Background: Catabolic utilization of sialic acid is essential for the pathogenesis of enteropathogens.
Results: NanR, CRP, and ManNAc-6P regulate the V. vulnificus nan cluster required for catabolism of Neu5Ac, a sialic acid.
Conclusion: This cooperative regulation leads to precise tuning of the nan cluster expression.
Significance: The results shed insight into the understanding of sialate metabolism central to host-microbe interactions.
Keywords: Bacteria, Bacterial Transcription, Gene Regulation, Microbiology, Sialic Acid, Transcription Regulation, Transcription Repressor, Vibrio vulnificus, nan Operon, ManNAc-6P
Abstract
The nan cluster of Vibrio vulnificus, a food-borne pathogen, consists of two divergently transcribed operons, nanTPSLAR and nanEK nagA, required for transport and catabolism of N-acetylneuraminic acid (Neu5Ac). A mutation of nanR abolished the extensive lag phase observed for the bacteria growing on Neu5Ac and increased transcription of nanTP and nanE, suggesting that NanR is a transcriptional repressor of both nan operons. Intracellular accumulation of Neu5Ac was dependent on the carbon source, implying that the nan operons are also subject to catabolite repression. Hence, cAMP receptor protein (CRP) appeared to activate and repress transcription of nanTPSLAR and nanEK nagA, respectively. Direct bindings of NanR and CRP to the nanTP-nanE intergenic DNA were demonstrated by EMSA. Two adjacent NanR-binding sites centered at +44.5 and −10 and a CRP-binding site centered at −60.5 from the transcription start site of nanTP were identified by DNase I protection assays. Mutagenesis approaches, in vitro transcription, and isothermal titration calorimetry experiments demonstrated that N-acetylmannosamine 6-phosphate specifically binds to NanR and functions as the inducer of the nan operons. The combined results propose a model in which NanR, CRP, and N-acetylmannosamine 6-phosphate cooperate for precise adjustment of the expression level of the V. vulnificus nan cluster.
Introduction
When bacteria invade the human gut, adverse environmental changes, such as increased competition for the specific nutrients imposed by the host cells and endogenous bacterial flora, are encountered. As such, the ability to acquire nutrients under these adverse environments is often crucial for bacteria to survive and multiply in the intestine (1, 2). In the intestine, as the presence of free glucose is quite limited, enteropathogenic bacteria must be able to use alternative nutrients to be a successful pathogen (3, 4). Epithelial surfaces of the intestine exposed to bacteria are covered by a viscous mucous layer where bacteria primarily colonize (for a recent review see Ref. 5). The mucous layer contains mucins that are highly glycosylated polymorphic glycoproteins (6). Up to 85% of mucins are carbohydrates in dry weight (7), indicating that the mucin sugars are important carbon and energy sources to support the survival and growth of infecting enteropathogens.
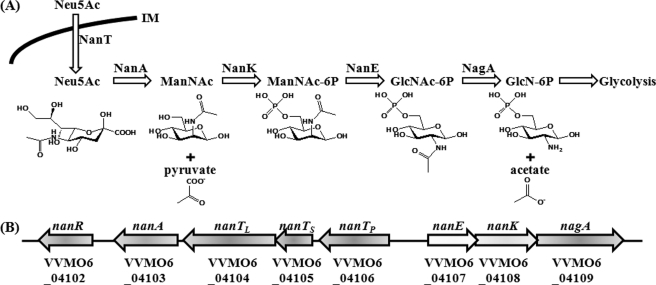
Sialic acid represents a family of related nine-carbon sugar acids that are notably found at the distal end of carbohydrate side chains of mucins (for recent reviews see Refs. 8–10). The most abundant sialic acid is N-acetylneuraminic acid (Neu5Ac),2 and therefore many intestinal commensal and pathogenic bacteria have evolved elaborate systems for the catabolic utilization of Neu5Ac. The bacteria employ different transporters to obtain Neu5Ac from the environments. For example, NanT of Escherichia coli belongs to the major facilitator superfamily, and SiaPQM of Haemophilus influenzae (or DctQPM of Vibrio cholerae) is a tripartite ATP-independent periplasmic transporter (9, 11). Regardless of the mode of Neu5Ac uptake, N-acetylneuraminate lyase (NanA) initiates the catabolism of Neu5Ac by cleaving it into pyruvate and N-acetylmannosamine (ManNAc) in most bacteria. ManNAc is ultimately converted into an intermediate of the central metabolism (fructose 6-phosphate) via the activities of various proteins, including NanK (N-acetylmannosamine kinase), NanE (N-acetylmannosamine-6-phosphate epimerase), and NagA (N-acetylglucosamine-6-phosphate deacetylase) as presented in Fig. 1A (8, 11).
FIGURE 1.
Sialic acid catabolism and the V. vulnificus nan cluster. A, schematic representation of the transport and catabolic pathway of sialic acid in bacteria is adopted from previously reported literature (8, 11). B, arrows represent the transcriptional directions and the coding regions of the V. vulnificus nan cluster. Gene names and locus tag numbers are based on the data base of the V. vulnificus MO6-24/O genome sequence (GenBankTM accession numbers CP002469 and CP002470; see Ref. 40). Proposed gene products are as follows: nanR, a putative transcriptional regulator; nanA, Neu5Ac lyase; nanTPSL, components of tripartite ATP-independent periplasmic type Neu5Ac transporter; nanE, N-acetylmannosamine-6-phosphate epimerase; nanK, N-acetylmannosamine kinase; nagA, N-acetylglucosamine-6-phosphate deacetylase.
Recently, to investigate the role of Neu5Ac catabolism in the pathogenesis of Vibrio vulnificus, a foodborne enteropathogen (12), a nanA mutant was constructed and used to evaluate phenotype changes. The nanA mutant was defective for intestinal colonization and significantly diminished in virulence in a mouse model (13). An independent study with V. cholerae demonstrated that the genes involved in the transport and catabolism of Neu5Ac are located in VPI-2 (Vibrio pathogenicity island 2), and the nanA mutant was also defective for colonization in the mouse intestine (14). These studies together suggested that the catabolic utilization of Neu5Ac is essential for the pathogenesis of the bacteria by ensuring growth and survival during infection (13, 14). Nevertheless, very little is known about the regulatory mechanism used by the bacteria to modulate the expression of the Vibrio nan clusters. Accordingly, in this study, the transcriptional units and promoters of the V. vulnificus nan genes were determined, and the roles of NanR, a putative transcription regulator, and CRP in the regulation of the nan genes were analyzed at the molecular level. In addition, ManNAc-6P, a Neu5Ac catabolic intermediate, was identified as an inducer that binds to NanR and alters its interaction with the nan promoter DNA.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, Cloning of the Nan Intergenic Region, and Culture Media
The strains and plasmids used in this study are listed in Table 1. The 299-bp nanTP-nanE intergenic DNA was amplified by PCR using primers NANT0903 and NANE0905 and cloned into pGEM-T easy (Promega) forming pBS0909. Unless noted otherwise, the V. vulnificus strains were grown in Luria-Bertani (LB) medium supplemented with 2.0% (w/v) NaCl (LBS) at 30 °C. When required, M9 (15), in which one of the carbon sources such as Neu5Ac (5 mm), xylose (10 mm), proline (10 mm), glycerol (0.2%, w/v), or glucose (0.2%, w/v) was supplemented, was used as a minimal medium. All chemicals were purchased from Sigma except ManNAc-6P, which was purchased from Carbosynth (Berkshire, UK).
TABLE 1.
Plasmids and bacterial strains used in this study
| Strain or plasmid | Relevant characteristicsa | Ref. or source |
|---|---|---|
| Bacterial strains | ||
| V. vulnificus | ||
| ATCC29307 | Wild type V. vulnificus, virulent | Laboratory collection |
| DI0201 | ATCC29307 with Δcrp | 19 |
| HG072 | ATCC29307 with nanA::nptI, Kmr, Rifr | 13 |
| BS0805 | ATCC29307 with ΔnanR::nptI, Kmr | This study |
| BS0902 | BS0805 with Δcrp, Kmr | This study |
| BS1012 | HG072 with ΔnanR, Kmr, Rifr | This study |
| BS1104 | ATCC29307 with ΔnanE | This study |
| E. coli | ||
| SM10λpir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu λ pir; Kmr; host for π-requiring plasmids; conjugal donor | 18 |
| BL21 (DE3) | F−, ompT, hsdS (rB−, mB−), gal (DE3) | Laboratory collection |
| Plasmids | ||
| pGEM-T easy | PCR product cloning vector; Apr | Promega |
| pBS0909 | pGEM-T easy with intergenic region between nanTP and nanE; Apr | This study |
| pDM4 | Suicide vector; oriR6K; Cmr | 17 |
| pBS1013 | pDM4 with ΔnanR; Cmr | This study |
| pBS0907 | pDM4 with Δcrp ; Cmr | This study |
| pBS1102 | pDM4 with ΔnanE ; Cmr | This study |
| pHIS-Parallel1 | Protein expression vector; Apr | 22 |
| pBS0820 | pHIS-Parallel1 with nanR; Apr | This study |
| pHK0201 | pRSET A with crp; Apr | 23 |
| pSH0505 | pET28a with V. vulnificus rpoD, Kmr | 25 |
| pRLG770 | General transcription vector; Apr | 27 |
| pBS0921 | pRLG770 with PnanE; Apr | This study |
a The following abbreviations are used: Apr, ampicillin-resistant; Cmr, chloramphenicol-resistant; Kmr, kanamycin-resistant; Rifr, rifampicin-resistant.
Generation of nanR, nanE, nanR crp, and nanR nanA Mutants
V. vulnificus nanR on chromosome was amplified and inactivated in vitro by deletion of about two-thirds (550-bp of 837-bp) of the nanR ORF using the PCR-mediated linker-scanning mutation method as described elsewhere (16). Briefly, pairs of primers NANR0801F and NANR0801R (for amplification of the 5′ amplicon) or NANR0802F and NANR0802R (for amplification of the 3′ amplicon) were designed and used as listed in Table 2. The ΔnanR, a 550-bp deleted nanR, was amplified by PCR using the mixture of both amplicons as the template and NANR0801F and NANR0802R as primers. Similar experimental procedures were adopted for amplification of the Δcrp and ΔnanE in vitro, except that primers CRP0901F, CRP0901R, CRP0902F, and CRP0902R (for 284-bp deleted crp) and NANE1001F, NANE1001R, NANE1002F, and NANE1002R (for 481-bp deleted nanE) were used, respectively, as indicated in Table 2. The resulting ΔnanR, Δcrp, and ΔnanE were ligated with SphI-SalI digested pDM4 (17) to form pBS1013, pBS0907, and pBS1102, respectively (Table 1).
TABLE 2.
Oligonucleotides used in this study
| Name | Oligonucleotide sequence, 5′ 3′a | Locationb | Use |
|---|---|---|---|
| NANT0903 | GTTGCAGCATTCGCAGAAAG | nanTP-nanE intergenic region | Amplification of nan intergenic region, primer extension |
| NANE0905 | CTCTTAGTATCGTATTGCGCTATGG | nanTP-nanE intergenic region | Amplification of nan intergenic region, primer extension |
| NANR0801F | CTGCGCAACAAAAAGCCGATGTC | nanR flanking region | Mutant construction |
| NANR0801R | GCGGATCCGGTTTGGCTCAAATCAA | nanR | Mutant construction |
| NANR0802F | CCGGATCCGCAAGAAACAGCAGAAGTA | nanR | Mutant construction |
| NANR0802R | CGCTAACGCCATATGAGAGAGTAAAGAGG | nanR flanking region | Mutant construction |
| CRP0901F | TAGCTTGCAGCCTTAGTTAACAGC | crp flanking region | Mutant construction |
| CRP0901R | CTGGATCCTTGATTTAGGTAAGAAAGG | crp | Mutant construction |
| CRP0902F | AAGGATCCAGATCAAGATCACTCG | crp | Mutant construction |
| CRP0902R | CCCTGAAGGTTCGAGAATGC | crp-flanking region | Mutant construction |
| NANE1001F | AAATGTGATCGCGAACAGAAATTCGTC | nanE-flanking region | Mutant construction |
| NANE1001R | GACGACCTAAGCCAGTTATACGTTTTCAACGCCTTC | nanE | Mutant construction |
| NANE1002F | AACTGGCTTAGGTCGTCGTTTAATTCTGCAACACAAGC | nanE | Mutant construction |
| NANE1002R | GCCGCTTGAGCATCATTGAGC | nanE-flanking region | Mutant construction |
| NANR0809 | CCATGGGTTCGCCGAAAAATTTATTAG | nanR | NanR overexpression |
| NANR0810 | CTCGAGTTAATTTGCCATTAAAGGAATG | nanR | NanR overexpression |
| NANTP_qRT_F | GCGTGCCGAATGCGAAACC | nanTP | qRT-PCR |
| NANTP_qRT_R | GATTCTCTTGCCCGTCAACTGC | nanTP | qRT-PCR |
| NANE_qRT_F | TTCAGCCTGTTGTCGGTAGCC | nanE | qRT-PCR |
| NANE_qRT_R | CCTTCAATACGCAGTGCCTTAGC | nanE | qRT-PCR |
a Regions of oligonucleotides not complementary to corresponding genes are underlined.
b Location indicate where the nucleotides were hybridized.
To generate the nanR mutant BS0805 by homologous recombination, E. coli SM10 λ pir, tra (containing pBS1013) (18), was used as a conjugal donor to V. vulnificus ATCC29307. Similarly, E. coli SM10 λpir, tra containing either pBS1102, pBS0907, or pBS1013 was used as a conjugal donor in conjunction with one of the recipients ATCC29307, BS0805, and the nanA mutant HG0702 (13) to generate the nanE mutant BS1104, nanR crp mutant BS0902, and nanR nanA mutant BS1012 as indicated in Table 1. The conjugation and isolation of the transconjugants were conducted using the method described previously (19).
RNA Purification and Transcript Analysis
Total cellular RNA was isolated from the wild type and nan mutants grown to A600 of 0.6 with LBS and M9, supplemented with or without 5 mm Neu5Ac, using an RNeasy® mini kit (Qiagen) (20). For quantitative real time PCR (qRT-PCR), cDNA was synthesized with iScriptTM cDNA synthesis kit (Bio-Rad), and real time PCR amplification of the cDNA was performed with the specific primer pairs for each nan gene (Table 2). Relative expression levels of the nan transcripts were calculated by using the 16 S rRNA expression level as the internal reference for normalization as described previously (21).
To determine the transcription start site of the nanTPSLAR operon, an end-labeled 20-base primer NANT0903 (Table 2) complementary to the coding region of nanTP was added to the RNA and then extended with SuperScript II RNase H− reverse transcriptase (Invitrogen). The cDNA products were purified and resolved on a sequencing gel alongside sequencing ladders generated from pBS0909 (Table 1) with the same primer used for the primer extension. A transcription start site of the nanEK nagA operon was also determined using similar methods, except that primer NANE0905 was used in place of NANT0903 (Table 2). The primer extension products were visualized using a phosphorimage analyzer (BAS1500, Fuji Photo Film Co. Ltd., Tokyo, Japan).
Overexpression and Purification of V. vulnificus NanR and CRP
The nanR coding region was amplified by a PCR using the primers NANR0809 and NANR0810 (Table 2) and was then subcloned into a His6 tagging expression vector, pHIS-Parallel1 (22), to result in pBS0820 (Table 1). The His-tagged NanR protein was then expressed in E. coli BL21 (DE3) and purified by affinity chromatography according to the manufacturer's protocol (Qiagen). Similarly, the expression and purification of the His-tagged CRP were carried out using pHK0201, carrying the V. vulnificus crp gene, as described elsewhere (23).
Electrophoretic Mobility Shift Assay (EMSA) and DNase I Protection Assay
The 299-bp nanTP-nanE intergenic region, extending from residues −138 to +161 from the transcription start site of nanTPSLAR operon, was amplified by a PCR using 32P-labeled NANT0903 and unlabeled NANT0905 as the primers (Table 2). The labeled 299-bp DNA (4 nm) fragment was incubated with varying concentrations of purified His-tagged NanR for 30 min at 37 °C in a 20-μl reaction mixture containing 1× binding buffer (24) and 1 μg of poly(dI-dC) (Sigma). The protein-DNA binding reactions with CRP were the same as those with NanR, except that the CRP binding buffer containing 1 mm cAMP was used as a 1× binding buffer (23). Electrophoretic analysis of the DNA·protein complexes have already been described (23).
The same 299-bp intergenic region was labeled by PCR amplification using a combination of 32P-labeled and unlabeled primers, NANT0903 and NANE0905, and used for the DNase I protection assays. The binding of NanR or CRP to the labeled DNA was performed as described above and DNase I digestion of the DNA·protein complexes followed the procedures previously described (20). After precipitation with ethanol, the digested DNA products were resolved on a sequencing gel alongside sequencing ladders of pBS0909 generated using NANT0903 as the primer. The gels were visualized as described above for the primer extension analysis.
Purification of V. vulnificus RNA Polymerase (RNAP) Core Enzyme and RpoD
The V. vulnificus core RNAP was purified by an immunoaffinity chromatography using the polyol-responsive monoclonal antibody 8RB13 (NeoClone Biotechnology) as described previously (25, 26). The overexpression and purification of His-tagged RpoD were fulfilled using pSH0505, carrying the V. vulnificus rpoD gene, as described elsewhere (25). Purified RNAP core enzyme and RpoD proteins were dialyzed with the storage buffer (25), and equimolar amounts of the RNAP coreenzyme and RpoD were mixed on ice to generate the V. vulnificus RNAP holoenzyme before use.
In Vitro Transcription Assays
The 319-bp DNA fragment containing nanEK nagA promoter (PnanE) region, isolated by digestion of pBS0909 with EcoRI, was recloned into pRLG770 that carries the rrnB terminator (27). The resulting plasmid pBS0921 (Table 1), as confirmed by DNA sequencing, was used as a template DNA to determine the effects of NanR and CRP on the transcription of PnanE in vitro. Mixtures of the supercoiled pBS0921 DNA (0.4 nm) and NanR (or CRP with 1 mm cAMP), with or without Neu5Ac and its catabolic intermediates, at the concentrations indicated in the figure legends, were preincubated at 37 °C for 40 min in a reaction buffer described previously (28). The radiolabeled nucleotide (2 μCi of [α-32P]UTP) and cold nucleotides (20 μm UTP and 400 μm each ATP, CTP, and GTP) were added to the mixture, and transcription was initiated by adding the V. vulnificus RNAP holoenzyme at the concentration of 0.25 nm. After incubation at 30 °C for 15 min, transcription was terminated by the addition of an equal volume of stop solution (25), electrophoresed on 7 m urea, 5.5% polyacrylamide gels, and processed as described for primer extension analysis.
Size-exclusion Chromatography and Isothermal Titration Calorimetry (ITC)
Purified NanR protein and ManNAc-6P were mixed in 1:100 molar ratio on ice for 2 h and subjected to size-exclusion chromatography using a Superdex 200 column (GE Healthcare) installed on an ÄKTA purifier FPLC system at room temperature with a flow rate of 0.35 ml/min. Apo-NanR was compared with the complex on the Superdex 200 column under the same condition. Aldolase (158 kDa) was used as a size reference protein. For ITC analyses, NanR protein, Neu5Ac, ManNAc-6P, and GlcN-6P were reconstituted in 0.5 mm DTT, 300 mm NaCl, and 50 mm Tris (pH 8.0). The calorimetric assays were performed as described previously (29). A 29 mm ligand solution in an injection syringe was titrated against 0.29 mm NanR in a reaction cell.
Data Analyses
Averages and standard errors of the mean (S.E.) were calculated from at least three independent trials. All data were analyzed by the Student's t test with the SAS program (SAS software; SAS Institute Inc., Cary, NC). Significance of differences between experimental groups was accepted at a p value of < 0.05.
RESULTS
Identification of the V. vulnificus nan Cluster
Many coding regions were identified immediately upstream and downstream of nanA from the V. vulnificus genome sequence (GenBankTM accession numbers AE016795 and AE016796). The postulated V. vulnificus nan cluster consists of the genes encoding a putative transcription regulator NanR, Neu5Ac transporters NanTPSL, and proteins involved in catabolic degradation of Neu5Ac such as NanA, NanE, NanK, and NagA (Fig. 1B). The deduced amino acid sequences of the coding regions of the V. vulnificus nan cluster were 73–97% identical to those of the nan cluster from V. cholera (data not shown). The coding regions of the V. vulnificus nan cluster are organized in the same orientation as in the V. cholerae nan cluster (Fig. 1B, data not shown) (14). All of this information suggested that the products of the nanTPSL, nanA, nanE, nanK, and nagA genes of the V. vulnificus nan cluster, are indeed involved in the transport and catabolic degradation of Neu5Ac as are the products of the V. cholerae nan cluster.
V. vulnificus nan Cluster Consists of Two Divergently Transcribed Operons
To analyze the expression pattern of the genes of the V. vulnificus nan cluster at the transcriptional level, the presence of transcripts of the intergenic regions of nanR, nanA, nanTPSL, nanE, nanK, and nagA was examined using reverse transcription-PCR methods. The results revealed that nanTPSL, nanA, and nanR were transcribed as a transcriptional unit, and another transcriptional unit consists of nanE, nanK, and nagA (data not shown). Therefore, it appeared that the V. vulnificus nan cluster consists of two operons, nanTPSLAR and nanEK nagA, that are divergently transcribed as presented in Fig. 1B. The relative positions and transcriptional directions of the genes of the V. vulnificus nan cluster differ from those of the E. coli and H. influenzae nan clusters, which are relatively well characterized at the molecular level (data not shown) (8, 30–32). These differences in genetic organization imply that the mechanisms for the regulation of the V. vulnificus nan cluster could be different from those of the E. coli and H. influenzae nan clusters.
Promoter Sequences for the nanTPSLAR and nanEK nagA Operon Overlap
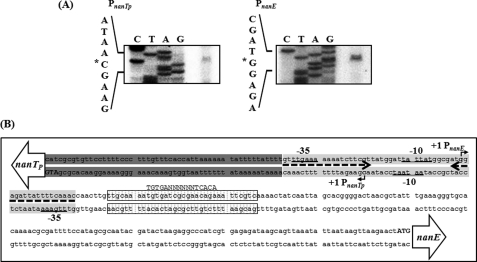
The transcription start sites of nanTPSLAR and nanEK nagA were determined by primer extension analyses. A reverse transcript was identified from each primer extension of the RNA isolated from the wild type cells (Fig. 2A). The 5′ end of the nanTPSLAR transcript is located 67 bp upstream of the translational initiation codon of nanTP and subsequently designated +1. The putative promoter constituting this transcription start site was named PnanTpto represent the nanTPSLAR promoter. The 5′ end of the nanEK nagA transcript is located 187 bp upstream of the start codon of nanE and is designated +1 of PnanE (the nanEK nagA promoter). Despite several attempts, no other transcription start sites were identified by primer extension analyses using different sets of primers hybridizing to the coding region of the nan cluster (data not shown). The sequences for −10 and −35 regions of both PnanTp and PnanE were assigned on the basis of similarity to consensus sequences of the E. coli σ70 promoter (Fig. 2B). It was noteworthy that the sequences for PnanTp and PnanE are partially overlapped, and four nucleotides of the −10 boxes are shared by each other (Fig. 2B).
FIGURE 2.
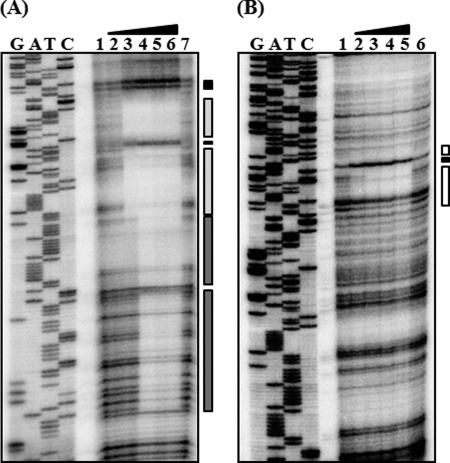
Transcription start sites of PnanTp and PnanE and sequence analysis of the regulatory region of the nan operons. A, TSSs, indicated by asterisks, were determined by primer extension of the RNA derived from the wild type (A600 0.6) grown with LBS. Lanes C, T, A, and G represent the nucleotide sequencing ladders of pBS0909. B, TSS of PnanTp and PnanE are indicated by bent arrows, and the positions of their putative −10 and −35 regions are underlined. The NanR- and CRP-binding sequences determined later in this study (Fig. 5) are presented as a shaded box and a dotted box, respectively. The conserved nucleotide sequences for the binding of CRP are indicated above the V. vulnificus DNA sequence in capital letters. The translation initiation codons (ATG in boldface) and coding regions of nanTP and nanE (open arrows) are also presented.
Construction of the nanR Mutants and Their Growth in Presence of Neu5Ac
To explore the role of NanR in the regulation of the V. vulnificus nan cluster, the nanR isogenic mutants were constructed by allelic exchanges (Table 1). Double crossovers, in which each wild type nanR on the chromosome was replaced with the ΔnanR allele, were confirmed using previously described methods (data not shown) (19). The growth of the nanR mutant BS0805 and its parental wild type was not significantly different in LBS (data not shown). However, the nanR mutant showed a significantly reduced lag phase (p < 0.05) compared with that of the wild type when cultured in M9 with Neu5Ac as a sole carbon source (supplemental Fig. S1A). Hence, disruption of nanR markedly increased growth on Neu5Ac, indicating that NanR functions as a repressor for the nan operons encoding proteins required for the transport and catabolism of Neu5Ac.
It has been known that the intracellularly accumulated Neu5Ac is toxic and inhibits the growth of E. coli (33). When Neu5Ac was added to the nanR mutant growing on glucose, growth of the mutant was not substantially inhibited, reflecting that Neu5Ac was not able to accumulate to the level of toxicity (supplemental Fig. S1B). This result can be explained if the genes for the catabolic degradation as well as transport of Neu5Ac were induced simultaneously by the nanR mutation, which provides further evidence for NanR acting as a repressor of both nan operons. Consistent with this assumption, growth of the nanR nanA double mutant, in which the NanA is inactivated and thereby catabolic degradation of Neu5Ac is blocked, was substantially inhibited by Neu5Ac (supplemental Fig. S1B). Growth inhibition by Neu5Ac was also dependent on the carbon source; larger inhibition zones by Neu5Ac were observed for the nanR nanA mutant growing on glycerol rather than glucose (supplemental Fig. S1B). This observation suggested that Neu5Ac accumulation was accelerated by growth on glycerol and that the expression of the nan genes, or at least nanTPSL, is subject to catabolite repression.
NanR Negatively Regulates Both the PnanTp and PnanE Activities
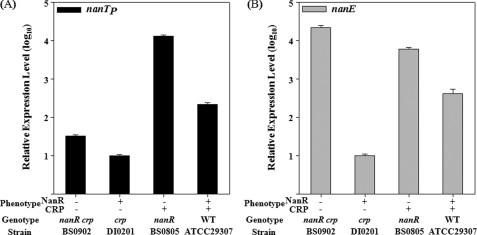
To examine the effect of NanR on PnanTp and PnanE, the transcripts of nanTP and nanE, the first genes of the nanTPSLAR and nanEK nagA operons, respectively, were quantitated in the same amount of total RNA isolated from the wild type and nanR mutant. Compared with the wild type, the nanR mutant revealed an almost 60-fold increase in nanTP expression and a 15-fold increase in nanE expression (Fig. 3, A and B). These results confirmed that both nan operons are under negative control of NanR and supported our previous observation that growth on Neu5Ac was accelerated by the mutation of nanR.
FIGURE 3.
Expression of nanTP and nanE in V. vulnificus with different genetic backgrounds. Cultures of the wild type, the nanR, crp, and nanR crp double mutants as indicated were grown with LBS, and samples removed at A600 of 0.6 were used to isolate total cellular RNA. The relative levels of nanTP (A) and nanE (B) expression were determined by qRT-PCR analyses and normalized to the 16 S rRNA expression level as presented the expression level of the crp mutants as 1 in log10. Error bars represent the S.E.
CRP Regulates the PnanTp and PnanE Activities Differentially
To investigate the role of CRP in the regulation of PnanTp, nanTP expression of the wild type and crp mutant was compared (Fig. 3A). Inactivation of crp resulted in reduction of nanTP expression, and the residual nanTP expression level of the crp mutant corresponded to only one-twentieth that of the wild type. This suggested that the CRP functions as an activator for PnanTp. This activation of PnanTp by CRP is consistent with our previous assumption that more Neu5Ac accumulates in the nanR nanA mutant growing on glycerol than on glucose (supplemental Fig. S1B).
Notably, nanE expression of the crp mutant decreased to only one-fortieth that of the wild type (Fig. 3B), and thus CRP appeared to elevate the PnanE activity. Despite this, CRP possibly stimulates nanE expression either directly by activating PnanE or indirectly by alleviating the NanR-mediated repression of PnanE. To distinguish these possibilities, the nanR crp double mutant was constructed, and nanE expression was quantitated (Table 1 and Fig. 3B). An additional inactivation of crp in the nanR mutant led to a 3.6-fold increase in nanE expression, indicating that CRP negatively regulates PnanE in the absence of NanR (Fig. 3B). Consequently, these results suggested that CRP activates PnanTp and represses PnanE and thereby regulates the nanTPSLAR and the nanEK nagA operons differentially.
NanR and CRP Function Cooperatively Rather than Sequentially to Regulate the nan Operons
The mutagenesis approaches implicated that both NanR and CRP played important roles in the regulation of the nan cluster. However, different mechanisms were still possible for the regulation of PnanTp by NanR and CRP. For example, CRP indirectly up-regulated the PnanTp activity by inhibiting nanR expression and thereby relieving the promoter from the negative control of NanR. To test this possibility, expression levels of nanTP of the nanR mutant and nanR crp double mutant were compared (Fig. 3A). An additional inactivation of crp in the nanR mutant resulted in about a 400-fold decrease in nanTP expression, indicating that the CRP activates PnanTp in the absence of NanR (Fig. 3A). Similarly, an additional mutation of nanR in the crp mutant led to a 3-fold increase in nanTP expression, indicating that NanR represses PnanTp in the absence of CRP (Fig. 3A). Consequently, it seems most likely that CRP and NanR regulate PnanTp cooperatively rather than sequentially in a regulatory cascade. However, it is also noteworthy that an additional mutation of nanR in the crp mutant led to an almost 2000-fold increase in nanE expression in the absence of CRP, whereas mutation of nanR resulted in only a 15-fold increase in the presence of CRP (Fig. 3B). This observation suggested that the repression of PnanE by NanR is affected by CRP.
NanR and CRP Specifically Bind to the PnanTp and PnanE Regulatory Region
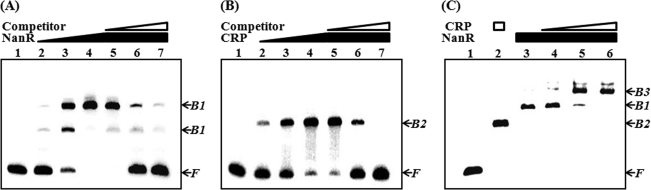
The 299-bp DNA fragment encompassing the nanTP-nanE intergenic region was incubated with increasing amounts of NanR and then subjected to electrophoresis. As seen in Fig. 4A, the addition of NanR resulted in a concentration-dependent ladder of two retarded bands, indicating that at least two binding sites with different affinities for NanR are present within the nanTP-nanE intergenic region. The binding of NanR was also specific, because assays were performed in the presence of 1 μg of poly(dI-dC) as a nonspecific competitor. In a second EMSA, the same but unlabeled 299-bp DNA fragment was used as a self-competitor to confirm the specific binding of NanR. The unlabeled 299-bp DNA competed for the binding of NanR in a dose-dependent manner (Fig. 4A, lanes 5–7), confirming that NanR binds specifically to the DNA. In similar DNA-binding assays, CRP also displayed specific binding to the nanTP-nanE intergenic region (Fig. 4B). These results indicated that NanR and CRP were able to bind to their operator within the nanTP-nanE intergenic region by themselves in the absence of the other in an independent manner. When both proteins were included in the binding reaction, they bound simultaneously to their operator and presented a slower moving band of DNA bound with both of the proteins as demonstrated by an EMSA in Fig. 4C.
FIGURE 4.
EMSA for binding of NanR and CRP to the nanTP-nanE intergenic region. A 299-bp DNA fragment of the nanTP-nanE intergenic region was radioactively labeled and used as a probe for DNA. The radiolabeled fragments (4 nm) were mixed with increasing amounts (0, 10, 50, and 100 nm in the lanes 1–4, respectively) of NanR (A) or CRP (B) and then resolved on a 5% polyacrylamide gel. For competition analysis, the same but unlabeled 299-bp DNA fragment was used as a self-competitor DNA. The labeled DNA probe was incubated with the self-competitor DNA (at 12.5, 37.5, and 75 nm, from lanes 5 to 7, respectively) prior to the addition of 200 nm NanR (A) or CRP (B). C, mixtures of 100 nm NanR and increasing amounts of CRP were added to the labeled DNA probe as indicated. Lane 1, no protein; lane 2, 200 nm CRP; lanes 3–6 (in the presence of 100 nm NanR), 0, 10, 50, 100 nm CRP, respectively. The positions of the unbound fragments (F), and the fragments retarded by NanR (B1), CRP (B2), or mixture of NanR and CRP (B3) are indicated by arrows.
Identification of Binding Sites for NanR and CRP Using DNase I Protection Analysis
When the sequences for the binding of NanR to the intergenic region of nanTP-nanE were mapped with NanR up to 100 nm, the NanR footprint extended from −37 to +18 relative to the transcription start site (TSS) of PnanTp (Figs. 5A and 2B). When increasing the NanR, another region extending from +70 to +19 was protected from DNase I digestion (Figs. 5A and 2B). This sequential protection with increasing NanR was consistent with the previous observation that at least two binding sites with different affinities for NanR are present in the intergenic region (Fig. 4A). The regions extending from −37 to +18 (centered at −10) and from +70 to +19 (centered at +44.5) were named NANRBI and NANRBII, to represent the NanR-binding sites I and II, respectively. Inspection of the sequences extending from −37 to +18 revealed a 19-bp (GTTTGAAAAAAATCTTCGT) inverted repeat. In contrast, the sequences from NANRBII, to which NanR bound with lower affinity, does not carry the 19-bp inverted repeat (Figs. 5A and 2B), indicating that the inverted repeat is important for NanR binding with higher affinity. Because the sequences of NANRBI overlap with the sequences of −10 and −35 regions of PnanE as well as PnanTp, bound NanR would be expected to prevent RNAP binding. This result supported our previous observation that NanR functions as a repressor for both promoters.
FIGURE 5.
Identification of binding sites for NanR and CRP. DNase I protection analysis of NanR (A) and CRP (B) binding to the intergenic region of nan operons. The 32P-labeled 299-bp DNA fragments were incubated with increasing amounts of NanR or CRP and then digested with DNase I. A, lanes 1 and 7, no NanR added; lanes 2–6, NanR at 50, 100, 200, 400, and 800 nm, respectively. B, lanes 1 and 6, no CRP added; lanes 2–5, CRP at 100, 200, 400, and 800 nm, respectively, with 1 mm cAMP. Lanes G, A, T, and C represent the nucleotide sequencing ladders of pBS0909. The regions protected by NanR are indicated by light (NANRBI) and dark (NANRBII) shaded boxes (A), and the regions protected by CRP are indicated by open boxes (B). The nucleotides showing enhanced cleavage are indicated by black boxes.
Similar DNase I protection assays were performed with CRP, and the DNase I footprinting revealed a clear protection pattern in the nanTP-nanE intergenic region extending from −45 to −76 (centered at −60.5) relative to TSS of PnanTp (Fig. 5B). The sequences of the protected region scored 87.5% similar to a consensus sequence for CRP binding (the TGTGAN6–8TCACA, see Ref. 34). This position for CRP binding indicated that the PnanTp is a class I CRP-dependent promoter. For class I CRP-dependent promoters, CRP-binding sites are centered at near integral turns of the helix (i.e. n × 10.5 bp) from TSS (35). These observations confirmed that CRP activates PnanTp directly, by binding to the upstream region of PnanTp. However, the CRP-binding site is located downstream of PnanE and centered at +40.5 from the TSS of PnanE. Therefore, from the standpoint of PnanE, CRP bound to the binding site could hinder RNAP movement and thereby repress its activity. This idea supported our earlier observation that CRP negatively regulates PnanE. Taken together, these results demonstrated that NanR and CRP control PnanTp and PnanE directly by binding to their respective operators independently.
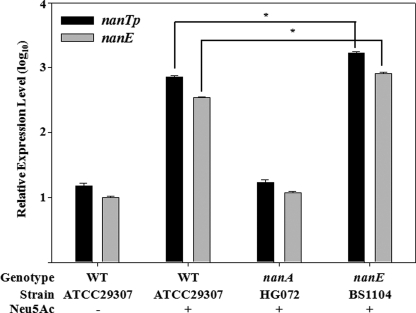
Neu5Ac Induction of the nan Operons Increased by the Mutation of nanE
Exogenous addition of Neu5Ac resulted in induction of the E. coli nanATEK operon and displaced NanR from its operator in vitro as demonstrated by EMSA (30). As shown in Fig. 6, addition of Neu5Ac to V. vulnificus growing on M9 with xylose and proline as sources of nutrient led to the induction of nanTP and nanE by 48- and 35-fold, respectively. However, it was still possible that intermediates of the Neu5Ac catabolic pathway, rather than Neu5Ac, influence induction of the nan genes. To test this possibility, the effects of either nanA or nanE mutation on the Neu5Ac-dependent induction of the nanTP and nanE were explored by the qRT-PCR analyses (Fig. 6). Neither nanTP nor nanE was induced by Neu5Ac in the nanA mutant, and their expression levels were indistinguishable from the wild type levels obtained in the absence of Neu5Ac (Fig. 6). Because Neu5Ac should accumulate in the nanA mutant, this result indicated that Neu5Ac is not the inducer for nanTP and nanE. In contrast, expression levels of both genes were significantly greater (p < 0.05) in the nanE mutant than in the wild type (Fig. 6). The enzyme converting ManNAc-6P to N-acetylglucosamine-6-phosphate (GlcNAc-6P) is lacking, and thus ManNAc or ManNAc-6P would be accumulated in the nanE mutant. These results implied that ManNAc or ManNAc-6P, the intermediates of Neu5Ac catabolism, could be an inducer for the induction of both nan operons.
FIGURE 6.
Effects of Neu5Ac on the nanTP and nanE expression of V. vulnificus with different genetic backgrounds. The wild type, the nanA, and nanE mutants were grown with M9 medium supplemented with 10 mm d-xylose and 10 mm l-proline in the presence or absence of Neu5Ac as indicated, and samples removed at A600 of 0.6 were used to isolate total cellular RNA. The relative levels of the nanTP and nanE transcripts were determined by qRT-PCR analyses and normalized to the 16 S rRNA expression level as presented the nanE transcript level of the wild type in the absence of Neu5Ac as 1 in log10. Error bars represent the S.E.
ManNAc-6P Specifically Alleviates the Repression by NanR
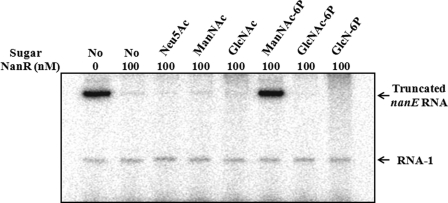
It was apparent that one of the Neu5Ac catabolic intermediates, rather than Neu5Ac, alleviates the NanR-mediated repression of the nan operons. To identify the inducer, effects of ManNAc and ManNAc-6P on the activity of PnanE were analyzed in the presence of purified NanR protein by in vitro transcription assays (Fig. 7). Consistent with our previous result, the PnanE activity was completely repressed in the presence of 100 nm NanR protein, and addition of Neu5Ac was not able to alleviate the NanR repression. In contrast, the addition of ManNAc-6P was able to relieve the NanR-mediated repression and induce the PnanE activity, indicating that ManNAc-6P is the inducer displacing the NanR from its operator. It is noteworthy that the addition of other catabolic intermediates of Neu5Ac, such as ManNAc, GlcNAc-6P, and glucosamine 6-phosphate (GlcN-6P), was unable to induce the PnanE activity in vitro at all (Fig. 7).
FIGURE 7.
Effects of Neu5Ac and its catabolic intermediates on the NanR repression of PnanE in vitro. Supercoiled plasmid pBS0921 containing the PnanE was used as a template DNA and transcribed in vitro in the presence of 100 nm NanR. Either Neu5Ac or one of its catabolic intermediates (1 mm) such as ManNAc, GlcNAc, ManNAc-6P, GlcNAc-6P, and GlcN-6P were added to the supercoiled template DNA as indicated. The 370-base PnanE-specific transcripts and vector-derived control transcripts RNA-1 are indicated.
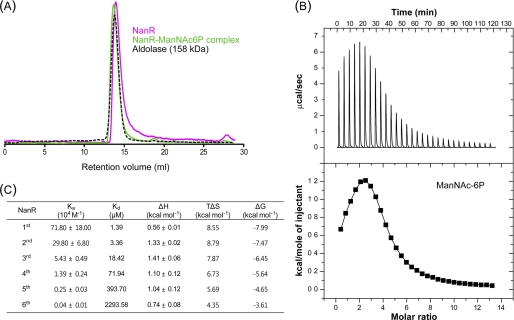
ManNAc-6P Interacts Directly with NanR
Size-exclusion chromatography showed that NanR exists mainly as a hexameric species in solution. The same oligomerization pattern was observed for the NanR·ManNAc-6P complex with a slight increase in molecular weight, suggesting that there may be a conformational change of NanR with the ligand (Fig. 8A). To further evaluate whether ManNAc-6P interacts directly with the NanR protein, ITC experiments were performed. The ITC results revealed that ManNAc-6P binds to NanR with a model of sequential binding to the hexameric NanR molecules with different binding affinities. The ligand binding affinities for the first to the sixth NanR molecules resulted in dissociation constants (Kd) ranging from 1.39 μm to 2.3 mm (Fig. 8, B and C). However, no interactions were observed between the NanR and other sugars such as Neu5Ac (supplemental Fig. S2A) and GlcN-6P (supplemental Fig. S2B). These results suggest that ManNAc-6P is a specific inducer to relieve NanR-mediated repression of the nan operons.
FIGURE 8.
ManNAc-6P binds specifically to NanR. A, NanR and ManNAc-6P complex exists as hexameric form in solution. Apo-NanR and complex are indicated by magenta and green, respectively. Aldolase (158 kDa) shown by the black dashed line was used as a size marker. B, typical isothermal titration calorimetric measurement of the interaction between the NanR protein and ManNAc-6P. The raw data are displayed in the upper panel, and integration plot of the data is displayed in the lower panel. C, thermodynamic parameters of the interaction between the NanR protein and ManNAc-6P. The representative ITC results were calculated using the best fit model (sequential binding sites model), and the heat data of ligand into the reaction buffer was subtracted from the reaction heat data between protein and ligand.
DISCUSSION
The diversity in the gene order and in the structure of nan clusters may be a common feature inherited among bacteria as proposed by Vimr et al. (8). Phylogenetic analyses indicated that the genes within the nan clusters show independent evolutionary histories (11). E. coli NanR and H. influenzae SiaR, major regulators of the nan clusters, belong to different families of transcription regulators. E. coli NanR and H. influenzae SiaR are members of the FadR/GntR and RpiR family, respectively (31). V. vulnificus NanR carries a highly conserved phosphosugar-binding (SIS) domain of the RpiR family (36), and thereby possibly falls into the RpiR family. However, the amino acid sequences of V. vulnificus NanR show low levels of identity (28%) to those of SiaR (data not shown). From this overall lack of similarity among NanR (SiaR) proteins, it is not surprising that the sialic acid catabolism of V. vulnificus is regulated through its own regulatory system that is different from that for E. coli and H. influenzae.
NanR and CRP negatively and positively regulate E. coli nan genes, where nanA and nanT encoding a sialic acid transporter and nanEK are transcribed in the same directions. Neu5Ac is an inducer and displaces NanR from its operators to induce the nan genes (30). In H. influenzae, CRP activates only the siaPT operon (encoding transporters), which is transcribed divergently from the nanEK siaR nagAB operon in the presence of cAMP (31, 32). In the absence of Neu5Ac, SiaR binds to the siaP-nanE intergenic region to repress both siaPT and nanEK siaR nagAB operons. It was surprising that SiaR changed from repressor to activator in the presence of GlcN-6P, a catabolic intermediate of Neu5Ac. The SiaR·GlcN-6P complex was presumed to bind to the siaPT-nanE intergenic region, with even greater affinity, to activate the siaPT and nan operons, and thus GlcN-6P is a coactivator rather than an inducer (32).
In this study, V. vulnificus NanR binds to the nanTP-nanE intergenic region to repress both nanTPSLAR and nanEK nagA operons in the absence of Neu5Ac. This negative feedback regulation of nanR could prevent overexpression of NanR and thus allow a basal level expression of transporters (NanTPSL), which is necessary for the initial uptake of Neu5Ac when the bacteria encounter Neu5Ac (Fig. 3 and supplemental Fig. S1). Increased induction of nanTP and nanE in the nanE mutant proposed that ManNAc-6P, rather than Neu5Ac, is an inducer for the nan cluster (Fig. 6). ManNAc-6P weakened binding of NanR to the nanTP-nanE intergenic DNA in vitro, whereas Neu5Ac and other catabolic intermediates of Neu5Ac had no effect on NanR binding to the DNA as determined by an EMSA (data not shown). Consistent with this, in vitro transcription assay revealed that only ManNAc-6P, not Neu5Ac and other Neu5Ac catabolic intermediates, was able to relieve the NanR-mediated repression of PnanE (Fig. 7). Furthermore, among the tested amino sugars, only ManNAc-6P specifically interacts with NanR (Fig. 8, B and C, and supplemental Fig. S2). These combined results confirmed that ManNAc-6P binds to NanR and induces the nan operons of V. vulnificus.
Many catabolic activities are typically regulated by the presence of the substrate acting as an inducer. V. vulnificus likely uses ManNAc-6P as an inducer because transported Neu5Ac can be toxic to cells unless it is utilized rapidly. If Neu5Ac is used as an inducer, transport of Neu5Ac into cells would occur even in the presence of a preferred sugar (e.g. glucose) in addition to Neu5Ac, which would result in toxic accumulation of the nonpreferred Neu5Ac, as opposed to catabolic degradation. In contrast, by using ManNAc-6P as an inducer, induction of the nan operons and high level Neu5Ac uptake would be initiated only when the preferred sugar is completely consumed, and catabolism of Neu5Ac has indeed progressed to yield a sufficient amount of ManNAc-6P.
It is noted that CRP regulates PnanTp and PnanE differentially (Fig. 3). The CRP-binding site is centered at −60.5 bp upstream from the transcription start site of nanTP, and this distance is typical for a class I CRP activation (Figs. 2B and Fig. 5B) (37). However, this CRP-binding site is centered +40.5 bp from the TSS of nanE, where the CRP represses PnanE, presumably by acting a roadblock for RNAP (38, 39). An in vitro transcription assay demonstrated that addition of CRP led to the reduction of the transcription from PnanE, confirming the CRP-dependent repression of PnanE (supplemental Fig. S3). For growth on Neu5Ac as a sole carbon source, a complete displacement of NanR from the operator is necessary for full induction of PnanTp and a strong expression of NanTPSL. Although PnanE is also induced by the lack of NanR on the operator, the induction would be rather moderate as a result of the CRP-dependent repression. The moderate (or adequate) expression of PnanE could keep the catabolic degradation rate of ManNAc-6P from outpacing the uptake rate of Neu5Ac, ensuring that a sufficient amount of the inducer is available in cells to maintain the full induction of PnanTp.
Interestingly, the PnanE activity in the wild type was higher than that of the crp mutant (Fig. 3B). This increased activity of PnanE by the presence of functional CRP might be the result of the position of the PnanTp −35 and −10 regions that entirely overlap the NanR-binding site required for repression of PnanE as well as PnanTp by NanR (Fig. 2B). As such, RNAP recruited by the CRP bound for activation of PnanTp could hinder the binding of NanR to its operator and eliminate the NanR-mediated repression of PnanE. The resulting inductive effect would be pronounced enough to mask the CRP-dependent roadblock effect and thereby CRP appeared to increase the PnanE activity.
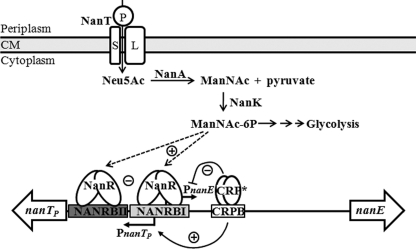
Fig. 9 summarizes our current understanding of the regulation of V. vulnificus nan cluster. When the bacteria are starved, RNAP recruited by CRP for activation of PnanTp could interfere with NanR binding to its operator and result in low but significant activity of PnanE as well as PnanTp. When a preferred sugar(s) is available, NanR alone, not CRP, could bind strongly to its operator and repress both nan operons tightly. When only Neu5Ac is available, ManNAc-6P displaces NanR from its operator, and additional activation by CRP renders PnanTp the strength of full induction. CRP also represses the PnanE as a roadblock to maintain a sufficient level of ManNAc-6P in cells, which is necessary for the full induction of PnanTp and efficient growth in this growth condition, presumably encountered in the host intestine.
FIGURE 9.
Proposed model for regulation of the V. vulnificus nan cluster by NanR, CRP, and ManNAc-6P. When the bacteria grow on Neu5Ac as a sole carbon source, a strong expression of PnanTp is necessary, which is possibly obtained by the class I activation by CRP in addition to ManNAc-6P-mediated displacement of NanR bound to its operator (NANRBI and MANRBII) overlapping the −35 and −10 region of PnanTp. PnanE is also induced by the lack of NanR on the operator; the induction would be rather moderate as a result of the CRP binding to its operator (CRPB) and thus acting as a roadblock for RNAP. The moderate expression of PnanE maintains ManNAc-6P sufficient for growth on Neu5Ac, which is probably encountered in the intestine. −, negative regulation; +, positive regulation; CM, cytoplasmic membrane.
Supplementary Material
Acknowledgments
We thank Dr. R. L. Gourse and members of his laboratory, University of Wisconsin, for advice with purifying V. vulnificus RNAP and performing in vitro transcription assays.
This work was supported by National Research Laboratory Grant R0A-2007-000-20039-0 through National Research Foundation funded by Ministry of Education, Science, and Technology (to S. H. C.) and by the Agriculture Research Center program of the Ministry for Food, Agriculture, Forestry and Fisheries, Republic of Korea.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- Neu5Ac
- N-acetylneuraminic acid
- CRP
- cAMP receptor protein
- ManNAc-6P
- N-acetylmannosamine 6-phosphate
- ManNAc
- N-acetylmannosamine
- qRT-PCR
- quantitative real time-PCR
- RNAP
- RNA polymerase
- TSS
- transcription start site
- GlcNAc-6P
- N-acetylglucosamine 6-phosphate
- GlcN-6P
- glucosamine 6-phosphate
- ITC
- isothermal titration calorimetry.
REFERENCES
- 1. Chang D. E., Smalley D. J., Tucker D. L., Leatham M. P., Norris W. E., Stevenson S. J., Anderson A. B., Grissom J. E., Laux D. C., Cohen P. S., Conway T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7427–7432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brown S. A., Palmer K. L., Whiteley M. (2008) Nat. Rev. Microbiol. 6, 657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hooper L. V., Midtvedt T., Gordon J. I. (2002) Annu. Rev. Nutr. 22, 283–307 [DOI] [PubMed] [Google Scholar]
- 4. Lång H., Jonson G., Holmgren J., Palva E. T. (1994) Infect. Immun. 62, 4781–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McGuckin M. A., Lindén S. K., Sutton P., Florin T. H. (2011) Nat. Rev. Microbiol. 9, 265–278 [DOI] [PubMed] [Google Scholar]
- 6. Larsson J. M., Karlsson H., Sjövall H., Hansson G. C. (2009) Glycobiology 19, 756–766 [DOI] [PubMed] [Google Scholar]
- 7. Wiggins R., Hicks S. J., Soothill P. W., Millar M. R., Corfield A. P. (2001) Sex Transm. Infect. 77, 402–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vimr E. R., Kalivoda K. A., Deszo E. L., Steenbergen S. M. (2004) Microbiol. Mol. Biol. Rev. 68, 132–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Severi E., Hood D. W., Thomas G. H. (2007) Microbiology 153, 2817–2822 [DOI] [PubMed] [Google Scholar]
- 10. Angata T., Varki A. (2002) Chem. Rev. 102, 439–469 [DOI] [PubMed] [Google Scholar]
- 11. Almagro-Moreno S., Boyd E. F. (2009) BMC Evol. Biol. 9, 118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jones M. K., Oliver J. D. (2009) Infect. Immun. 77, 1723–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jeong H. G., Oh M. H., Kim B. S., Lee M. Y., Han H. J., Choi S. H. (2009) Infect. Immun. 77, 3209–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Almagro-Moreno S., Boyd E. F. (2009) Infect. Immun. 77, 3807–3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Section A2.2, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 16. Lee J. H., Kim M. W., Kim B. S., Kim S. M., Lee B. C., Kim T. S., Choi S. H. (2007) J. Microbiol. 45, 146–152 [PubMed] [Google Scholar]
- 17. Milton D. L., O'Toole R., Horstedt P., Wolf-Watz H. (1996) J. Bacteriol. 178, 1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Miller V. L., Mekalanos J. J. (1988) J. Bacteriol. 170, 2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jeong H. S., Lee M. H., Lee K. H., Park S. J., Choi S. H. (2003) J. Biol. Chem. 278, 45072–45081 [DOI] [PubMed] [Google Scholar]
- 20. Lee J. H., Choi S. H. (2006) Mol. Microbiol. 60, 513–524 [DOI] [PubMed] [Google Scholar]
- 21. Oh M. H., Lee S. M., Lee D. H., Choi S. H. (2009) Infect. Immun. 77, 1208–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sheffield P., Garrard S., Derewenda Z. (1999) Protein Expr. Purif. 15, 34–39 [DOI] [PubMed] [Google Scholar]
- 23. Choi H. K., Park N. Y., Kim D. I., Chung H. J., Ryu S., Choi S. H. (2002) J. Biol. Chem. 277, 47292–47299 [DOI] [PubMed] [Google Scholar]
- 24. Rhee J. E., Jeong H. G., Lee J. H., Choi S. H. (2006) J. Bacteriol. 188, 6490–6497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee H. J., Park S. J., Choi S. H., Lee K. H. (2008) J. Biol. Chem. 283, 30438–30450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bergendahl V., Thompson N. E., Foley K. M., Olson B. M., Burgess R. R. (2003) Protein Expr. Purif. 31, 155–160 [DOI] [PubMed] [Google Scholar]
- 27. Ross W., Thompson J. F., Newlands J. T., Gourse R. L. (1990) EMBO J. 9, 3733–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Newlands J. T., Gaal T., Mecsas J., Gourse R. L. (1993) J. Bacteriol. 175, 661–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim Y., Kim B. S., Park Y. J., Choi W. C., Hwang J., Kang B. S., Oh T. K., Choi S. H., Kim M. H. (2010) J. Biol. Chem. 285, 14020–14030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kalivoda K. A., Steenbergen S. M., Vimr E. R., Plumbridge J. (2003) J. Bacteriol. 185, 4806–4815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Johnston J. W., Zaleski A., Allen S., Mootz J. M., Armbruster D., Gibson B. W., Apicella M. A., Munson R. S., Jr. (2007) Mol. Microbiol. 66, 26–39 [DOI] [PubMed] [Google Scholar]
- 32. Johnston J. W., Shamsulddin H., Miller A. F., Apicella M. A. (2010) BMC Microbiol. 10, 240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vimr E. R., Troy F. A. (1985) J. Bacteriol. 164, 845–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Botsford J. L., Harman J. G. (1992) Microbiol. Rev. 56, 100–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ebright R. H. (1993) Mol. Microbiol. 8, 797–802 [DOI] [PubMed] [Google Scholar]
- 36. Bateman A. (1999) Trends Biochem. Sci. 24, 94–95 [DOI] [PubMed] [Google Scholar]
- 37. Barnard A., Wolfe A., Busby S. (2004) Curr. Opin. Microbiol. 7, 102–108 [DOI] [PubMed] [Google Scholar]
- 38. Santangelo T. J., Artsimovitch I. (2011) Nat. Rev. Microbiol. 9, 319–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Toulmé F., Mosrin-Huaman C., Artsimovitch I., Rahmouni A. R. (2005) J. Mol. Biol. 351, 39–51 [DOI] [PubMed] [Google Scholar]
- 40. Park J. H., Cho Y. J., Chun J., Seok Y. J., Lee J. K., Kim K. S., Lee K. H., Park S. J., Choi S. H. (2011) J. Bacteriol. 193, 2062–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.