Background: CYLD is a deubiquitinating enzyme (DUB) that hydrolyzes Lys-63-linked polyubiquitin chains that are attached covalently to cellular proteins.
Results: CYLD knock-out mice have increased numbers of regulatory T cells (Tregs) in peripheral lymphoid organs but not in the thymus.
Conclusion: CYLD regulates lysine 63-linked ubiquitination of Smad7 to control the development of peripheral Tregs.
Significance: TGF-β signaling in T cells is regulated by lysine 63-Linked ubiquitination.
Keywords: Deubiquitination, Development, Signaling, Transforming Growth Factor Beta (TGFbeta), Ubiquitination, CYLD, Smad7, Tregs
Abstract
CYLD is a lysine 63-deubiquitinating enzyme that inhibits NF-κB and JNK signaling. Here, we show that CYLD knock-out mice have markedly increased numbers of regulatory T cells (Tregs) in peripheral lymphoid organs but not in the thymus. In vitro stimulation of CYLD-deficient naive T cells with anti-CD3/28 in the presence of TGF-β led to a marked increase in the number of Foxp3-expressing T cells when compared with stimulated naive control CD4+ cells. Under endogenous conditions, CYLD formed a complex with Smad7 that facilitated CYLD deubiquitination of Smad7 at lysine 360 and 374 residues. Moreover, this site-specific ubiquitination of Smad7 was required for activation of TAK1 and p38 kinases. Finally, knockdown of Smad7 or inhibition of p38 activity in primary T cells impaired Treg differentiation. Together, our results show that CYLD regulates TGF-β signaling function in T cells and the development of Tregs through deubiquitination of Smad7.
Introduction
Regulatory T (Treg)2 cells are essential for maintaining immune tolerance, limiting chronic inflammation, and preventing autoimmune diseases (1, 2). This specialized subpopulation of CD4+ T cells expresses the transcription factor forkhead box P3 (Foxp3) (3, 4). The importance of Foxp3-expressing Treg cells in immune system homeostasis is highlighted by the development of a profound autoimmune-like lymphoproliferative disease in Foxp3-deficient humans and mice (4, 5). Transforming growth factor-β (TGF-β) induces the expression of Foxp3 in CD4+ T cells and the development of Treg cells in peripheral lymphoid tissue (6, 7). Mice deficient in TGF-β die prematurely due to severe inflammation that appears in part to be a consequence of reduced Treg number and function (8). Although the roles of TGF-β and Foxp3 are well appreciated, the intracellular signaling mechanisms that control the peripheral development of Treg cells are still not fully understood.
In eukaryotic cells, the covalent attachment of ubiquitin to target proteins is important for many types of cellular events including protein degradation, DNA repair, cell cycle control, and receptor-mediated endocytotic cellular signaling (9, 10). More recently, the role of ubiquitination in regulating proximal signaling events is becoming better appreciated. Protein ubiquitination is reversible and highly regulated by ubiquitin ligases that catalyze the attachment of ubiquitin to target proteins (11, 12) and deubiquitinating enzymes that specifically cleave ubiquitin linkages (13).
Cylindromatosis (CYLD) is a deubiquitinating enzyme of the ovarian tumor family, which consists of proteases that disassemble Lys-63-linked polyubiquitin chains from various cellular proteins (14, 15). CYLD was first described as a tumor suppressor gene that is mutated in patients with familial cylindromatosis, a disease characterized by numerous tumors of hair follicles and sweat glands of the head and neck (16). Recent in vitro studies and the development of CYLD knock-out (Cyld−/−) mice have suggested an important role for CYLD in immune function and tumorigenesis (17, 18). CYLD mediates the hydrolysis of lysine 63-linked polyubiquitin chains from TNF receptor-associated factors (TRAFs), TGF-β activated kinase-1 (TAK1), and IKKγ, the regulatory subunit of the IKK complex (14, 19). As a consequence, CYLD has been implicated in the regulation of the NF-κB and JNK signaling pathways (20, 21).
In this study, we verify that CYLD-deficient mice exhibit increased numbers of CD4+CD25+ T cells in peripheral lymph nodes. In an analysis of the mechanism of this finding, we noted first that TGF-β-stimulated T cells lacking CYLD exhibit increased TAK1 and p38 mitogen-activated protein kinase (MAP kinase) activity. We then traced this finding to the fact that the absence of CYLD leads to excess ubiquitination and increased activity of Smad7, a TGF-β-induced signaling molecule that controls TAK1 kinase activity. Finally, we established that increased TAK1 activity facilitates downstream signaling events, such as binding of AP-1 to the Foxp3 promoter, that could explain enhanced Treg development. These studies thus define a new role for ubiquitination in Treg development and somewhat surprisingly show that the key target of such ubiquitination is Smad7.
EXPERIMENTAL PROCEDURES
Mice
CYLD-deficient mice (22) and wild type mice were bred in the mouse-specific pathogen-free animal facility at the NIAID, National Institutes of Health. All animal experiments were approved by the review board of the NIAID Animal Care and Use Committee.
Cell Lines, Plasmids, and Reagents
HeLa and HEK 293 cells were purchased from the American Type Culture Collection (Manassas, VA). Human Smad7 plasmids (FLAG-Smad7, GST-Smad7, GST-Smad7-N, and GST-Smad7-C) were obtained from Seong-Jin Kim (NCI, National Institutes of Health) (23). Smad7-K360R, Smad7-K368R, Smad7-K374R, and Smad7-K402R were generated by substituting the lysine residues at amino acid positions 360, 368, 374, and 402 with arginine using the QuikChange XL site-directed mutagenesis kit (Stratagene). The mutagenic primers are available upon request. CYLD and CYLD K870H were obtained from Dr. Guozhi Zhu (NCI, National Institutes of Health). pRK5-HA-tagged ubiquitin and pRK5-HA-tagged Lys-63-only ubiquitin were obtained from Ted Dawson (Johns Hopkins University, Baltimore, MD). Antibodies against Smad7, GST, HA, and ubiquitin were purchased from Santa Cruz Biotechnology. Anti-phospho-p38 (Thr-180/Tyr-182), anti-p38, and anti-TAK1 antibodies were obtained from Cell Signaling Technology. Anti-β-actin and anti-FLAG antibodies were purchased from Sigma; antibodies against CYLD, CD3, CD28, and CD25 were purchased from Pharmingen. Recombinant human TGF-β was obtained from R&D Systems. p38 inhibitor SB 203580 was purchased from Calbiochem.
Lymphocyte Preparation
Splenocytes, lymph node cells (five lymph nodes per mouse), or thymocytes were passed through a 70-μm nylon cell strainer (BD Falcon) to make a single cell suspension. Red blood cells were lysed with ACK lysis buffer (Lonza). One million cells were stained per condition, and 10,000 cells were analyzed by flow cytometry.
Lentiviral Transduction
Primary T cells and HeLa cells were transduced with SMARTvector shRNA lentiviral particles (Thermo Scientific) containing the Smad7 targeting sequences using Amaxa Nucleofector kit (Lonza) and following the manufacturer's protocol. Smad7-deficient HeLa cells stably expressing Smad7 shRNA were selected and maintained in medium containing puromycin (10 μg/ml, Invitrogen).
T Cell Isolation and Analysis
CD4+ T cells and CD4+CD25− T cells were purified with cell isolation kits from Miltenyi Biotec using protocols provided by the manufacturer. For in vitro differentiation, purified CD4+CD25− T cells were cultured in the presence of plate-bound anti-CD3 and anti-CD28 antibodies. Cells were cultured at an initial density of 5 × 105 cells/well, and at the end of the culture period, the cells were harvested, washed, stained with a Treg Foxp3 staining kit (e-Bioscience), and analyzed on a FACSCalibur (BD Biosciences). Treg suppression was performed using methods previously described (24). In brief, WT or CYLD−/−CD4+CD25− cells (5 × 104), CD4+CD25+ cells (5 × 104), or CD4+CD25− cells (5 × 104) were co-cultured with the indicated number of CD4+CD25+ cells (2.5 × 104) in 96-well plates (0.2 ml) in the presence of antigen-presenting cells (5 × 104) and 0.5 μg/ml anti-CD3 for 72 h/37 °C with 7% CO2. Cultures were pulsed with [3H]thymidine for the last 6 h of culture. All results are expressed as the mean cpm of triplicate cultures. The S.D. was always <10% of the mean.
Cell Transfection
HEK 293 cells or HeLa cells were transfected with the indicated expression vectors using Lipofectamine 2000 (Invitrogen). After 24 h, the cells were harvested and lysed in radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm PMSF, 1 mm EDTA, 1% Triton X-100, 1% sodium deoxycholate, and 0.1% SDS) with EDTA-free protease inhibitor mixture (Roche Applied Science).
Immunoprecipitation and Immunoblotting
Cells were lysed in lysis buffer (20 mm Tris, pH 7.5, 150 mm NaCl, 1% Triton X-100, 1 mm EDTA, 30 mm NaF, 2 mm sodium pyrophosphate, supplemented with protease inhibitor mixture (Roche Applied Science) and 1 mm N-ethylmaleimide). Lysates were normalized for protein concentration and denatured in sample buffer, and 25 μg of lysate were loaded in each well, resolved by SDS-PAGE, and immunoblotted with the appropriate antibodies. Immunoblots were visualized using horseradish peroxidase-conjugated secondary Ab and ECL (GE Biosciences). For immunoprecipitations, lysates were normalized for protein concentration and incubated with the appropriate Abs and protein A beads at 4 °C for 16 h. After extensive washing, the bead-bound complexes were eluted in sample buffer, resolved by SDS-PAGE, and immunoblotted with the appropriate monoclonal Abs. In some experiments, non-covalent protein-protein interactions were disrupted by heating the lysates at 95 °C in the presence of 1% SDS and diluted with lysis buffer to a final concentration of 0.1% SDS.
Reverse Transcription-Polymerase Chain Reaction
Total cellular RNA was extracted from cells using TRIZOL reagent (Invitrogen) according to the manufacturer's instructions. cDNAs were reverse-transcribed from 1 μg of total RNA in a 20-μl reaction using standard methods.
Electrophoretic Mobility Shift Assay (EMSA)
Whole-cell extracts were prepared using whole-cell lysis buffer as described above. The AP1 probe was designed to correspond to the sequence of the AP1-binding sites in the FoxP3 promoter. Binding reactions for AP-1 and SP-1 were performed using the Trans-AM kit (Active Motif) according to the manufacturer's instructions.
RESULTS
CYLD Deficiency Leads to Increased Development of Peripheral Treg Cells
To elucidate the physiological functions of CYLD, we previously generated and analyzed CYLD knock-out mice (22). We found that Cyld−/− mice were born healthy at normal Mendelian ratios and showed no overt phenotype at early stages of development. We did, however, observe inflammatory infiltration of numerous tissues and increased susceptibility to colitis-associated tumorigenesis in older Cyld−/− mice. This is in contrast to mice deficient in A20, another enzyme with an N-terminal ubiquitin hydrolase domain that also removes Lys-63-linked ubiquitin from NF-κB essential modulator (NEMO), TRAF2, and TRAF6. A20−/− mice fail to down-regulate TNF-α-induced NF-κB activity and die prematurely at 6 weeks of age from chronic inflammation and cellular death (25). Although A20 and CYLD target some of the same substrates in the NF-κB signaling pathway, the phenotypic and cellular differences between these two genetically altered mice suggest that CYLD may negatively regulate additional targets that prevent the development of severe inflammation.
The transcription factor Foxp3 is required for the development of Tregs and plays a vital role in their function to maintain immune cell homeostasis (4, 5). To investigate whether the absence of severe inflammation in the Cyld−/− mice could be attributed to differences in Tregs, we measured the expression of Foxp3 in T cells from the thymus and spleen of 6-week-old mice. We previously reported that there are no differences in thymic or splenic cellularity, or alterations in the lymphocyte subsets, between Cyld−/− and control mice (22). In this study, we show that the proportion of CD4+CD25+ Foxp3+ Tregs was significantly higher in the spleen of Cyld−/− mice when compared with control mice (Fig. 1, A and B), whereas the number of these cells in the thymus was not appreciably different between Cyld−/− and control mice (Fig. 1, C and D). Thus, the loss of CYLD does not seem to affect the development of central Treg cells but may have a previously unrecognized role in the differentiation of naive CD4+ T cells into Foxp3-expressing Treg cells in the periphery. Helios, as an Ikaros transcription factor, differentiates thymus-derived Fopx3+ Tregs from peripherally induced Foxp3+ Tregs. We further checked Helios expression in both thymocytes and splenic cells from CYLD-deficient mice and control mice. The results showed that Helios expression in thymus and spleen is comparable between wild type and CYLD-deficient mice (supplemental Fig. S1). These data indicated that increased Foxp3+ Tregs in CYLD-deficient spleen might be due to peripheral induction.
FIGURE 1.
The number of Tregs is increased in the spleen of CYLD-deficient mice. A, wild type and CYLD-deficient CD4+ cells from splenocytes were analyzed for Foxp3 expression by flow cytometry. APC, allophycocyanin; PE, phycoerythrin. B, graph showing the mean and distribution of CD4+ Foxp3+ splenic T cells from wild type and CYLD-deficient mice. Twelve mice per group were analyzed. **, p < 0.01. C and D, FACS staining of mesenteric lymph nodes (MLN) performed as in A and B. E and F, FACS staining of thymocytes performed as in A and B. *, p < 0.05. Error bars represent the mean ± S.D. p values were calculated by using Student's t test. G and H, proliferative and suppressive function of CYLD-deficient CD4+ T cells. Wild type or CYLD-deficient CD4+CD25− cells and CD4+CD25+ cells, or CD4+CD25− cells co-cultured with CD4+CD25+ cells, were stimulated with antigen-presenting cells and anti-CD3. Cultures were incubated for 72 h and pulsed with [3H]TdR for the last 6 h of culture. One representative experiment of two separate experiments is shown.
To investigate the capacity of Cyld−/− Treg cells to suppress the activation of control CD4+CD25− responder T cells, we isolated CD4+CD25− and CD4+CD25high cells from Cyld−/− and control mice. The suppressor and responder cells were co-cultured with control antigen-presenting cells and stimulated with anti-CD3. CYLD-deficient CD4+CD25+ T cells suppressed the in vitro proliferation of control CD4+CD25− T cells, measured by [3H]TdR incorporation, across a wide range of suppressor-to-effector ratios. In addition, control CD4+CD25+ T cells efficiently suppressed the proliferation of CYLD-deficient responder T cells (Fig. 1, E and F). Taken together, these findings suggest that although CYLD may have a role in the development of immune cell homeostasis, it does not affect the suppressor or effector capacity of T cells.
CYLD-deficient T Cells Have Increased Responsiveness to TGF-β
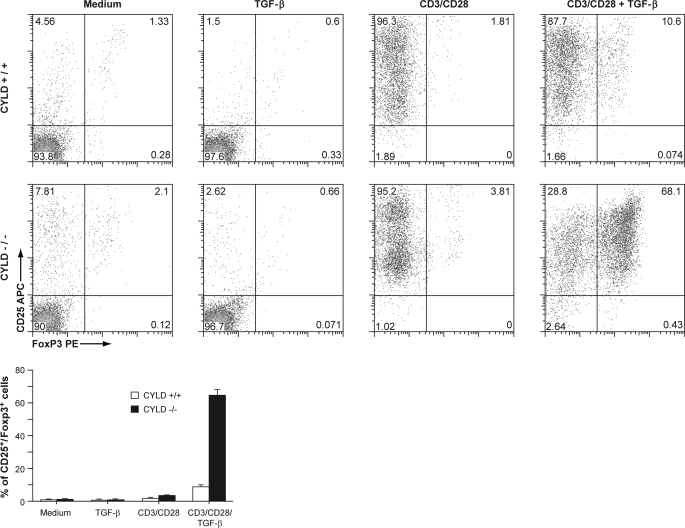
TGF-β plays an important role in the differentiation of naive CD4+ T cells into Tregs (6). To evaluate whether CYLD is involved in the development of Tregs, we isolated CD4+CD25− T cells and stimulated them in vitro with anti-CD3/28 and 10 ng/ml TGF-β in the absence of IL-2 for induction under suboptimal conditions. After 5 days of in vitro culture, we assessed the expression of Foxp3 and CD25 by flow cytometry. In the absence of either TGF-β or anti-CD3/28, most of the control and CYLD-deficient T cells remained Foxp3-negative (Fig. 2). However, when cultured in the presence of both anti-CD3/28 and TGF-β, nearly 70% of naive T cells deficient in CYLD expressed Foxp3, in contrast to control T cells, which remained predominantly Foxp3-negative. The lack of expression of Foxp3 in the control cells was not due to failed activation because 96% of control cells stimulated with anti-CD3/28 and TGF-β expressed CD25. In addition, neither the final concentration of cytokine IL-2 nor the cell number was appreciably different between activated control T cells and activated CYLD-deficient T cells. These results indicate that naive T cells deficient in CYLD have an increased capacity to differentiate into Foxp3-expressing cells following TGF-β stimulation.
FIGURE 2.
CYLD-deficient CD4 naive T cells demonstrate increased responsiveness to TGF-β in vitro. Purified wild type and CYLD-deficient CD4+CD25− T cells were cultured with TGF-β alone, in the presence of plate-bound anti-CD3/CD28 antibodies alone, or with TGF-β and anti-CD3/CD28 antibodies. At the end of the culture period, the cells were analyzed by flow cytometry. The experiment was repeated four times. Bottom panel, bar graph demonstrating the mean and distribution of CD25+ Foxp3+ T cells.
CYLD Regulates TGF-β Signaling
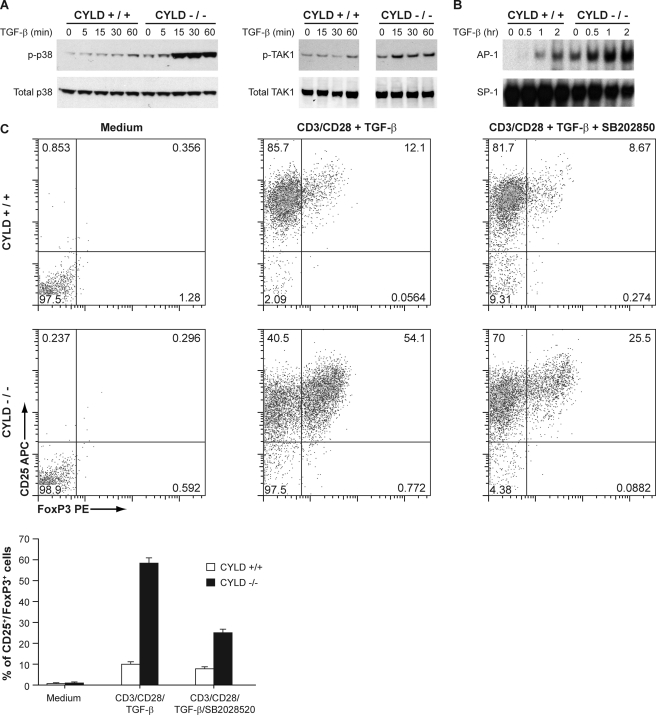
To further elucidate the mechanism by which CYLD negatively regulates the development of peripheral Tregs, we examined the effect of CYLD deficiency on TGF-β signaling events. Stimulation of naive wild type CD4+ T cells resulted in activation of TAK1 and p38 (Fig. 3A). Interestingly, the activity of both TAK1 and p38 was markedly enhanced in Cyld−/− cells. The basal Foxp3 promoter contains several AP-1-binding sites, and AP-1 has been implicated in the induction of Foxp3 (26). We therefore examined whether the loss of CYLD also promoted the activation of AP-1. Indeed, substantially higher levels of AP-1 activation, as determined by EMSA, were measured in the nuclear lysates prepared from TGF-β-stimulated Cyld−/− CD4+ T cells (Fig. 3B). These results suggest that CYLD regulates the sustained activation of TAK1, p38, and AP-1 as well as the induction of Foxp3 in CD4+ T cells and provide insight into the role of CYLD in regulating peripheral Tregs. SB 203580 is a pyridinyl imidazole that suppresses the activation of p38 and is useful for studying the physiological roles and targets of p38. Purified CD4+CD25− T cells were stimulated with anti-CD3/28 and 10 ng/ml TGF-β in the presence or absence of SB 203580 for 5 days, and the expression of Foxp3 and CD25 was assessed by flow cytometry. Inhibition of p38 impaired differentiation of both WT and Cyld−/− Tregs, but with a greater reduction of Cyld−/− Tregs (Fig. 3C).
FIGURE 3.
TAK1, p38, and AP-1 activities in response to TGF-β were enhanced in CYLD-deficient CD4+ T cells. A, cellular extracts prepared from TGF-β-stimulated splenic CD4+ T cells were analyzed by immunoblotting for the expression of phospho-TAK1 (p-TAK1) and phospho-p38 (p-p38). B, cellular extracts prepared from TGF-β-stimulated splenic CD4+ T cells were subjected to EMSA for AP-1 and SP-1 binding activity. C, purified wild type and CYLD-deficient CD4+CD25− T cells were cultured with medium alone, in the presence of plate-bound anti-CD3/CD28 antibodies with TGF-β, or in the presence of anti-CD3/CD28 antibodies with TGF-β and SB 203580. At the end of the culture period, the cells were analyzed by flow cytometry. The experiment was repeated three times. Bottom panel, bar graph demonstrating the mean and distribution of CD25+ Foxp3+ T cells.
Smad7 regulates TGF-β-induced TAK1 activity. It has recently been demonstrated that Smad7 can inhibit TNF-induced NF-κB activity by binding to the adaptors TAB2 and TAB3 and preventing TAK1 forming a complex with TRAF2 (23). We therefore addressed whether Smad7 can also modulate the activity of TAK1 in the TGF-β signaling pathway. We transduced primary CD4+ cells with a retroviral vector encoding Smad7-specific small hairpin RNA (shSmad7) (Fig. 4A). Following TGF-β stimulation, Smad7 knockdown cells demonstrated greatly reduced levels of phospho-TAK1, which was associated with highly reduced p38 phosphorylation (Fig. 4B). Similar findings were observed in HeLa cells transduced with a retroviral vector encoding Smad7-specific small hairpin RNA (shSmad7) (supplemental S2). These results indicate that Smad7 plays an important role in modulating TAK1 activity.
FIGURE 4.
Smad7-deficient primary T cells exhibit impaired TAK1 and p38 activities. A, RT-PCR analysis of Smad7 expression in primary CD4+ T cells transduced with non-targeted or Smad7 (S7)-targeted shRNA. B, immunoblot analysis of phospho-TAK1 (p-TAK1) and phospho-p38 (p-p38) levels in TGF-β-stimulated CD4+ non-targeted cells or TGF-β-stimulated CD4+ Smad7 knockdown cells. C and D, interaction of Smad7 and CYLD in CD4+ T cells was demonstrated by immunoprecipitation (IP) of endogenous Smad7 followed by immunoblotting (IB) with antibodies against CYLD (C) or the reverse immunoprecipitation of endogenous CYLD and immunoblotting with antibodies against Smad7 (D). Western blotting of the lysates (WB) showed that equal amounts of protein were used for immunoprecipitation.
We next examined whether CYLD and Smad7 can form a complex under endogenous conditions. Following TGF-β stimulation in WT primary CD4+ cells, interaction between Smad7 and CYLD was readily detected by co-immunoprecipitation using anti-Smad7 antibody and immunoblotting with anti-CYLD antibody. Similarly, CYLD-Smad7 interaction was clearly detected in the reverse immunoprecipitation with an anti-CYLD antibody followed by immunoblot analysis with anti-Smad7 antibody (Fig. 4, C and D). These results establish a physiological interaction between Smad7 and CYLD.
CYLD Regulates Smad7 Polyubiquitination
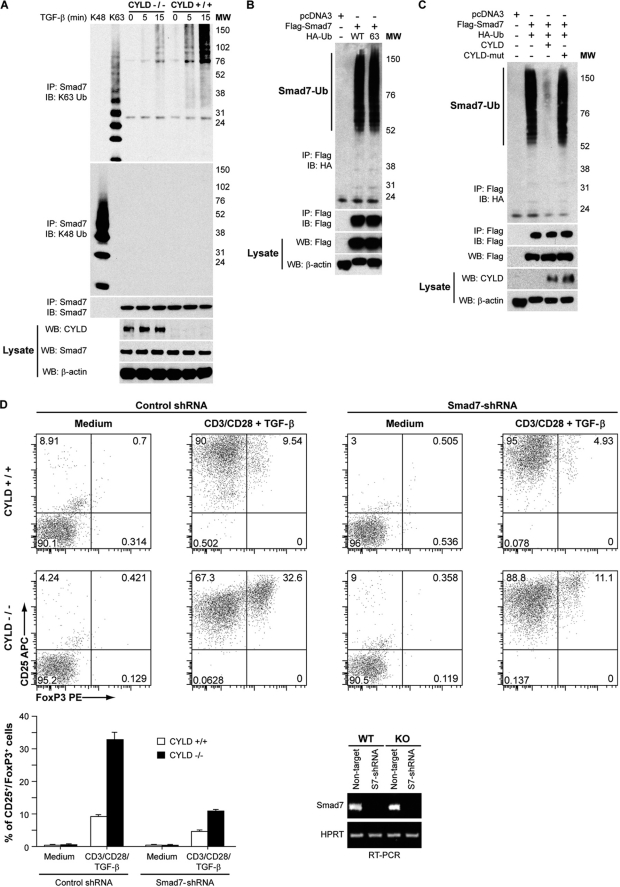
To further investigate the interaction between CYLD and Smad7, we analyzed the ubiquitination of endogenous Smad7 in TGF-β-stimulated T cells. The loss of CYLD resulted in a considerable elevation in lysine-63 (but not lysine-48)-ubiquitinated Smad7 in TGF-β-stimulated T cells (Fig. 5A), providing evidence for the involvement of CYLD in the modulation of Smad7 ubiquitination in vivo. We next overexpressed Smad7 in HEK 293 cells with ubiquitin mutant constructs possessing a lysine residue only at position 63 and found that Smad7 was lysine-63-ubiquitinated to a similar level with mutant and wild type ubiquitin (Fig. 5B). To determine whether Smad7 is a functional target of CYLD, we analyzed the direct effect of CYLD on Smad7 ubiquitination. Importantly, the Lys-63-linked polyubiquitination of transfected Smad7 was effectively inhibited by wild type CYLD, but not by mutant CYLD lacking enzyme activity (Fig. 5C). Diminished induction of Foxp3 was previously reported in Id3−/− CD4+ T cells with reduced levels of Smad7 (27). We transduced purified CD4+CD25− T cells with Smad7-specific small hairpin RNA (shSmad7) and stimulated the cells with anti-CD3/28 and 10 ng/ml TGF-β for 5 days. Treg cells were assessed by flow cytometric analysis of Fox3p expression. Knockdown of Smad7 expression impaired both WT and Cyld−/− Treg differentiation, but with a greater reduction of Cyld−/− Tregs (Fig. 5D).
FIGURE 5.
Smad7 ubiquitination in response to TGF-β is enhanced in CYLD-deficient cells. A, Smad7 polyubiquitination in TGF-β-stimulated splenic wild type or CYLD-deficient CD4+ T cells was analyzed by immunoprecipitation (IP) of the proteins from denatured cellular lysates followed by immunoblotting (IB) with an anti-Lys-63-linked ubiquitin Ab (K63 Ub) or anti-K48-linked ubiquitin Ab (K48 Ub). The amount of immunoprecipitated Smad7 was determined by immunoblotting with anti-Smad7. WB, Western blotting; MW, molecular weight markers. B, cell lysates from HEK 293 cells transfected with pcDNA3, FLAG-Smad7, HA-ubiquitin, or HA-tagged Lys-63-only ubiquitin as indicated were immunoprecipitated as in A. The amount of polyubiquitinated FLAG-Smad7 in the immunoprecipitate was determined by immunoblotting with anti-HA, and the amount of immunoprecipitated FLAG-Smad7 was determined by immunoblotting with anti-FLAG. FLAG-Smad7 and β-actin levels in the lysate were determined by Western blotting. C, HEK 293 cells were transfected with pcDNA3, FLAG-Smad7, HA-tagged wild type ubiquitin, CYLD, and CYLD-mut lacking enzymatic activity as indicated. The cells were harvested, and the lysates were immunoprecipitated as in A. The amount of polyubiquitinated FLAG-Smad7 in the immunoprecipitate was determined by immunoblotting with anti-HA, and the amount of immunoprecipitated FLAG-Smad7 was determined by immunoblotting with anti-FLAG. The amounts of FLAG-Smad7, CYLD, and β-actin in the lysate were determined by Western blotting. D, transduced wild type and CYLD-deficient CD4+CD25− T cells were cultured with medium alone or in the presence of plate-bound anti-CD3/CD28 antibodies with TGF-β. At the end of the culture period, the cells were analyzed for CD25 and Foxp3 expression by flow cytometry. The experiment was repeated three times. Bottom left panel, bar graph demonstrating the mean and distribution of CD25+ Foxp3+ T cells. Bottom right panel, RT-PCR analysis of Smad7 expression in both WT and Cyld−/− primary CD4+CD25− T cells transduced with non-targeted or Smad7 (S7)-targeted shRNA.
Smad7 Polyubiquitination Plays a Critical Role in TGF-β-induced TAK1 Activity and MAPK Signaling
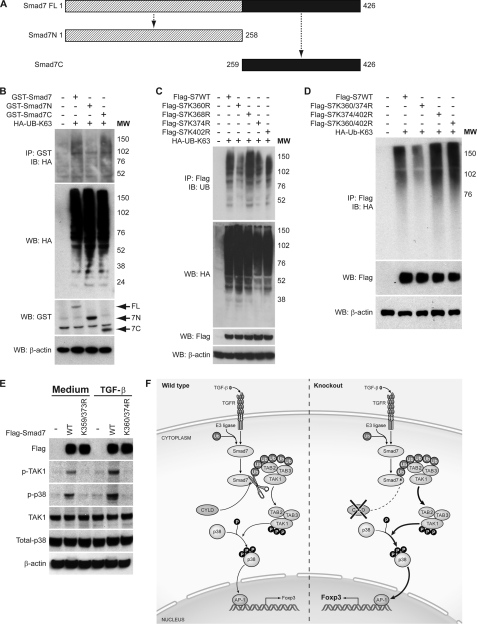
To localize the region within Smad7 that is the target of lysine-63 linked polyubiquitination, we analyzed ubiquitination of GST-tagged Smad7 deletion mutants by immunoprecipitation. A mutant Smad7 construct consisting of 258 amino acids from the N terminus was not polyubiquitinated (Fig. 6, A and B). In contrast, a deletion construction consisting of the remainder of the C terminus of Smad7 was lysine-63-ubiquitinated at levels comparable with wild type Smad7, indicating that the ubiquitination site of Smad7 is probably located between amino acids 259 and 426. There are four lysine residues in this C-terminal region of the protein; thus, one or more of these lysines are likely involved in Smad7 polyubiquitination. To test this, lysine-to-arginine substitutions were introduced into the FLAG-Smad7 constructs to generate mutants harboring Lys/Arg substitutions at these residues, both alone and in combination. Following co-transfection of HA-tagged Lys-63-linked polyubiquitin and FLAG-tagged Smad7 into HEK 293 cells, cellular lysates were probed with anti-FLAG antibody followed by immunoblot analysis with anti-HA antibody. Although all of the Smad7 single lysine mutants were polyubiquitinated, the Smad7-K360R and Smad7-K374R single lysine mutants were ubiquitinated at reduced levels when compared with wild type Smad7 (Fig. 6C). Furthermore, combined mutation of these two lysines (K360R/K374R) significantly decreased Smad7 polyubiquitination (Fig. 6D). These results identify Lys-360 and Lys-374 as targets of Smad7 polyubiquitination.
FIGURE 6.
Lys-63-linked polyubiquitination of Smad7 at its C terminus regulates TAK1 and p38 MAPK activities in response to TGF-β. A, schematic representation of the structure of wild type Smad7 and N- and C-terminal truncation mutants. B, cell lysates were prepared from HEK 293 cells transfected with GST-tagged Smad7, GST-tagged Smad7 deletion mutants (GST-Smad7N and GST-Smad7C), and HA-tagged Lys-63-only ubiquitin (HA-UB-K63) as indicated. The lysates were subjected to immunoprecipitation (IP) as in Fig. 5A. The amount of polyubiquitinated GST-Smad7 in the immunoprecipitate was determined by immunoblotting (IB) with anti-HA antibody. The amounts of ubiquitin, GST-Smad7, and β-actin in the lysate were determined by Western blotting (WB). MW, molecular weight markers. C, cell lysates from HEK 293 cells transfected with FLAG-Smad7, FLAG-Smad7-K360R (Flag-S7K360R), FLAG-Smad7-K368R (Flag-S7K368R), FLAG-Smad7-K374R (Flag-S7K374R), FLAG-Smad7-K402R (Flag-S7K402R), and HA-tagged Lys-63-only ubiquitin (HA-UB-K63) were subjected to immunoprecipitation as in Fig. 5A. The amount of polyubiquitinated FLAG-Smad7 in the immunoprecipitate was determined by immunoblotting with anti-HA. The amounts of ubiquitin, FLAG-Smad7, and β-actin in the lysate were determined by Western blotting. D, cell lysates from HEK 293 cells transfected with FLAG-Smad7, FLAG-Smad7-K360R/K374R (Flag-S7K360/374R), FLAG-Smad7-K374R/K402R (Flag-S7K374/402R), FLAG-Smad7-K360R/K402R (Flag-S7K360/402R), and HA-tagged Lys-63-only ubiquitin (HA-Ub-K63) as indicated were harvested and immunoprecipitated as in Fig. 5A. The amount of polyubiquitinated FLAG-Smad7 in the immunoprecipitate was determined by immunoblotting with anti-HA. FLAG-Smad7 and β-actin in the lysate were determined by Western blotting. E, HeLa Smad7 knockdown cells were reconstituted with FLAG-Smad7 or FLAG-Smad7-K360R/K374R (K360/374R). Following TGF-β stimulation, phospho-TAK1 (p-TAK1) and phospho-p38 (p-p38) levels were determined by immunoblotting. K359/373R, K359R/K373R. F, proposed model of the role of CYLD in TGF-β signaling and Treg development. TGFR, TGF receptor; Ub, ubiquitin; P, phosphorylated.
To understand the functional significance of Smad7 polyubiquitination, we next determined whether ubiquitination at Lys-360 and Lys-374 affected the phosphorylation of TAK1 and its downstream target p38. These experiments were performed using HeLa-shSmad7 cells reconstituted with RNA-resistant wild type Smad7 or a Smad7-K360R/K374R mutant (Fig. 6E). Endogenous phospho-TAK1 and phospho-p38 were readily detected in HeLa cells reconstituted with wild type Smad7. Furthermore, following TGF-β stimulation of these cells, the level of endogenous phospho-TAK1 and phospho-p38 increased. In contrast, phosphorylation of TAK1 and p38 was not detected in HeLa cells reconstituted with the ubiquitination-defective Smad7 mutant (Fig. 6E, lanes 3 and 5). The differences in TAK1 and p38 phosphorylation were not due to alterations in the overall levels of Smad7 (Fig. 6E, upper panel); therefore, these findings indicate that Smad7 ubiquitination is required for TGF-β-induced TAK1 activity.
DISCUSSION
The deubiquitinating protein CYLD has previously been shown to be a key negative regulator of the NF-κB and JNK signaling pathways following stimulation by a variety of immune receptors (18, 21, 22). In this study, we present evidence that CYLD also plays an important role in TGF-β signaling, and in doing so, up-regulates the development of peripheral (induced) Foxp3-expressing Tregs. These studies thus provide a mechanistic explanation of the previous observation by Lee et al. (28) that CYLD−/− mice manifest increased numbers of Foxp3+ CD4+ T cells. However, in the study by Lee et al. (28), both peripheral and thymic compartments contained increased Tregs, whereas in the present study, the increased Tregs were noted mainly in the peripheral compartment. The reason for this discrepancy is unclear. However, under the influence of TGF-β, one would expect that CD4+CD25− T cells convert into induced Treg at peripheral sites. Furthermore, the numbers of peripheral CD4+CD25+ T cells expressing Helios were comparable between CYLD−/− and control mice. Helios has previously been reported to be a specific marker of thymus-derived Tregs (26); therefore, the increased numbers of Tregs in the peripheral lymphoid tissue of CYLD−/− mice are not simply due to the expansion of thymus-derived Tregs. Also of interest, Lee et al. (28) found that although CYLD−/− CARMA1−/− double KO mice retained enhanced NF-κB function characteristic of CYLD−/− mice, they exhibited a marked decrease in Treg T cells. This indicates that the signaling pathway mediated by CARMA1 affecting Treg development is independent of the CYLD-dependent pathway explored here.
To better understand the role of CYLD on Treg development, we focused on TAK1, recognizing that the presence of TAK1 is necessary for the induction of Foxp3+ regulatory T cells following TGF-β signaling (29, 30). We found that TGF-β activation of TAK1 and p38 in T cell receptor-stimulated CD4+ T cells is markedly enhanced in cells deficient in CYLD. Moreover, Cyld−/− cells exhibited increased activation of AP-1, and in previous studies, increased activation of NF-κB, factors implicated in the induction of Foxp3. Taken together, these findings indicated that CYLD negatively regulates the activation of TAK1, p38, and AP-1.
Further studies to elucidate the mechanism of CYLD regulation of the TAK1 effect on Treg development was based on the fact that TGF-β signaling has been shown to recruit TAB2 and TAB3 to TGF-β receptor 1 via TRAF6 (31). This links TGF-β signaling to TAK1 and the TGF-β signaling protein, Smad7. Smad7 binds to TAB2 and TAB3 at their carboxyl termini, which contain ubiquitin-binding domains (23). Exploring these facts, we showed that TGF-β induced Lys-63-linked polyubiquitination of Smad7 and that this step is necessary for TAK1 phosphorylation as well as for downstream p38 and AP1 activation. Furthermore, we showed that Lys-63-linked polyubiquitination of Smad7 is required for TGF-β-induced TAK1 activity because phosphorylation of TAK1 and p38 was not observed following TGF-β stimulation of HeLa-shSmad7 cells reconstituted with mutant Smad7-K360R/K374R. Finally, we found that Smad7 is Lys-63-polyubiquitinated in both T cells and HeLa cells following TGF-β stimulation and that CYLD binds to Smad7 in T cells under endogenous conditions to modulate the polyubiquitination of Smad7. On the basis of these data, we propose that that CYLD deficiency leads to the accumulation of Lys-63-linked ubiquitinated Smad7 in TGF-β-stimulated T cells, which results in ubiquitin-dependent recruitment and activation of TAK1 (Fig. 6F). This hypothesis is supported by a previous study in T cells showing that CYLD physically interacts with TAK1 and inhibits its ubiquitination and catalytic activity following T cell receptor stimulation (19).
The A20 protein shares the ability to remove Lys-63-linked ubiquitin molecules from target proteins such as TRAF6, TRAF2, IKKγ, and TAK1; however, in contrast to mice deficient in CYLD, A20−/− mice develop severe inflammation and cachexia and die prematurely. The fact that this does not occur in CYLD-deficient mice could be attributed to the fact that enhanced TGF-β signaling and increased numbers of peripheral Tregs may limit inflammation in these mice. In addition, the phenotypic differences between A20- and CYLD-deficient mice can also be explained by the fact that CYLD is a specific Lys-63 polyubiquitin hydrolase, whereas A20 displays Lys-48 polyubiquitin E3 ligase activity for the protein RIP in addition to its Lys-63-linked deubiquitination function (32). TNF-α stimulation of A20-deficient cells leads to failure to terminate TNFα-induced NF-κB responses and increased susceptibility to TNFα-mediated apoptosis. Thus, despite sharing common protein targets for Lys-63-linked deubiquitination, the E3 ligase function of A20 leads to specific regulation of the TNF-α signaling pathway.
The precise downstream mechanism by which TAK1 signaling affects Foxp3 expression remains to be fully elucidated. One possibility is that, as shown here, TAK1 activation leads to activation of AP-1, a known Foxp3 transcription factor. However, this is not likely to be the complete story as IL-2 signaling also leads to activation of AP-1 (33). TAK1 phosphorylation has previously been shown to activate the IKK complex. Although we did not notice increased NF-κB activity in CYLD−/− T cells following TGF-β stimulation in a 1-h time course experiment, it is possible that such activity could be seen at later time points, thereby leading to transcriptional activity of c-Rel and p65, which have been shown to be involved in Foxp3 induction.
Previous studies have shown that Smad7 binds to Smurf2 to form an E3 ubiquitin-ligase complex in the nucleus that is exported into the cytosolic compartment to target the TGF-β receptor for lysine 48-linked polyubiquitination and degradation (34). This suggests that the ubiquitination status of Smad7 likely determines its precise role in the TGF-β signaling pathway. Our findings indicate that Lys-63-linked ubiquitin conjugation of Smad7 is important for TGF-β-induced TAK1 kinase activity in T cells and the development of Tregs.
Supplementary Material
Acknowledgments
We thank Warren Strober and Jonathan Ashwell for helpful discussions.
This work was supported by the Intramural Research Program of the National Institutes of Health through the NIAID.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- Treg
- regulatory T
- TRAF
- TNF receptor-associated factor
- TAK1
- TGF-β activated kinase-1
- IKK
- IκB kinase
- Ab
- antibody.
REFERENCES
- 1. Sakaguchi S., Yamaguchi T., Nomura T., Ono M. (2008) Cell 133, 775–787 [DOI] [PubMed] [Google Scholar]
- 2. Sather B. D., Treuting P., Perdue N., Miazgowicz M., Fontenot J. D., Rudensky A. Y., Campbell D. J. (2007) J. Exp. Med. 204, 1335–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hori S., Nomura T., Sakaguchi S. (2003) Science 299, 1057–1061 12522256 [Google Scholar]
- 4. Fontenot J. D., Gavin M. A., Rudensky A. Y. (2003) Nat. Immunol. 4, 330–336 [DOI] [PubMed] [Google Scholar]
- 5. Khattri R., Cox T., Yasayko S. A., Ramsdell F. (2003) Nat. Immunol. 4, 337–342 [DOI] [PubMed] [Google Scholar]
- 6. Chen W., Jin W., Hardegen N., Lei K. J., Li L., Marinos N., McGrady G., Wahl S. M. (2003) J. Exp. Med. 198, 1875–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cobbold S. P., Castejon R., Adams E., Zelenika D., Graca L., Humm S., Waldmann H. (2004) J. Immunol. 172, 6003–6010 [DOI] [PubMed] [Google Scholar]
- 8. Prud'homme G. J., Piccirillo C. A. (2000) J. Autoimmun 14, 23–42 [DOI] [PubMed] [Google Scholar]
- 9. Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 10. Mukhopadhyay D., Riezman H. (2007) Science 315, 201–205 [DOI] [PubMed] [Google Scholar]
- 11. Jentsch S., Seufert W., Sommer T., Reins H. A. (1990) Trends Biochem. Sci. 15, 195–198 [DOI] [PubMed] [Google Scholar]
- 12. Tanaka K., Suzuki T., Chiba T. (1998) Mol. Cells 8, 503–512 [PubMed] [Google Scholar]
- 13. Nijman S. M., Luna-Vargas M. P., Velds A., Brummelkamp T. R., Dirac A. M., Sixma T. K., Bernards R. (2005) Cell 123, 773–786 [DOI] [PubMed] [Google Scholar]
- 14. Kovalenko A., Chable-Bessia C., Cantarella G., Israël A., Wallach D., Courtois G. (2003) Nature 424, 801–805 [DOI] [PubMed] [Google Scholar]
- 15. Trompouki E., Hatzivassiliou E., Tsichritzis T., Farmer H., Ashworth A., Mosialos G. (2003) Nature 424, 793–796 [DOI] [PubMed] [Google Scholar]
- 16. Bignell G. R., Warren W., Seal S., Takahashi M., Rapley E., Barfoot R., Green H., Brown C., Biggs P. J., Lakhani S. R., Jones C., Hansen J., Blair E., Hofmann B., Siebert R., Turner G., Evans D. G., Schrander-Stumpel C., Beemer F. A., van Den Ouweland A., Halley D., Delpech B., Cleveland M. G., Leigh I., Leisti J., Rasmussen S. (2000) Nat. Genet. 25, 160–165 [DOI] [PubMed] [Google Scholar]
- 17. Glittenberg M., Ligoxygakis P. (2007) Fly 1, 330–332 [DOI] [PubMed] [Google Scholar]
- 18. Sun S. C. (2008) Nat. Rev. Immunol. 8, 501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reiley W. W., Jin W., Lee A. J., Wright A., Wu X., Tewalt E. F., Leonard T. O., Norbury C. C., Fitzpatrick L., Zhang M., Sun S. C. (2007) J. Exp. Med. 204, 1475–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun S. C. (2010) Cell Death Differ. 17, 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reiley W., Zhang M., Sun S. C. (2004) J. Biol. Chem. 279, 55161–55167 [DOI] [PubMed] [Google Scholar]
- 22. Zhang J., Stirling B., Temmerman S. T., Ma C. A., Fuss I. J., Derry J. M., Jain A. (2006) J. Clin. Invest. 116, 3042–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hong S., Lim S., Li A. G., Lee C., Lee Y. S., Lee E. K., Park S. H., Wang X. J., Kim S. J. (2007) Nat. Immunol. 8, 504–513 [DOI] [PubMed] [Google Scholar]
- 24. Thornton A. M., Piccirillo C. A., Shevach E. M. (2004) Eur. J. Immunol. 34, 366–376 [DOI] [PubMed] [Google Scholar]
- 25. Boone D. L., Turer E. E., Lee E. G., Ahmad R. C., Wheeler M. T., Tsui C., Hurley P., Chien M., Chai S., Hitotsumatsu O., McNally E., Pickart C., Ma A. (2004) Nat. Immunol. 5, 1052–1060 [DOI] [PubMed] [Google Scholar]
- 26. Mantel P. Y., Ouaked N., Rückert B., Karagiannidis C., Welz R., Blaser K., Schmidt-Weber C. B. (2006) J. Immunol. 176, 3593–3602 [DOI] [PubMed] [Google Scholar]
- 27. Maruyama T., Li J., Vaque J. P., Konkel J. E., Wang W., Zhang B., Zhang P., Zamarron B. F., Yu D., Wu Y., Zhuang Y., Gutkind J. S., Chen W. (2011) Nat. Immunol. 12, 86–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee A. J., Wu X., Cheng H., Zhou X., Cheng X., Sun S. C. (2010) J. Biol. Chem. 285, 15696–15703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wan Y. Y., Chi H., Xie M., Schneider M. D., Flavell R. A. (2006) Nat. Immunol. 7, 851–858 [DOI] [PubMed] [Google Scholar]
- 30. Juszczynski P., Ouyang J., Monti S., Rodig S. J., Takeyama K., Abramson J., Chen W., Kutok J. L., Rabinovich G. A., Shipp M. A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13134–13139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yamashita M., Fatyol K., Jin C., Wang X., Liu Z., Zhang Y. E. (2008) Mol. Cell 31, 918–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wertz I. E., O'Rourke K. M., Zhou H., Eby M., Aravind L., Seshagiri S., Wu P., Wiesmann C., Baker R., Boone D. L., Ma A., Koonin E. V., Dixit V. M. (2004) Nature 430, 694–699 [DOI] [PubMed] [Google Scholar]
- 33. Wu Y., Borde M., Heissmeyer V., Feuerer M., Lapan A. D., Stroud J. C., Bates D. L., Guo L., Han A., Ziegler S. F., Mathis D., Benoist C., Chen L., Rao A. (2006) Cell 126, 375–387 [DOI] [PubMed] [Google Scholar]
- 34. Kavsak P., Rasmussen R. K., Causing C. G., Bonni S., Zhu H., Thomsen G. H., Wrana J. L. (2000) Mol. Cell 6, 1365–1375 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.