounts of PP5−/−, PP5+/−, or PP5+/+ tissues or cells as indicated using TRIzol reagent (Invitrogen). The RNA was precipitated with ethanol and resuspended in 100 μl of nuclease-free water. Then 0.75 μg of RNA was used to synthesize first strand cDNA at 42 °C with murine leukemia virus reverse transcriptase using random hexamers as primers (GeneAmp RNA PCR kit Applied Biosystems). The cDNA generated was then used as a template for PCR. PP5 was detected using the primer pairs that amplify exon 1 and exon 8: 5′-GCT TTG CGG CAT GGC GAT GGC-3′ and 5′-CAG CAC TTT GCC ATT GAT AC-3′. GAPDH was amplified as a control with the primers pair: 5′-GCC CAT CAC CAT CTT CCA G-3′ and 5′-TGA GCC CTT CCA CAA TGC C-3′ (35 cycles of 95 °C for 1 min; 60 °C 1:00 min followed by 72 °C for 2 min). Equal aliquots from each reaction were then separated by electrophoresis (1% agarose) and visualized by staining with ethidium bromide.
Immunoblotting
Western analysis was performed essentially as described previously using polyclonal rabbit antibodies generated against a synthetic 15-amino acid peptide identical to unique regions contained in the near C-terminal regions of PP1, PP2A, or PP5 (33, 34). Briefly, cells or tissues were washed twice with ice-cold PBS and homogenized in cell lysis buffer. The extract was then subjected to centrifugation at 13,000 × g for 5 min, and an aliquot of the supernatant was removed for protein determination. The remaining supernatant was added to an equal volume of 2× sample buffer (120 mm Tris-HCl, pH 7.4, 200 mm dithiothreitol, 20% glycerol, 4% SDS, and 0.02% bromphenol blue). Protein was determined using a Bio-Rad protein quantitation assay, with bovine serum albumin as standard. Typically 25–50 μg of protein was then separated by electrophoresis on 10% SDS-polyacrylamide gels. The gel was electrophoretically transferred to Immobilon-P (Millipore, Billerica, MA) and blocked for 1 h with 100 mm Tris-HCl (pH 7.5), containing 150 mm NaCl, 0.1% Tween 20, and 5% nonfat milk. To detect proteins, the membranes were incubated with the indicated antibody diluted in Tris-HCl (pH 7.6), 150 mm NaCl, 0.2% Tween 20 (TBST) containing 2% bovine serum albumin for 18 h at 4 °C. The membrane was then washed, and the primary antibody was detected employing ECL Western blotting detection reagents (Amersham Biosciences), following the protocols of the manufacturer. Antibodies for actin were purchased from Sigma-Aldrich. Antibodies to p53, phospho-p53 (S15/18), Chk1, phospho-Chk1 (Ser-317), and phospho-Chk1 (Ser-345) were purchased from Cell Signaling Technology, Inc. Antibodies to Cdc25A were purchased from Abcam. Dr. Brian Wadzinski (Vanderbilt University) generously provided polyclonal antibodies to PP4 and PP6. Procedures associated with antisense oligonucleotides targeting PP5 and PP1γ1 have been described previously (34–36).
RESULTS
Generation of a Flanked by loxP (flox) Conditional Allele of PP5 and PP5-deficient Mice
A Cre/loxP-conditional allele of PP5 was generated in mice. To produce these mice, Southern analysis was used to screen a genomic bacterial artificial chromosome library for PP5. Three bacterial artificial chromosome-plasmids containing exons 1 and 2 were identified, and a ∼10-kb fragment containing exon 1 (surrounded both 5′ and 3′ by >4 kb) was produced by digestion with BamHI. loxP sites were placed on each side of regions surrounding exon 1, which upon digestion with Cre recombinase achieves deletion of the translation initiation methionine and the following 74 amino acids (Fig. 1). The Neo-TK cassette (containing a Neo-resistant gene for positive selection and thymidine kinase (TK) gene for negative selection) was introduced to aid the identification of ES cells containing the desired targeting vector. Enhanced GFP was added immediately downstream of the third loxP site to allow the PP5 promoter in PP5−/− cells to drive enhanced GFP expression following Cre-mediated recombination (Fig. 1).
Following transfection and selection of 129/X1A-ES cells, recombination into the PP5 locus was confirmed by Southern analysis with probes generated from sequences outside the targeted regions. Following digestion with SpeI, detection of a 6.2-kb band with an external 3′ probe indicated appropriate recombination, whereas an 18.5-kb SpeI digestion product indicated the presence of the wild-type allele (Fig. 1, B and C). Detection of a 6.5-kb band following digestion with DrdI, which can be detected with an external 5′ probe and a probe targeting exon 1, confirmed appropriate recombination (supplemental Fig. S1). An ES cell clone with this tri-loxP configuration was subsequently used to generate chimeric mice. Four in ten of the chimeric mice transmitted the targeted PP5 allele through the germ line.
PP5 Is Not Essential for Growth or Embryonic Development
Excision of the selection marker and PP5 disruption were achieved selectively in vivo, using MeuCre40 transgenic mice (supplemental Fig. S3). The MeuCre40 line reliably generates the partial mosaic Cre-loxP recombination pattern in the early embryo, allowing efficient removal of the Neo-TK selection marker in vivo (31). PCR analysis was used to identify mice containing the PP5flox conditional allele (Fig. 2). Surprisingly, heterozygous (+/−) mating not only produced homozygous PP5flox mice, viable homozygous mice lacking exon 1 (PP5−/−) were also generated. A second line of PP5−/− mice was then generated by mating with a strain (ACTB-Cre), in which the Cre recombinase is under the human β-actin gene promoter and expressed in all cells of the embryo by the blastocyst stage. Northern analysis of PP5−/− and PP5+/+ (wild-type) cells did not reveal truncated mRNA species, and both PCR and Western analysis of cells and tissues derived from PP5−/− animals revealed no detectable PP5 (Fig. 2, D and E). These studies indicate that the deletion of exon 1 is indeed sufficient to produce PP5-deficient mice. Furthermore, following the disruption of PP5, the levels of PP1, PP2A, PP4, and PP6 did not increase in the PP5−/− animals (Fig. 2F), suggesting that the viability of the PP5−/− mice was not due to compensation by another PPP family phosphatase.
Phenotypic Differences between PP5+/+ and PP5−/− Animals Are Modest
Although backcrossing our PP5−/− lines, the phenotypic characterization of an additional line produced by Lexicon Genetics, at F2/N2, was deposited on-line. All three PP5−/− lines have been examined extensively, both by ourselves and others. All of the PP5−/− lines reported are viable and fertile, with no discernable aberrant phenotypes observed in young (<8 weeks) animals. After further backcrossing on the C57BL/6J background, analysis of the genotyping data at weaning from F6 to F12 revealed that the PP5−/− animals were slightly underrepresented (Table 1). Analysis of 1,086 mice produced ratios of 1:2:0.72, with PP5−/− animals representing 19.15% (25% expected) of the viable offspring. Litters from PP5−/− /PP5−/− mating were also smaller, averaging 2.9 ± 0.9 pups/litter. In addition, and in contrast to MEFs generated from PP5+/+ mice, MEFs generated from PP5−/− embryos failed to thrive in conventional MEF medium, demonstrating erratic rates of growth and phenotypic characteristics. Within a few days, the PP5−/− MEFs failed to adhere to the plates and died. Nonetheless, PP5 is not essential for normal growth, because viable PP5−/− mice that are fertile and have no detectable differences at birth were produced. In addition, PP5−/− MEFs readily proliferate in culture (with consistent rates of growth and maintaining a typical fibroblast appearance) when they are cultured in enriched medium (see “Experimental Procedures”) following transformation with SV40-large T-antigen. Under these conditions, the PP5−/− cell lines are stable through passage 7, and the MEF data reported here were generated from at least three independently derived cell lines (passages 3–6). At this time, it is not clear whether all of the components added to the culture media are necessary for the survival of the PP5−/− MEFs.
TABLE 1.
Genotypic ratios of PP5 heterozygous intercrosses
| Viable mice | Wild type; PP5+/+ number | Heterozygous; PP5+/− number | Knock-out; PP5−/− number |
|---|---|---|---|
| % | % | % | |
| Expected distribution | 271 (25) | 543 (50) | 271.5 (25) |
| Experimental distribution | 304 (27.9) | 574 (52.9) | 208 (19.2)a |
a Statistically significant difference between PP5−/− and PP5+/+ expected and experimental distribution (p < 0.05) by χ2 test.
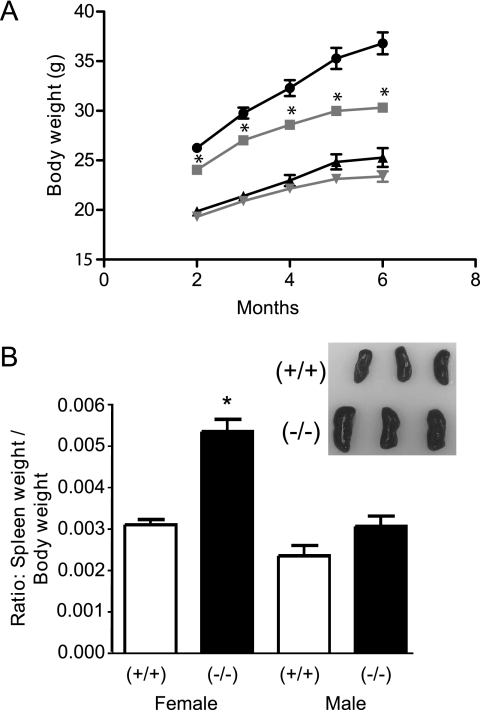
In fully backcrossed animals, two phenotypic differences were observed in nonstressed mice (Fig. 3). Male PP5−/− mice produced from PP5+/−/PP5+/− matings weigh less than their littermate controls (statistically different after ∼2 months of age). A trend toward smaller mice was also observed in PP5−/− female mice. However, this trend was not statistically different by the end of the study (Fig. 3A). In addition, upon autopsy, splenomegaly was observed in female PP5−/− animals (noted after ∼1 year of age). How the lack of PP5 contributes to these observations is not yet clear, and investigators willing to explore possible mechanisms are encouraged to contact the corresponding author to obtain the PP5−/− mice for further studies.
FIGURE 3.
Phenotypic differences between PP5−/− and PP5+/+ mice. A, comparison of body weights between PP5−/− (gray lines) and PP5+/+ (black lines) mice with time. Circles and squares represent male mice; triangles and inverted triangles represent females. B, comparison of spleen weight in male and female mice at ∼1 year of age plotted as a ratio of spleen weight to total body weight. The data shown are the means ± S.E., n = 10–22. The asterisk indicates a statistically significant difference between PP5−/− and PP5+/+ animals (p < 0.05) by two-way ANOVA (A) and one-way ANOVA (B).
PP5−/− MEFs Have Increased Sensitivity to UV Irradiation, Camptothecin, and Hydroxyurea
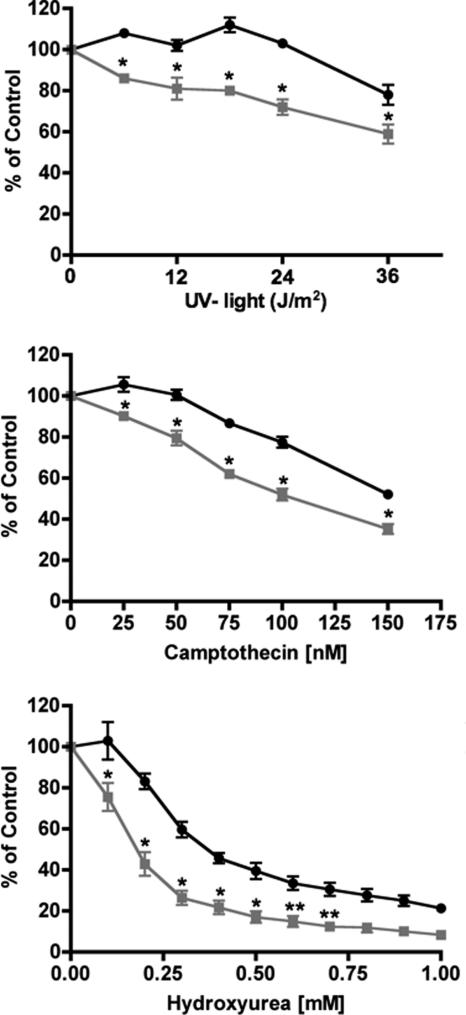
In human cells, the overexpression of PP5 provides resistance to UV-induced apoptosis, and PP5 can affect ATR-mediated checkpoint signaling (12, 14, 22). Therefore, we next conducted dose-response studies to determine whether the PP5-deficient 13.5-day MEFs demonstrated altered sensitivity to UV irradiation. Measuring cell viability 24 h after treatment as an outcome, MEFs from littermate PP5−/− or PP5+/+ mice were exposed to UV light. The PP5−/− MEFs were more sensitive to UV light. Exposures that had no effect on PP5+/+ MEFs killed ∼25% of the PP5−/− cells by 24 h (Fig. 4). The PP5−/− MEFs were also more sensitive to both camptothecin and hydroxyurea. However, other agents that damage DNA and arrest the cell cycle (i.e. cisplatin) demonstrated equal potency in both the wild-type and PP5-deficient MEFs (supplemental Fig. S4).
FIGURE 4.
Sensitivity of MEFs to UV light, camptothecin, or hydroxyurea. MEFs produced from PP5+/+ (circles) or PP5−/− (squares) animals were treated as indicated. After 24 h, cell viability was measured as described under “Experimental Procedures.” The data represent the means ± S.E. from three separate experiments in which four replicate plates for each condition and cell type were processed simultaneously. * (p < 0.01) and ** (p < 0.05) denote statistically significant differences (two-way ANOVA).
PP5−/− MEFs Have Increased and Prolonged Phosphorylation of Chk1 (Ser-345) and p53 (Ser-18; Equivalent to Ser-15 in Humans) following Exposure to UV Light
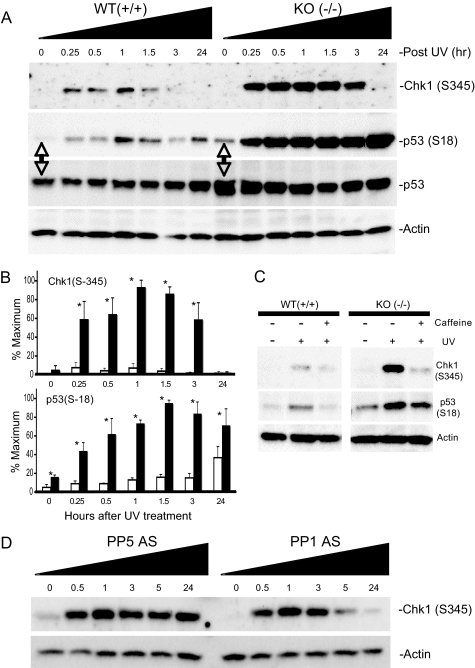
The kinase activity of ATR is activated in response to UV light, and PP5 forms a complex with ATR following exposure to UV light (22). When activated, ATR phosphorylates a number of proteins that participate in the DNA damage response, and ATR-mediated phosphorylation of Chk1 at Ser-317 and Ser-345 activates Chk1-kinase activity and represents a hallmark of ATR-activation (1, 5, 37, 38). In turn, Chk1 phosphorylates p53 (Ser-15/18 and Ser-20/23) and Cdc25A. ATR can also directly phosphorylate p53 at several residues (1, 38). In both PP5−/− and PP5+/+ MEFs, UV light (24 J/m2) produced a transient increase in Chk1 phosphorylation at Ser-345, with phosphorylation levels increasing rapidly (apparent by 15 min) peaking after ∼1 h and then returned to basal levels (Fig. 5). Comparison of the UV response in PP5+/+ and PP5−/− MEFs revealed that Ser-345 phosphorylation was increased, and the duration that Ser-345 remained phosphorylated was longer, in the PP5−/− MEFs. In both PP5+/+ and PP5−/− MEFs, without exposure to UV light (time 0), Chk1 phosphorylation at Ser-345 was not detected. By 24 h, phosphorylation returned to basal levels. Chk1, Rad17, and H2AX protein levels were similar in the PP5+/+ and PP5−/− MEFs, and PP5 was not required for UV-induced phosphorylation of Rad17 (Ser-645) or H2AX (Ser-139) (supplemental Fig. S5). The phosphorylation of Chk1 at Ser-317 was observed in both PP5+/+ and PP5−/− MEFs.
FIGURE 5.
Enhanced phosphorylation of Chk1 and p53 in PP5−/− MEFs or HeLa cells following exposure to UV light. MEFs (embryonic day 13.5) derived from PP5+/+ (WT) or PP5−/− (KO) littermate embryos or HeLa cells, as indicated below, were exposed to 24 J/m2 UV light. At the times indicated, the cells were harvested and phospho-Chk1 (Ser-345), phospho-p53 (Ser-18), p53, and actin protein levels were detected by Western analysis. A, representative images of three separate experiments conducted with MEFs. B, quantitation of image data from separate experiments comparing the phosphorylation of Chk1 (Ser-345) in the top panel and p53 (Ser-18) in the bottom panel. Means ± S.E., n = 3; the asterisk denotes a statistically significant difference (p < 0.05). C, representative image illustrating the effect of caffeine on UV light-induced phosphorylation of Chk1 (Ser-345) and p53 (Ser-18). PP5+/+ (WT) or PP5−/− (KO) MEFs were treated with 1 mm caffeine and then exposed to UV light (24 J/m2). After 30 min, the cells were harvested and processed for Western analysis. Phosphospecific antibodies that recognize the phosphorylated forms of Chk1 (Ser-345) or p53 (Ser-18) were used to detect changes in phosphorylation. Actin levels were measured as a control for loading. D, enhanced phosphorylation of Chk1 in following exposure to UV light. HeLa cells treated with antisense oligonucleotides targeting PP5 (ISIS 15534) or PP1 (ISIS 14436), as indicated. After 48 h, the cells were exposed to 24 J/m2 UV light. At the times indicated, the cells were harvested, and phospho-Chk1 (Ser-345) and actin protein levels were detected by Western analysis. The images shown are representative images of three or more separate experiments.
Analysis of p53 in aliquots from the same samples revealed that the phosphorylation of p53 at Ser-18 was also increased in the PP5−/− MEFs. In comparison with PP5+/+ cells, Ser-18 phosphorylation caused by UV light was more robust at all time points in the PP5−/− cells. In addition, Ser-18 phosphorylation was observed even in the untreated PP5−/− cells. The level of p53 protein was also elevated in the PP5−/− cells at all time points examined (Fig. 5). When the cells were pretreated with caffeine at concentrations sufficient to inhibit the activation of ATR and ATM, as known from previous studies of UV-induced phosphorylation in the wild-type cells, the phosphorylation of both Chk1(Ser-345) and p53 (Ser-18) was decreased in the PP5+/+ MEFs (Fig. 5C). In the PP5-deficient cells, UV-induced Chk1 (Ser-345) hyperphosphorylation was also markedly suppressed by caffeine. In contrast, the basal and UV-induced hyperphosphorylation of p53 (Ser-18) was only slightly suppressed in the caffeine-treated PP5−/− cells.
Suppression of PP5 and PP1 in HeLa Cells Confirms a Specific Role for PP5 in the Regulation of Chk1 Phosphorylation at Ser-345
Similar to observations made in the MEFs, in HeLa cells UV light induces a transient increase in Chk1 phosphorylation (39). Using previously characterized antisense oligonucleotides to suppress the expression of PP5 (34, 36) or PP1γ1 (35), we next examined the specificity of PP5 in UV light-induced Chk1 phosphorylation. As seen in Fig. 5D, similar to the observations made in the PP5−/− MEFs, the suppression of PP5, but not PP1γ1, was associated with enhanced and prolonged Chk1 phosphorylation (Ser-345).
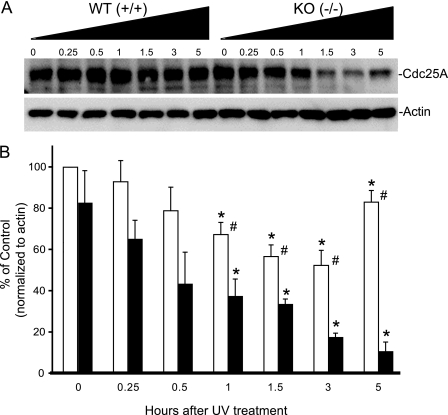
PP5−/− MEFs Have Decreased Levels of Cdc25A following Exposure to UV Light
In response to sublethal UV exposure, phosphorylation-activated Chk1 also phosphorylates Cdc25A, leading to its ubiquitination and degradation (40, 41). When damaged DNA has been repaired, Chk1 activity returns to basal levels, and Cdc25A levels increase, allowing cell cycle progression to continue. Consistent with previous reports, UV light (24 J/m2) produced a transient decrease in Cdc25A levels in the wild-type MEFs, which is apparent after ∼1 h and returns to a basal state after ∼5 h. In the PP5−/− MEFs, following UV exposure, the level of Cdc25A is less, and the rebound noted after 5 h in the wild-type animals has not yet occurred (Fig. 6). By 24 h, the levels of Cdc25A in the PP5+/+ and PP5−/− MEFs were similar (not shown).
FIGURE 6.
Changes in Cdc25A levels in MEFs following treatment with UV light. MEFs produced from wild-type PP5+/+ or PP5 disrupted PP5−/− mice were treated with UV light as in Fig. 5. At the time indicated, the cells were harvested and processed for Western analysis using an antibody that recognizes Cdc25A. A, representative images illustrating changes in Cdc25A protein levels. B, quantitation of images from three separate experiments (mean ± S.E., n = 3); the asterisk indicates a statistically significant difference between UV treatment and untreated controls (p < 0.05); # indicates a statistically significant difference between PP5+/+ and PP5−/− MEFs receiving identical treatment (p < 0.05) by two-way ANOVA.
DISCUSSION
We have produced a Cre/loxP-conditional mouse line to help evaluate the roles of Ser/Thr protein phosphatase 5 (PP5). Intercrosses with lines (i.e. MeuCre40 and ACTB-Cre) that express the Cre recombinase during early embryonic development (31, 32) produced PP5-deficient mice that were viable and fertile, demonstrating that PP5 is not necessary for survival. After backcrossing on the C57BL/6J background, minor phenotypic differences between PP5+/+ and PP5−/− mice that were not noticed in preliminary studies conducted with (F2/N2) PP5-disrupted mice became apparent (Table 1 and Fig. 3). The under-representation of PP5−/− live births from PP5+/− heterozygous intercrosses and the small litter size of PP5−/− homozygous intercrosses indicate that PP5 provides a slight advantage during embryonic development. A decrease in the weight of male PP5−/− mice that became significant after 2 months of age was also observed, and autopsies of mice at the age of 1 year revealed splenomegaly in female PP5−/− animals. At this time, it is not clear how the disruption of PP5 contributes to these observations.
Previous studies conducted with human cells, employing siRNA or antisense oligonucleotides to suppress PP5 protein levels, indicate that PP5 participates in the regulation of checkpoint signaling mechanisms that allow a cell to respond appropriately following exposure to ionizing radiation, reactive oxygen species, genotoxic chemicals, and UV light (for review see Ref. 14). In response to ionizing radiation, PP5 interacts with ATM, and initial studies with F2/N2 backcrossed PP5−/− mice confirmed many of the proposed roles for PP5 in ATM signaling previously identified in human cells with antisense oligonucleotides targeting PP5 (23, 29).
In cultured human cells, PP5 has been reported to be required for ATR-mediated checkpoint activation. Following UV irradiation, PP5 associates with ATR in both cancer and noncancer cell lines (22), and in cancer cells (i.e. A549 and HeLa) the suppression of PP5 does not affect ATR association at sites of DNA damage or the interaction of ATR with ATRIP (22). Our observations are consistent with these previously reported studies. However, the development of viable and fertile PP5-deficient mice indicates that not all aspects of ATR signaling are dependent on PP5, because ATR deletion in mice results in embryonic lethality (4) and Cre-mediated deletion of ATR in tissues of adult mice produces marked phenotypes that are not observed in the PP5−/− mice (37).
Therefore, we examined the role of PP5 in UV light-induced signaling in more detail. ATR activation has broad cellular effects that help slow cell cycle progression, affect replication fork stability, and help regulate fork restart. Dose-response studies using MEFs from littermate PP5−/− or PP5+/+ mice exposed to UV light (which damages DNA), camptothecin (which arrests S phase replication via the inhibition DNA topoisomerase activity), or hydroxyurea (which arrests replication and damages DNA) revealed that PP5−/− MEFs are more sensitive to all three treatments that are all known to activate ATR. This observation is consistent with the increased sensitivity to stress-induced apoptosis produced by siRNA or antisense oligonucleotide-mediated suppression of PP5 in human cells (14, 20, 22, 34) and suggests that PP5 may act to help suppress pathways leading to death, while the damage is assessed and repaired. However, other agents that damage DNA and arrest cell cycle progression (i.e. cisplatin; supplemental Fig. S4) demonstrated equal potency in both the wild-type and PP5-deficient MEFs, suggesting that PP5 does not directly act on apoptotic mechanisms. In addition, we observed no evidence that ATR activation was prevented in the PP5−/− MEFs. For example, UV light-induced phosphorylation of Rad17 at Ser-645, which is dependent on ATR (42), was observed in the PP5−/− MEFs (supplemental Fig. S5). Therefore, we next examined events downstream of ATR activation.
In response to UV light, ATR phosphorylates Chk1 (at both Ser-317 and Ser-345), which activates the kinase activity of Chk1 (5, 43, 44). In turn, Chk1 phosphorylates p53 (at Ser-15/18). Chk1 also phosphorylates Cdc25A, leading to ubiquitin-mediated proteolysis and a prolonged cell cycle (40, 45–47). ATR can also directly phosphorylate p53 at Ser-15/18, which has been implicated in the control of both cell cycle regulation and apoptosis (37, 48). Dose-response studies revealed that in PP5−/− cells UV-induced Chk1 phosphorylation at Ser-345 was both enhanced and prolonged. Without UV light exposure, increased basal Ser-345 phosphorylation was not observed, and the lack of PP5 did not prevent Chk1 from returning to a basal state of low phosphorylation by 24 h. Caffeine, at a concentration sufficient to inhibit the kinase activity of both ATR and ATM, effectively suppressed Chk1 Ser-345 phosphorylation in response to UV light in both the PP5+/+ and PP5−/− MEFs. Cdc25A degradation, which represents a hallmark of ATR-mediated Chk1 activation, was also more pronounced in the PP5−/− MEFs, as compared with the PP5+/+ controls. In HeLa cells, the suppression of PP5 but not PP1γ1 produced a prolonged phosphorylation of Chk1 at Ser-345. Together, these observations refine the role of PP5 in UV light-induced response originally proposed by Zhang et al. (22), placing PP5 downstream of ATR and upstream of Chk1. This may indicate that the elevated levels of PP5 observed in some human cancers may contribute to tumor development by inducing a compensatory mechanism that counters the specific action of ATR. Still, the observation that Chk1 (Ser-345) phosphorylation returns to normal levels in the PP5−/− MEFs after 24 h indicates that PP5 is not the only phosphatase that acts on Chk1 at Ser-345. This observation is supported by elegant studies that indicate that Chk1 phosphorylation by ATR can also be regulated by PP2A, with PP2A acting in a ATR-Chk1-PP2A regulatory circuit that functions to keep Chk1 in a low activity state during an unperturbed cell division cycle while allowing Chk1 to remain primed to rapidly respond to genomic stress (41). Our data support and build upon this model, suggesting that PP5 acts as a governor of this Chk1 (Ser-345) phosphorylation-dependent circuit, acting to dampen, rather than prevent, spikes in Chk1 activity produced by genomic damage. This may help cells gauge the magnitude of the DNA damage. Alternatively, PP5 has been reported to be capable of interacting with a PP2AA/PP2Ac/PP2Ab53 heterotrimer (49), and our data cannot exclude the possibility that PP5 is affecting one or a subset of the >20 holoenzymes that share the same PP2AA/PP2Ac scaffold/catalytic components.
The relationship between PP5 and p53 is less clear. Our studies revealed that the genetic disruption of PP5 is associated with increased p53 protein levels and enhanced p53 phosphorylation (at Ser-18; equivalent to Ser-15 in humans). In response to UV treatment, p53 accumulates and becomes hyperphosphorylated at Ser-18 in the PP5−/− MEFs. The same observations have been made in human cells using antisense oligonucleotides to suppress the expression of PP5, where changes in the phosphorylation at other sites (i.e. Ser-6, Ser-37, and Ser-392; human numbering) were also not observed (33, 34). Unlike Chk1 (Ser-345) phosphorylation, caffeine suppressed some, but not all, p53 phosphorylation at Ser-18, and both Ser-18 phosphorylation and p53 protein levels were elevated in the PP5−/− MEFs prior to treatment with UV light. This suggests that, in addition to suppressing Chk1-mediated phosphorylation of p53 (Ser-18), PP5 may also acts to suppress a second pathway leading to Ser-18 phosphorylation and increased p53 protein levels. Because many convergent pathways control p53 (Ser-15/18) phosphorylation, substantial future effort will be needed to determine precisely how PP5 affects p53 signaling. In addition, it should also be noted that to maintain the PP5−/− MEFs in culture, they were transfected with the SV40 large T-antigen. SV40 large T-antigen evolved to bind and inactivate p53, preventing apoptosis in virally infected cells. The observations made are not likely due to SV40 large T-antigen alone, because littermate control MEFs were also SV40 large T-antigen transformed and treated in an identical manner. In addition, similar observations were made using antisense oligonucleotides to suppress PP5 expression in human cell lines that were not SV40-tranformed (33, 34). Still, at this time our data does not rule out the possibility that PP5, via a currently obscure mechanism, affects SV40 large T-antigen suppression of a p53 response.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants CA60750 (to R. E. H.) and T32 HL076125 (to J. W. L.). This work was also supported by the Olle Engkvist Byggmästare Foundation, the Diabetes Research and Wellness Foundation, Berth von Kantzow's Foundation, Golje's Memorial Foundation, the Eva and Oscar Ahrén's Foundation, and the Karolinska Institutet (to Å. S.) and the Swedish Society for Medical Research (to H. O.). Some of the studies for this investigation were conducted in a facility constructed with support from Research Facilities Improvement Program Grant C06 RR11174 from the United States National Center for Research Resources.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text and Figs. S1–S5.
- MEF
- mouse embryonic fibroblast
- ES
- embryonic stem
- Neo
- neomycin
- ANOVA
- analysis of variance.
REFERENCES
- 1. Reinhardt H. C., Yaffe M. B. (2009) Curr. Opin. Cell Biol. 21, 245–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhou B. B., Elledge S. J. (2000) Nature 408, 433–439 [DOI] [PubMed] [Google Scholar]
- 3. Xu Y., Baltimore D. (1996) Genes Dev. 10, 2401–2410 [DOI] [PubMed] [Google Scholar]
- 4. Brown E. J., Baltimore D. (2000) Genes Dev. 14, 397–402 [PMC free article] [PubMed] [Google Scholar]
- 5. Smits V. A., Warmerdam D. O., Martin Y., Freire R. (2010) Front. Biosci. 15, 840–853 [DOI] [PubMed] [Google Scholar]
- 6. Lovejoy C. A., Cortez D. (2009) DNA Repair 8, 1004–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Swingle M., Ni L., Honkanen R. E. (2007) Methods Mol. Biol. 365, 23–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Swingle M. R., Amable L., Lawhorn B. G., Buck S. B., Burke C. P., Ratti P., Fischer K. L., Boger D. L., Honkanen R. E. (2009) J. Pharmacol. Exp. Ther. 331, 45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Swingle M. R., Honkanen R. E., Ciszak E. M. (2004) J. Biol. Chem. 279, 33992–33999 [DOI] [PubMed] [Google Scholar]
- 10. Ghobrial I. M., McCormick D. J., Kaufmann S. H., Leontovich A. A., Loegering D. A., Dai N. T., Krajnik K. L., Stenson M. J., Melhem M. F., Novak A. J., Ansell S. M., Witzig T. E. (2005) Blood 105, 3722–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shirato H., Shima H., Nakagama H., Fukuda H., Watanabe Y., Ogawa K., Matsuda Y., Kikuchi K. (2000) Int. J. Oncol. 17, 909–912 [DOI] [PubMed] [Google Scholar]
- 12. Golden T., Aragon I. V., Rutland B., Tucker J. A., Shevde L. A., Samant R. S., Zhou G., Amable L., Skarra D., Honkanen R. E. (2008) Biochim. Biophys. Acta 1782, 259–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Golden T., Aragon I. V., Zhou G., Cooper S. R., Dean N. M., Honkanen R. E. (2004) Cancer Lett. 215, 95–100 [DOI] [PubMed] [Google Scholar]
- 14. Golden T., Swingle M., Honkanen R. E. (2008) Cancer Metastasis Rev. 27, 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fukuda H., Tsuchiya N., Hara-Fujita K., Takagi S., Nagao M., Nakagama H. (2007) J. Cell Biochem. 101, 321–330 [DOI] [PubMed] [Google Scholar]
- 16. Vaughan C. K., Mollapour M., Smith J. R., Truman A., Hu B., Good V. M., Panaretou B., Neckers L., Clarke P. A., Workman P., Piper P. W., Prodromou C., Pearl L. H. (2008) Mol. Cell 31, 886–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shao J., Hartson S. D., Matts R. L. (2002) Biochemistry 41, 6770–6779 [DOI] [PubMed] [Google Scholar]
- 18. Skarra D. V., Goudreault M., Choi H., Mullin M., Nesvizhskii A. I., Gingras A. C., Honkanen R. E. (2011) Proteomics 11, 1508–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mkaddem S. B., Werts C., Goujon J. M., Bens M., Pedruzzi E., Ogier-Denis E., Vandewalle A. (2009) J. Biol. Chem. 284, 12541–12549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou G., Golden T., Aragon I. V., Honkanen R. E. (2004) J. Biol. Chem. 279, 46595–46605 [DOI] [PubMed] [Google Scholar]
- 21. Huang S., Shu L., Easton J., Harwood F. C., Germain G. S., Ichijo H., Houghton P. J. (2004) J. Biol. Chem. 279, 36490–36496 [DOI] [PubMed] [Google Scholar]
- 22. Zhang J., Bao S., Furumai R., Kucera K. S., Ali A., Dean N. M., Wang X. F. (2005) Mol. Cell Biol. 25, 9910–9919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ali A., Zhang J., Bao S., Liu I., Otterness D., Dean N. M., Abraham R. T., Wang X. F. (2004) Genes Dev. 18, 249–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wechsler T., Chen B. P., Harper R., Morotomi-Yano K., Huang B. C., Meek K., Cleaver J. E., Chen D. J., Wabl M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 1247–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Morita K., Saitoh M., Tobiume K., Matsuura H., Enomoto S., Nishitoh H., Ichijo H. (2001) EMBO J. 20, 6028–6036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. von Kriegsheim A., Pitt A., Grindlay G. J., Kolch W., Dhillon A. S. (2006) Nat. Cell Biol. 8, 1011–1016 [DOI] [PubMed] [Google Scholar]
- 27. Kutuzov M. A., Andreeva A. V., Voyno-Yasenetskaya T. A. (2005) J. Biol. Chem. 280, 25388–25395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kang Y., Cheong H. M., Lee J. H., Song P. I., Lee K. H., Kim S. Y., Jun J. Y., You H. J. Biochem. Biophys. Res. Commun. 404, 476–481 [DOI] [PubMed] [Google Scholar]
- 29. Yong W., Bao S., Chen H., Li D., Sánchez E. R., Shou W. (2007) J. Biol. Chem. 282, 14690–14694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Amable L. (2009) Generation of a Knockout Mouse Model to Determine the in Vivo Function of Protein Phosphatase 5 (PP5), Ph.D. dissertation, University of South Alabama [Google Scholar]
- 31. Leneuve P., Colnot S., Hamard G., Francis F., Niwa-Kawakita M., Giovannini M., Holzenberger M. (2003) Nucleic Acids Res. 31, e21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lewandoski M., Meyers E. N., Martin G. R. (1997) Cold Spring Harbor Symp. Quant. Biol. 62, 159–168 [PubMed] [Google Scholar]
- 33. Zuo Z., Urban G., Scammell J. G., Dean N. M., McLean T. K., Aragon I., Honkanen R. E. (1999) Biochemistry 38, 8849–8857 [DOI] [PubMed] [Google Scholar]
- 34. Zuo Z., Dean N. M., Honkanen R. E. (1998) J. Biol. Chem. 273, 12250–12258 [DOI] [PubMed] [Google Scholar]
- 35. Cheng A., Dean N. M., Honkanen R. E. (2000) J. Biol. Chem. 275, 1846–1854 [DOI] [PubMed] [Google Scholar]
- 36. Golden T. A., Honkanen R. E. (2003) Methods Enzymol. 366, 372–390 [DOI] [PubMed] [Google Scholar]
- 37. Brown E. J., Baltimore D. (2003) Genes Dev. 17, 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen Y., Sanchez Y. (2004) DNA Repair 3, 1025–1032 [DOI] [PubMed] [Google Scholar]
- 39. Zhao H., Piwnica-Worms H. (2001) Mol. Cell Biol. 21, 4129–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xiao Z., Chen Z., Gunasekera A. H., Sowin T. J., Rosenberg S. H., Fesik S., Zhang H. (2003) J. Biol. Chem. 278, 21767–21773 [DOI] [PubMed] [Google Scholar]
- 41. Leung-Pineda V., Ryan C. E., Piwnica-Worms H. (2006) Mol. Cell Biol. 26, 7529–7538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Post S., Weng Y. C., Cimprich K., Chen L. B., Xu Y., Lee E. Y. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13102–13107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu Q., Guntuku S., Cui X. S., Matsuoka S., Cortez D., Tamai K., Luo G., Carattini-Rivera S., DeMayo F., Bradley A., Donehower L. A., Elledge S. J. (2000) Genes Dev. 14, 1448–1459 [PMC free article] [PubMed] [Google Scholar]
- 44. Jiang K., Pereira E., Maxfield M., Russell B., Goudelock D. M., Sanchez Y. (2003) J. Biol. Chem. 278, 25207–25217 [DOI] [PubMed] [Google Scholar]
- 45. Shimuta K., Nakajo N., Uto K., Hayano Y., Okazaki K., Sagata N. (2002) EMBO J. 21, 3694–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sørensen C. S., Syljuåsen R. G., Falck J., Schroeder T., Rönnstrand L., Khanna K. K., Zhou B. B., Bartek J., Lukas J. (2003) Cancer Cell 3, 247–258 [DOI] [PubMed] [Google Scholar]
- 47. Chen M. S., Ryan C. E., Piwnica-Worms H. (2003) Mol. Cell Biol. 23, 7488–7497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tibbetts R. S., Brumbaugh K. M., Williams J. M., Sarkaria J. N., Cliby W. A., Shieh S. Y., Taya Y., Prives C., Abraham R. T. (1999) Genes Dev. 13, 152–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lubert E. J., Hong Y., Sarge K. D. (2001) J. Biol. Chem. 276, 38582–38587 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.