Background: Post-translational modulation of preexisting translational factors is a well-established cardiac hypertrophic mechanism.
Results: The abundance of NFATc4, Nkx2.5 and GATA4 is up-regulated by newly-expressed protein kinase D3 (PKD3) to induce pathological cardiac hypertrophy.
Conclusion: PKD3 is a pivotal mediator of cardiac hypertrophic signaling cascade.
Significance: PKD3 is a potential new drug target for pathological cardiac hypertrophy.
Keywords: Calcineurin, Cardiac Hypertrophy, Histone Deacetylase, NFAT Transcription Factor, Protein Kinase D (PKD)
Abstract
Fetal cardiac gene reactivation is a hallmark of pathological cardiac hypertrophy (PCH) driven by cardiac transcription factors (TFs) such as nuclear factor of activated T-cells (NFATs). Nuclear import of dephosphorylated NFATs catalyzed by calcineurin (CaN) is a well-established hypertrophic mechanism. Here we report that NFATc4 expression is also up-regulated by newly expressed protein kinase D3 (PKD3) to induce PCH. In both in vitro and in vivo cardiac hypertrophic models, the normally undetectable PKD3 was profoundly up-regulated by isoproterenol followed by overt expression of cardiac TFs including NFATc4, NK family of transcription factor 2.5 (Nkx2.5), GATA4 and myocyte enhancer factor 2 (MEF2). Using gene silencing approaches, we demonstrate PKD3 is required for increasing the expression of NFATc4, Nkx2.5, and GATA4 while PKD1 is required for the increase in MEF2D expression. Upstream induction of PKD3 is driven by nuclear entry of CaN-activated NFATc1 and c3 but not c4. Therefore, PKD3 is a pivotal mediator of the CaN-NFATc1/c3-PKD3-NFATc4 hypertrophic signaling cascade and a potential new drug target for the PCH.
Introduction
Pathological cardiac hypertrophy (PCH)3 is a major predictor of human cardiovascular morbidity and mortality (1) and partly mediated by fetal cardiac gene reactivation in response to diverse stimuli including persistent hypertension, myocardial infarction, valvular heart disease, and diabetes (2–4). Reactivation of fetal cardiac genes such as atrial natriuretic factor (ANF) and β-myosin heavy chain (βMHC) is driven by a number of hypertrophic transcription factors (TFs). Some of the major TFs identified in animal and human hypertrophied hearts are myocyte enhancer factor 2 (MEF2), NK family of transcription factor 2.5 (Nkx2.5), GATA4, nuclear factor of activated T-cells (NFAT) (5), forkhead box (FOXO), serum response factor (SRF), and Ca2+/cAMP response element-binding protein (CREB) (6–8). Studies on the role of these TFs in PCH have mostly focused on post-translational modulation of pre-existing TFs in heart cells. Alterations in phosphorylation, acetylation, and redox status of many of these TFs have been extensively studied in PCH (9, 10).
Most TFs do not become activated until they form a transcription activation complex (TAC) with other coactivators (11, 12). When incoming transcription stimuli cease, the TAC becomes degraded, disassembled or can form co-repressor complexes (CoRC) with various histone deacetylases (HDAC) (13) or other co-repressors such as transducer of regulated CREB-binding protein (TORC). In cardiomyocytes undergoing hypertrophy, some TFs tend to work in close association, suggesting formation of super-TACs. For instance, a TF triad comprising Nkx2.5, GATA4 and SRF was identified in cardiac myocytes as well as smooth muscle cells (6). Upon activation, NFATc4 can further recruit GATA4 and MEF2 (14), and likely forms a super-TAC. Examples of cardiac CoRCs include one formed by MEF2, HDAC5 and MEF2-interacting transcriptional repressor (MITR) (15), and another formed by Nkx2.5, HDAC5 and CAMTA2 (16). The presence of TACs, super-TACs and CoRCs explains why genetic overexpression or knock-out of a single molecule is sufficient to prevent or induce PCH. Thus, alteration in the abundance of any molecule in a cardiac hypertrophic signaling pathway can perturb the transcriptional balance resulting in PCH. This is what was mainly addressed in the present study.
In the normal adult heart, the transcription factors important for cardiac development and growth are inhibited by multiple mechanisms (17). First, the expression of some transcription factors is kept at low abundance. Second, many of the TFs tend to form CoRCs with HDACs and thus also remain in an inactivated state due to constant deacetylation. Third, NFATs and perhaps other TFs, are kept out of the nucleus and inactivated by sustained phosphorylation (3). At last, the compact conformation of normal adult cardiac chromatin is maintained through HDAC-mediated deacetylation of the core histone proteins. Therefore, the initiation of cardiac hypertrophy in adult hearts requires at least nuclear export of the HDACs and/or nuclear import of NFAT-like TFs. Studies in the past two decades have established that nuclear export of class II HADCs (HDAC-4, -5, -7, -9) is primarily mediated by phosphorylation catalyzed by the HDAC kinases including Ca2+/calmodulin-activated kinase II (CaMKIIδ) (18) and protein kinase D1 (PKD1, known as PKCμ) (19). In comparison, activation of NFATs is achieved by CaN-catalyzed dephosphorylation (20). Interestingly, it was recently reported that nuclear export of HDACs play a permissive role in full disinhibition of some NFAT isoforms (21).
In this study, we report that the expression up-regulation of MEF2, GATA4, Nkx2.5, and NFATc4 is a new mechanism contributing to the induction of PCH. We reveal that expression up-regulation of these cardiac TFs was differentially reduced by inhibition of protein kinase D3 (a.k.a. PKCν) and PKD1. Functional inhibition of PKD3 prevented PCH even though this kinase is not normally detected in normal adult cardiomyocytes (22). We used isoproterenol as an inducer of PCH since clinical studies reveal higher plasma norepinephrine is related to a subsequent risk of cardiovascular mortality (23). The validity of isoproterenol-induced PCH as well as cell death models has been well established (24–26).
The PKD family comprises three isozymes: PKD1, PKD2, and PKD3. In the heart, PKD1 has been best characterized for its role in regulating myocardial contraction, cardiac hypertrophy and remodeling (27, 28). PKD1 is a typical HDAC5 kinase and thus more relevant to HDAC5-MEF2 hypertrophic signaling (19, 29). Mice with PKD1 ablation show substantial resistance to cardiac hypertrophy (30). Unexpectedly, a recent study reported a newly developed PKD1-selective inhibitor failed to attenuate PCH in whole animal models (31). One possible explanation for such a discrepancy is that PKD3 could substitute for PKD1 as a HDAC5 kinase as documented in non-cardiac cells (32). Our new results shown herein indicate PKD3 is indeed required for PCH but works through a different mechanism.
EXPERIMENTAL PROCEDURES
Isolation and Culture of Ventricular Myocytes
Sprague-Dawley (SD) rats were used throughout experiments, and animals were purchased from the Military Academy of the Medical Science Laboratory Animal Center (Beijing, China). All animal protocols used in this study comply with the Guide for the Care and Use of Laboratory Animals, published by the National Institutes of Health (NIH Pub. No. 85-23, revised 1996). Isolation of adult rat ventricular myocytes (ARVM) was performed by Langendorff perfusion with a buffer containing low Ca2+, collagenase and protease as described (26). Isolation and culture of neonatal rat ventricular myocytes (NRVM) were conducted using the overnight trypsin-collagenase digestion method as described (26). All experiments with NRVMs were performed on 2–4 days cultures when synchronously contracting cells were observed. The purity of the neonatal myocytes was confirmed by anti-α-actin antibody.
Plasmid Construction
To obtain PKD3 mRNA, neonatal myocytes were treated with 10 μm isoproterenol for 48 h, and mRNA was extracted from cell lysates using Trizol reagent (Invitrogen). The PKD3 cDNA was generated by reverse transcription-polymerase chain reaction (RT-PCR) with the following primer sets: 5′-GCGGGATCCATGTCTGCAAATAATTCCCCTCCA-3′ (forward); 5′-GCCGCTCGAGCTAAGGACCTTCCTCCATGTCATCC-3′ (reverse). The PCR product was digested with BamH1/Xho1 and subcloned into the pCDNA3.1 plasmid. The PKD1 plasmid was constructed in Dr. Tang's previous laboratory in the University of Maryland School of Medicine as described (28).
Small RNA Interference (siRNA)
siRNA was performed using standard methods as described in one of our recent reports (33). All nucleotides were synthesized and 2′ O-methyl modified by GenePharma (Shanghai, China). For each gene at least two siRNA sequences were designed targeting the coding regions and selected based on silencing efficacy by quantitative real time PCR (qPCR). The siRNA sequences including that of negative control (NC) used in this study are shown in the supplemental Table S1. For transient transfection, cells were incubated for 6 h in serum-free Dulbecco's modified Eagle's medium (DMEM, Invitrogen) supplemented with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells were cultured for further 36 h before treatment with normal saline (NS) vehicle, ISO for 48 h in a 37 °C incubator with 5% CO2-95% air.
RT-PCR and qPCR
Total RNA was extracted from cultured neonatal myocytes or isolated adult myocytes using Trizol reagent (Invitrogen). For reverse transcription, 1.0 μg of RNA was used, and reactions were carried out using a reverse transcription system (Promega). Primers were synthesized by Invitrogen (Beijing, China) and listed in supplemental Table S2. RT-PCR was performed using a Genemate thermal cycler (Jinge Instr, Hangzhou, China) and qPCR was performed using SYBR Green Master Mix (Takara Bio) in a Bio-Rad IQ5 detection system. The cycle threshold (CT) values were automatically determined in triplicates and averaged. Abundance of target genes were normalized to that of 18 S rRNA using the formula of 2−ΔCT where ΔCT = CT of target genes − CT of 18 S rRNA. The ratios were expressed as fold change and the mean value for expression of each gene in control cells was defined as 1. These experiments were performed in triplicate for statistical analysis.
Western Blotting
Cell lysates were resolved in 10% SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore). Nuclear protein extraction was performed using a Nuclear and Cytoplasmic Protein Extraction kit according to the manufacturer's instruction (KeyGen Biotech, China). Target proteins were reacted with their respective 1° antibodies and then incubated with a peroxidase-conjugated 2° antibody. Antibodies used for Western blot are shown in supplemental Table S3. GAPDH was used as a loading control. Band densities were quantified using the NIH ImageJ software.
Immunofluorescent Staining
Cardiomyocytes were seeded on laminin-coated glass coverslips and fixed in 4% paraformaldehyde followed by permeabilization with 0.5% Triton X-100. After blocking in 1% BSA-containing PBS, cells were incubated with 1° antibody and subsequently with appropriate 2° antibodies (Invitrogen). Antibodies used for indirect immunofluorescence are shown in supplemental Table S3. Images were analyzed on a TCS-SP confocal laser microscope (Leica, Germany). For surface area determination ImageJ was used, and at least 50 individualized cells were analyzed for each experiment.
NFAT-Luciferase Assays
Neonatal myocytes were co-transfected with pGL4.30 [luc2P/NFAT-RE/Hygro] plasmid and control plasmid pGL7.4 expressing Renilla reniformis luciferase reporter gene. Transient transfection of pcDNA3.1 vector, pcDNA3.1-PKD3 and pcDNA3.1-PKD1 were performed using the electroporation nucleofector kit (Amaxa Biosystems). After 24 h of incubation in 12-well plates, cells were treated with or without isoproterenol for further 48 h. Cells were lysed and luciferase assay was conducted with the Dual Luciferase kit (Promega) according to the manufacturer's instructions. Luciferase activity was measured using a TR717 microplate luminometer (Applied Biosystems). Data were expressed as fold change (average relative light units of induced cells/average relative light units of control cells).
In Vivo Cardiac Hypertrophy Model and Histology
SD rats (150–220 g) were injected with saline vehicle, ISO (5 mg kg−1 d−1, s.c.) or isoproterenol plus oral verapamil (10 mg kg−1 d−1, dissolved in the drinking water). Verapamil was started 48 h before isoproterenol. Animals were sacrificed 7 days later for assessment of cardiac hypertrophy. Sodium pentobarbital (30 mg kg−1, intraperitoneal, Abbott Lab.) was used to anesthetize animals, and hearts were excised, weighed, fixed in PBS-buffered formalin, embedded inparaffin, and sectioned at 4 μm for histology. Sections were stained with hematoxylin and eosin, and photographs were acquired with an Olympus IX71 inverted microscope. Isolated ventricular myocytes were made for mRNA preparation.
Statistical Analysis
Results are expressed as means ± S.E. unless noted otherwise. One-way ANOVA followed by Newman-Keuls test, Student's t test or Pearson's chi-square test were performed as implemented in IgorPro (Wavemetrics) (34). A value of p < 0.05 was accepted as statistically significant.
RESULTS
PKD3 Expression Is Up-regulated by Isoproterenol in Adult and Neonatal Cardiomyocytes
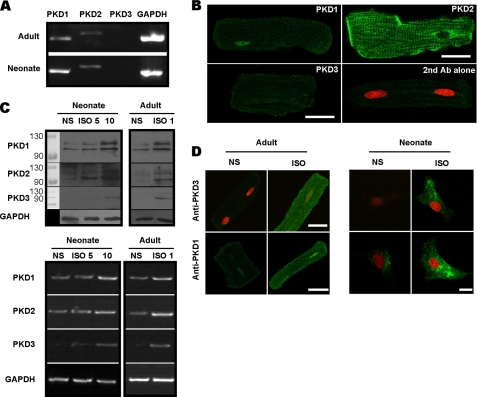
We screened for the presence of the three PKD isoforms in unstimulated cardiac myocytes and examined how these PKD isoenzymes were induced by hypertrophic agents such as isoproterenol. Using RT-PCR, we readily detected mRNAs for PKD1 and PKD2 in both neonatal and adult cardiomyocytes (Fig. 1A). Consistent with previous reports (22), mRNA for PKD3 was essentially undetectable in adult cells. The presence of PKD1 and PKD2 in adult cardiac myocytes was further demonstrated at the protein level by immunofluorescence. As shown in Fig. 1B, PKD1 signal is prominently concentrated in the nucleus while PKD2 is mostly located at the plasma membrane and T-tubules. Unstimulated adult myocytes did not appear to express PKD3 at the protein level since the fluorescent signal of adult myocytes stained with anti-PKD3 plus appropriate secondary antibody was similar to that stained with second antibody alone (Fig. 1B). The antibodies used in this study were carefully chosen for their high specificity based on several experimental criteria including gene silencing of endogenous PKDs and overexpression of exogenous PKD isoforms as detailed in supplemental Fig. S1.
FIGURE 1.
The presence of PKD isoforms and their induction by ISO in cardiac myocytes. A, RT-PCR detection of mRNAs for PKD1 and PKD2 but not PKD3 in isolated adult (upper) and cultured neonatal (lower) myocytes. GAPDH was used as internal control. B, immunolocalization of each PKD isoform in adult myocytes as detected using a confocal microscopy. Cells were fixed and incubated with primary antibodies followed by reaction with Alexa-488 conjugated secondary antibody (green). Propidium iodide (PI) was used as a counterstain for nuclei (red). Note the fluorescent signal conferred by anti-PKD3 is as weak as that stained with second antibody alone, indicating PKD3 is absent in the adult myocyte. Scale bars = 25 μm. C, Western blot (upper) and RT-PCR (lower) assays showing expression up-regulation of three PKD isoforms by ISO in adult and neonatal myocytes. Adult cells were treated with 1 μm ISO for 24 h and neonate was treated with 5 and 10 μm ISO for 48 h. GAPDH was used as loading control. D, expression up-regulation of PKD3 and PKD1 by ISO in adult and neonatal myocytes as detected with a confocal microscopy. Immunofluoresent procedures were the same as that described in B. Scale bars, 25 μm.
We further investigated how isoproterenol would induce expression of the three PKD isoforms. Using Western blot and RT-PCR assays, we found isoproterenol treatment readily up-regulated all three PKD isoforms in neonatal and adult myocytes with PKD3 being particularly increased (Fig. 1C). Indirect immunofluorescence also demonstrated PKD3 and PKD1 were up-regulated in adult and neonatal myocytes (Fig. 1D). Interestingly, PKD3 protein up-regulated in adult myocytes was not concentrated in the nucleus but mostly localized in plasma membranes and cytoplasm, arguing against its proposed function as a HDAC5 kinase in cardiac muscles, in contrast to PKD1.
PKD3 Is Required for Isoproterenol-induced Fetal Cardiac Gene Reactivation and Hypertrophy in Neonatal Myocytes
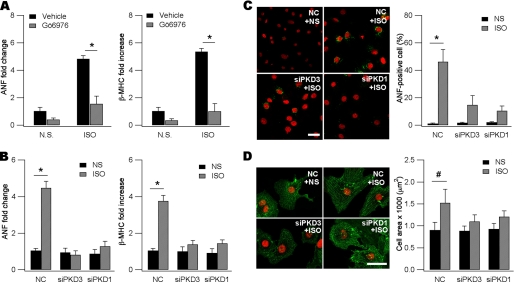
To test how PKD3 engages in fetal cardiac gene reactivation and hypertrophy, we adopted a classic in vitro neonatal hypertrophic model (26). Because ANF and βMHC are sensitive markers of pathological hypertrophy, we assayed for their expression using qPCR. Stimulation with isoproterenol readily induced expression of the two fetal cardiac genes by ∼5-fold compared with vehicle DMSO (Fig. 2A). In the presence of Gö 6976 (2 μm), a known pan-PKD selective inhibitor (35), isoproterenol failed to up-regulate the two hypertrophic marker genes (Fig. 2A). As positive controls, H89 (2 μm) and Rp-8-Br-cAMP (10 μm) were used to inhibit PKA and both agents blunted isoproterenol-induced up-regulation of ANF and βMHC (supplemental Fig. S2). These results are consistent with the idea that isoproterenol-induced fetal cardiac gene reactivation was mediated by certain isoforms of PKDs.
FIGURE 2.
PKD3 and PKD1 are both required for ISO-induced fetal gene reactivation and hypertrophy. A, induction of ANF (left) and βMHC (right) mRNAs by ISO (10 μm, 48 h) was blunted by 2 μm Gö 6976 (n = 3, *, p < 0.01, ANOVA). Gö 6976 was dissolved in DMSO (vehicle) as stock in 2 mm, applied 30 min before ISO treatment, and remained in the culture throughout the experiment. mRNAs were detected with qPCR and 18 S rRNA as an internal standard. The mean value for expression of each gene in DMSO-treated cells is defined as 1. NS, normal saline. B, ISO-induced ANF (left) and βMHC (right) mRNAs were prevented by gene silencing of PKD3 and PKD1 (n = 3, *, p < 0.01, ANOVA). Note in the absence of ISO, siPKD3, or siPKD1 did not affect baseline ANF and βMHC. Cells were transfected with negative control (NC) siRNA, siPKD3, or siPKD1 followed by NS or ISO application. The mean value for mRNAs of each gene in NS- and NC-treated cells is defined as 1. C, ISO-induced ANF expression was reduced by knockdown of PKD3 or PKD1. Cells were fixed and incubated with anti-ANF followed by reaction with Alexa-488 conjugated second antibody (green). Shown on the right panel is quantitative analysis (*, p < 0.01, Pearson's chi-square test). Data are expressed as means ± S.D. For each experiment at least 50 cells were counted. Scale bar, 25 μm. D, ISO-induced cardiac myocyte hypertrophy was blunted by gene silencing of PKD3 or PKD1. Cells were fixed and incubated with anti-α-actinin followed by reaction with Alexa-488-conjugated second antibody (green). Note larger cell size and highly- organized sarcomeres in ISO-treated myocytes. In the absence of ISO, siPKD3, or siPKD1 did not affect baseline cell morphology. Scale bar, 25 μm. Shown on the right panel is quantitative analysis (n = 75–130, #, p < 0.05, ANOVA). Data are expressed as means ± S.D.
To assess how each of the PKD isoforms was differentially involved in PCH, we used siRNA gene silencing techniques. In our system we could achieve >80% transfection efficiency as estimated by expression of a FAM-conjugated siRNA (supplemental Fig. S3). Maximum silencing efficacy (∼70%) required combinatorial use of two siRNA sequences for each PKD gene. Using qPCR assays we demonstrated the siPKD sequences used in this study were isoform-specific since knockdown of a given PKD isoform did not significantly affect expression of other two isoforms (supplemental Table S4).
As shown in Fig. 2B, gene silencing of either PKD1 or PKD3 blunted isoproterenol-induced expression of ANF and βMHC whereas silencing PKD2 was ineffective (supplemental Fig. S4). In the absence of isoproterenol, knockdown of PKD1 or PKD3 alone did not significantly affect expression of the two fetal cardiac genes (Fig. 2B). We also monitored ANF at the protein level using indirect immunofluorescence. Propidium iodide (PI) was used to counterstain the nucleus. As shown in Fig. 2C, isoproterenol could induce ANF expression in ∼50% cells whereas only <1% cells transfected with NC siRNA expressed ANF in the absence of isoproterenol. The number of ANF-positive cells induced by isoproterenol was substantially decreased by knockdown of either PKD3 or PKD1 (Fig. 2C).
We directly measured cell size and examined sarcomere assembly to ensure cardiac hypertrophy was indeed induced. To this end, myocytes were stained for α-actinin and the nucleus was again counterstained with PI. Remaining in culture for 48 h, normal saline-treated myocytes transfected with NC siRNA appeared smaller, round-shaped with soft edges and disorganized sarcomeres (Fig. 2D). The average cross-sectional area was ∼1000 μm2. By comparison, stimulation with isoproterenol for 48 h resulted in the appearance of highly-organized sarcomeres and increased cell size (Fig. 2D). Typically, isoproterenol increased cell cross-sectional area to 1500 μm2 in our experimental conditions. Gene silencing of PKD1 or PKD3 markedly attenuated all the isoproterenol-induced alterations (Fig. 2D). In the absence of isoproterenol stimulation, knockdown of PKD1 or PKD3 did not decrease cross-sectional cell area (Fig. 2D). These results indicate PKD1 and PKD3 are both required for morphological alterations of the pathological hypertrophy.
PKD3 and PKD1 Are Differentially Required for the Increased Expression of Hypertrophic Transcription Factors
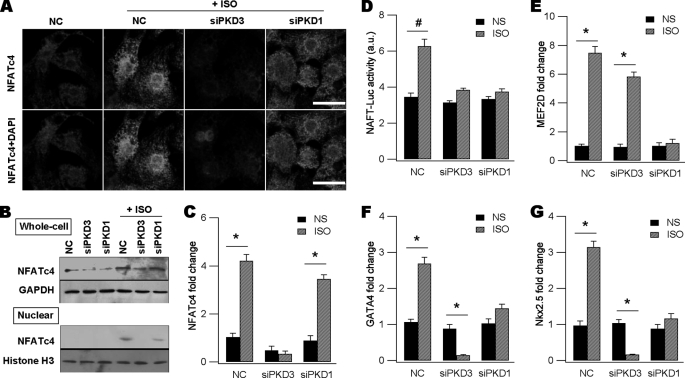
Because previous studies suggested formation of TACs, super-TACs and CoRCs, we reasoned alteration of the abundance of certain hypertrophic TFs might be another mechanism inducing pathological hypertrophy. We chose four endogenous TFs as hypertrophic markers and probed how abundance of these TFs was altered by PKDs to drive fetal gene reactivation. NFATc4 is a master hypertrophic TF present in heart, and dual-regulated by phosphorylation (20) and acetylation (21) in PCH. Using indirect immunofluorescent techniques (Fig. 3A), Western blot analysis of whole cell lysates (Fig. 3B, upper panel) and qPCR assays (Fig. 3C), we revealed isoproterenol indeed enhanced overall expression levels of NFATc4 compared with NC myocytes. Gene silencing of PKD3 markedly depressed NFATc4 expression up-regulation raised by isoproterenol whereas knockdown of PKD1 only had a minor effect (Fig. 3, A–C). This is better reflected by qPCR assays showing isoproterenol evoked a 4-fold increase in NFATc4 mRNA and this was largely blunted by knockdown of PKD3 but not significantly by that of PKD1 (Fig. 3C).
FIGURE 3.
PKD3 and PKD1 differentially upregulate expression of hypertrophic transcription factors. A, fluorescent detection of endogenous NFATc4 protein in each group as indicated. Cells were transfected with NC siRNA, siPKD3, or siPKD1 followed by ISO stimulation. Cells were fixed and incubated with anti-NFATc4 followed by reaction with Alexa-546-conjugated second antibody (red). DAPI was used to counterstain the nucleus. Note overall NFATc4 intensity is much weaker in NS- or siPKD3-treated groups. Scale bars, 20 μm. B, Western blot assays showing ISO-induced expression of NFATc4 in whole-cell lysates (upper) and nuclear appearance of NFATc4 in nuclear protein extracts (lower). Both effects of ISO were largely attenuated by siPKD3. For Western blot of whole cell lysates GAPDH was used as loading control while for that of nuclear protein extracts, histone H3 was used. C, ISO-induced NFATc4 mRNA measured by qPCR was reduced by knockdown of PKD3 but not by PKD1 (n = 3, *, p < 0.01, ANOVA). D, ISO-induced NFAT-Luc activity was decreased by silencing of either PKD3 or PKD1 (n = 3, #, p < 0.05, ANOVA). E, ISO-induced MEF2D mRNA was reduced by knockdown of PKD1 but not by PKD3 (n = 3, *, p < 0.01, # p < 0.05, ANOVA). F and G, ISO-induced GATA4 (F) and Nkx2.5 (G) mRNAs were blunted by siPKD1 or siPKD3 (n = 3, *, p < 0.01, #, p < 0.05, ANOVA). Note in the absence of ISO, siPKD3 or siPKD1 did not affect baseline expression of GATA4 and Nkx2.5.
Another consequence of isoproterenol stimulation was to promote nuclear entry of NFATc4 (Fig. 3A) as reported in many previous studies. The basal nuclear NFATc4 was ∼30% (n = 3) of the total cellular NFATc4 and the value was increased to ∼50% by isoproterenol treatment (n = 3, p < 0.01). PKD1 silencing decreased the value to 37% (n = 3) without changing isoproterenol-induced NFATc4 abundance, as shown in Fig. 3C. These results were further supported by Western blot analysis of nuclear protein extracts showing isoproterenol-induced NFATc4 signals were decreased by PKD1 knockdown and nearly abolished by PKD3 silencing (Fig. 3B, lower panel).
The above results indicate gene silencing of PKD1 or PKD3 had different impacts on isoproterenol-induced NFATc4 expression and nuclear accumulation. However, inhibition of the two kinases would result in similar functional consequence, i.e. abrogating the NFATc4-driven transcription. This conclusion was supported by the NFAT-Luc assays showing isoproterenol-elicited NFAT-Luc activity was significantly blunted by gene silencing of either PKD3 or PKD1 (Fig. 3D).
MEF2D is another dominant hypertrophic TF and normally forms a CoRC with class II HDACs, Cain and MITR (15). We next examined whether this TF was up-regulated by isoproterenol and how PKD3 and PKD1 were involved in its transcriptional regulation. Results from these experiments indicated incubation of neonatal myocytes with isoproterenol for 48 h significantly increased MEF2D mRNA by ∼7-fold, and this was blunted by PKD1 silencing only. Knockdown of PKD3 had a marginal effect (Fig. 3E). Thus, we concluded MEF2D expression up-regulation is predominantly controlled by PKD1 but not by PKD3.
Nkx2.5 and GATA4 are also hypertrophic TFs essential for pathological hypertrophy. Previous studies showed Nkx2.5, GATA4 and SRF form a TF triad in cardiac myocytes (6). We then tested whether Nkx2.5 and GATA4 were up-regulated by isoproterenol and the role of PKD3 and PKD1 in the process. As expected, isoproterenol increased the two TFs significantly and gene silencing of either PKD isoform was effective in blunting the isoproterenol action (Fig. 3, F and G). However, silencing PKD1 only brought the two TFs back to basal levels whereas silencing PKD3 decreased the two TFs to a level much lower than baseline values (Fig. 3, F and G). These results indicate PKD3 is more powerful in upregulating expression of Nkx2.5 and GATA4 induced by isoproterenol.
Expression Up-regulation of Hypertrophic Transcription Factors in Cardiac Hypertrophy in Vivo
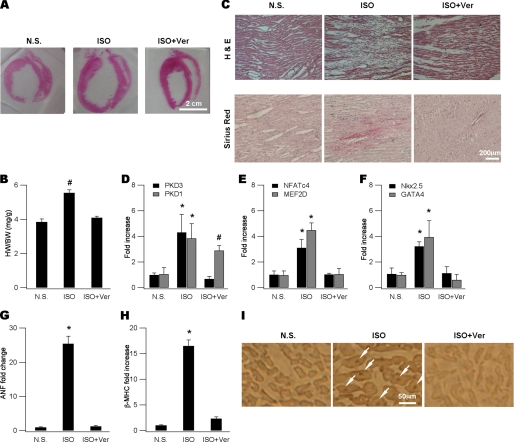
We used an in vivo cardiac hypertrophy model to test our findings made primarily in neonatal settings. We could reliably induce cardiac hypertrophy in rats by injecting isoproterenol for 5 days and animals were sacrificed on day 7 for examination (26). The isoproterenol-induced cardiac hypertrophy model well recapitulates the cardiac infarction model in many pathological and metabolic abnormalities (25). Cardiac hypertrophy was evident in isoproterenol-treated animals as manifested by enlarged heart size (Fig. 4A) and increased heart-to-body ratio (HW/BW) (Fig. 4B). Histological examination revealed profound loss of cardiomyocytes (Fig. 4C, upper panels) that were replaced by fibrosis (Fig. 4C, lower panels). Consistent with our findings in neonatal myocytes, two protein kinases PKD3 and PKD1 (Fig. 4D), four TFs NFATc4, MEF2D (Fig. 4E), Nkx2.5 and GATA4 (Fig. 4F), and two fetal genes ANF (Fig. 4G) and βMHC (Fig. 4H) in isolated cardiac myocytes were all up-regulated by ISO injection. Furthermore, nuclear entry of NFATc4 as demonstrated by immunohistochemistry methods was obviously induced by isoproterenol administration (Fig. 4I). Interestingly, except for PKD1 expression level, all isoproterenol-induced cardiac alterations were significantly blunted (Fig. 4, A–I) by co-administration of verapamil, an L-type calcium channel inhibitor that has been shown to depress cardiac hypertrophy in both animal models (36) and human (37).
FIGURE 4.
PKD3-mediated hypertrophic signaling in in vivo cardiac hypertrophy. A, representative gross heart pictures showing ISO-induced hypertrophy and its blockade by verapamil (Ver). ISO was injected at a dose of 5 mg kg−1 d−1 and Ver (10 mg kg−1 d−1) was administrated by dissolving in drinking water. Animals were sacrificed on day 7 and heart sections were made for histology and immunohistochemistry. Isolated ventricular myocytes were made for mRNA determination. B, comparison of HW/BW ratios (mg/g) for animals shown in (A) (n = 5, #, p < 0.05 versus N.S., ANOVA). C, ISO injection resulted in prominent cell loss (upper, H and E) replaced with fibrosis (lower, Sirus Red) and these were attenuated by verapamil. D, mRNAs of PKD3 and PKD1 were increased by isoproterenol and prevented by co-administration with verapamil (n = 5, *, p < 0.01, #, p < 0.05 versus N.S., ANOVA). E and F, mRNAs of NFATc4, MEF2D (E), GATA4 and Nkx2.5 (F) were elevated by isoproterenol and prevented by co-administration with verapamil (n = 5, *, p < 0.01 versus N.S., ANOVA). G and H, mRNAs of ANF (G) and βMHC (H) were up-regulated by isoproterenol and prevented by co-administration of verapamil (n = 5, *, p < 0.01 versus N.S., ANOVA). I, in situ detection of nuclear NFATc4 proteins (arrows in the middle panel) using immunohistochemistry in an isoproterenol-treated animal and this was not observed in verapamil-coapplied animals (right).
PKD3 Expression Up-regulation Requires Activated Calcinurin in Neonatal Myocytes
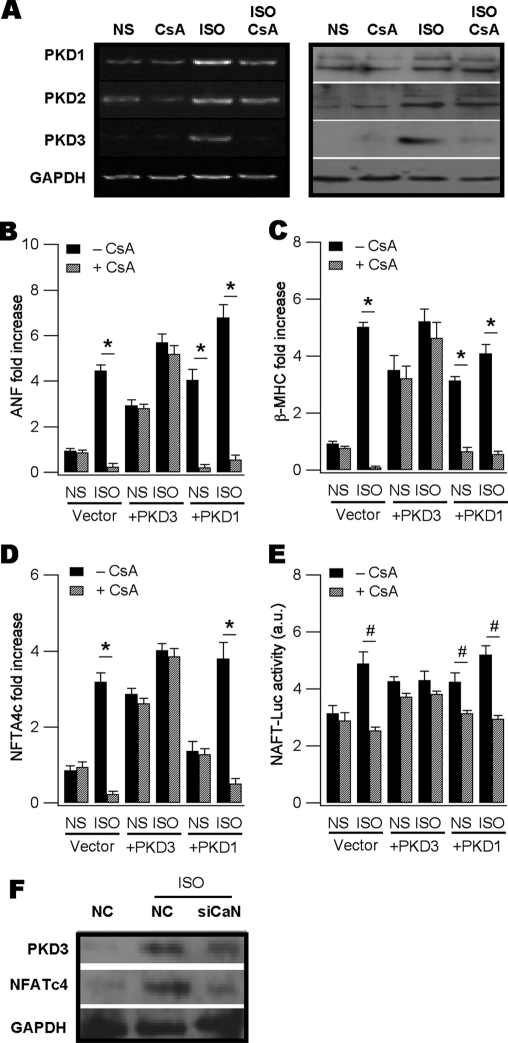
We next addressed how PKD3 was up-regulated by isoproterenol using the neonatal hypertrophic model. We hypothesized PKD3 expression up-regulation by isoproterenol is mediated by CaN and we predicted this would be prevented by the CaN inhibitor cyclosporine (CsA). This prediction was borne out as revealed by RT-PCR (Fig. 5A, left panel) and Western blot assays (Fig. 5A, right panel). By comparison, endogenous and isoproterenol-induced PKD1 and PKD2 expression was not noticeably affected by CsA treatment (Fig. 5A). Consistently, qPCR assays indicated CsA significantly inhibited isoproterenol-up-regulated genes including ANF, βMHC and NAFTc4 (Fig. 5, B–D, vector panels). CsA also prevented isoproterenol-induced enhancement of NFAT-Luc activity (Fig. 5E, vector panel).
FIGURE 5.
PKD3 is a downstream target of calcineurin. A, RT-PCR (left panel) and Western blot (right panel) assays showing ISO up-regulated all PKD isoforms in the neonatal myocytes but only PKD3 up-regulated was abolished by CsA pre-treatment. CsA (2 μm) was applied 30 min before ISO addition and remained throughout the experiment. B and C, ISO-induced ANF and βMHC mRNAs in vector-transfected cells were decreased by CsA treatment (n = 3, *, p < 0.01, ANOVA, vector panels). Basal ANF and βMHC were also up-regulated by overexpressing PKD3 (+PKD3 panels) or PKD1 (+PKD3 panels). Addition of ISO conferred small additive effects. CsA failed to attenuate ANF and βMHC induction by overexpressing PKD3 or PKD3 plus ISO (+PKD3 panels) but blunted those by PKD1 or PKD1 plus ISO (+PKD1 panels). D and E, ISO-induced NFATc4 expression (D) and NFAT-Luc activity (E) was attenuated by CsA treatment (n = 3, #, p < 0.05, ANOVA, vector panels). Overexpressing PKD3 up-regulated NFATc4 expression and enhanced NFAT-Luc activity (n = 3, #, p < 0.05, ANOVA) and CsA failed to prevent these effects. Overexpressing PKD1 increased NFAT-Luc activity and this effect was blunted by CsA (E). All mRNAs were measured by qPCR with 18 S rRNA as control and NFAT-driven transcription activity was estimated using the dual luciferase assay kit. F, Western blot showing gene silencing of CaN blunted ISO-induced expression up-regulation of PKD3 and NFATc4 at the protein level.
If the above hypothesis is correct, we would predict forced expression of PKD3 be sufficient to induce the hypertrophic response and CsA would no longer abrogate the PKD3-induced hypertrophic reprogramming. In our system the transfection efficiency was ∼50% in live cardiomyocytes as indicated by expressing the enhanced green fluorescent protein (EGFP) (supplemental Fig. S5). RT-PCR assays revealed mRNAs of PKD3 and PKD1 overexpressed were 3.24 + 0.26 (n = 3) and 3.43 + 0.2 (n = 3)-fold higher compared with vector transfection alone. As expected, overexpression of PKD3 alone was sufficient to up-regulate mRNAs of ANF, βMHC and NAFTc4, along with enhanced NFTA-Luc activity, while CsA did not impose any significant inhibition to these PKD3-induced effects (Fig. 5, B–E, +PKD3 panels). Addition of isoproterenol to PKD3-expressing cells only resulted in a small additive effect in the above-mentioned hypertrophic parameters. In contrast, although overexpression of PKD1 could also induce various hypertrophic responses as PKD3 did, most of these PKD1-induced hypertrophic markers were sensitive to CsA treatment (Fig. 5, B, C, E, +PKD1 panels).
To demonstrate the CsA effect on PKD3 induction by isoproterenol was specific to CaN, we knocked down the phosphatase using siRNA techniques and found CaN silence profoundly blunted isoproterenol-induced increases in the expression of PKD3 accompanied by NFATc4 down-regulation (Fig. 5F). These results indicated PKD3 expression up-regulation by isoproterenol requires activated CaN and overexpressing PKD3 alone is sufficient to induce hypertrophy by working on yet to be identified downstream targets.
PKD3 Expression Up-regulation Is Driven by Multiple NFATs Other Than NFATc4 in Neonatal Myocytes
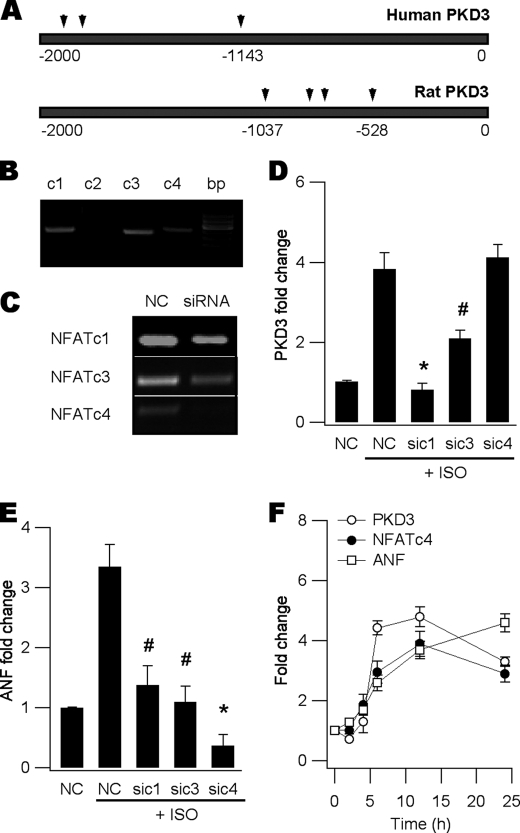
Finally, we wished to address how activated CaN induced PKD3 expression. Because activated CaN is well known to dephosphorylate and subsequently activate Ca2+-sensitive NFATc1-c4 in various cell types, we postulated some of the NFAT members were most likely candidates. Using the online software TESS (38), we identified multiple putative NFAT binding sites in the promoter regions of rat and human PKD3 (Fig. 6A). We then screened our neonatal myocyte mRNAs using RT-PCR and found 3 out of the 4 CaN-regulated NFATs, i.e. NFATc1, c3 and c4 were present (Fig. 6B). Despite NFATc2 being prominent in mouse heart (39), it was not detectable in rat cardiac myocytes and was excluded from further experiments. Using siRNA gene silencing (Fig. 6C), we revealed knockdown of NFATc1 or c3 but not c4 significantly blunted isoproterenol-induced PKD3 expression (Fig. 6, D and E). To outline the chronological order between PKD3 induction, hypertrophic TF expression and fetal gene reactivation, a time course study was performed using PKD3, NFATc4, and ANF as indexes. These studies indicated induction of PKD3 by isoproterenol was prior to that of NFATc4 and ANF (Fig. 6F). Thus, we conclude the increased expression of PKD3 was largely driven by CaN-regulated NFATc1 and c3, and required for subsequent expression of the hypertrophic TFs and fetal gene reactivation.
FIGURE 6.
PKD3 expression is driven by NFATc1 and c3. A, analysis of PKD3 promoter regions of human (upper) and rat (lower) using the online TESS software. Several putative NFAT binding sites were predicted at −1134, −1915, and −1945 in human, and −528, −821, −798, −1037 in rat PKD3. B, RT-PCR screening of NFAT isoforms in neonatal myocytes showing the presence of NFATc1, c3 and c4 but not c2. C, RT-PCR verification of siRNA gene silencing efficacy. D, ISO-induced PKD3 expression up-regulation was blunted by silencing NFATc1 and c3 but not c4 (n = 3, *, p < 0.01, # p < 0.05 versus ISO-NC, ANOVA). E, ISO-induced ANF expression increase was blocked by gene silencing of NFATc1, c3 and c4 (n = 3, *, p < 0.01, #, p < 0.05 versus ISO-NC, ANOVA). F, time course study showing increased expression of PKD3 peaked at ∼6 h following ISO application whereas expression up-regulation of NFATc4 and ANF substantially lagged behind.
DISCUSSION
We have pinpointed PKD3 as a gatekeeper of cardiac hypertrophy by allowing the increases of the expression of a cohort of cardiac hypertrophic TFs including NFATc4, GATA4, and Nkx2.5. These new results help us understand in depth the classical CaN-NFAT signaling pathway. The CaN-NFATc1/c3-PKD3-NFATc4 signaling cascade can be summarized as follows: 1) in response to hypertrophic stimuli, PKD3 is up-regulated by activated CaN through nuclear import of NFATc1 and c3. 2) Newly synthesized PKD3 provokes further expression of NFATc4, GATA4, and Nkx2.5, which in turn become activated by dephosphorylation and/or acetylation. 3) In parallel, activated PKD1 (perhaps along with other HDAC kinases) primarily promotes MEF2D expression and activation by inducing class II HDAC phosphorylation and nuclear export. 4) Cooperative activation of these hypertrophic TFs drives fetal gene reactivation and induces pathological hypertrophy (40). The identity of downstream targets along the PKD3-mediated hypertrophic pathway is under active investigation.
Our results clearly indicate that, in addition to post-translational modification of the TFs pre-existed in cardiac myocytes, up-regulation of TF abundance is also critical to pathological hypertrophy. We demonstrate PKD3 and PKD1 are differentially required for expression up-regulation of various hypertrophic TFs, at least in isoproterenol-induced hypertrophy models. Although the two kinases were shown to functionally complement each other's biological roles in non-cardiac settings (32), this is apparently not the case with cardiac myocytes.
It is surprising that isoproterenol could upregulate MEF2D through a PKD1-dependent mechanism as revealed in this study. Previous studies established PKD1 mainly phosphorylates HDAC5 and this is typically activated by Gαq-coupled receptor agonists but not by Gαs-type agonists such as isoproterenol. Nevertheless, our new results are corroborated by a recent report indicating isoproterenol could induce PCH by stimulating secretion of the autocrine/paracrine Gαq agonist endothelin-1 (41).
Drugs targeting critical molecules within hypertrophic signaling pathways, including endothelin-1 receptor blockers and HDAC inhibitors, are being tested for their potential therapeutic use in cardiac hypertrophy and heart failure. Inhibitors targeting PKD1 such as BPDKi have been developed but this aminobipyridyl compound failed to blunt animal cardiac hypertrophy (31). Our new results call for further tests on PKD3-selective inhibitors or pan-PKD inhibitors to determine their efficacy in blunting cardiac hypertrophy and heart failure. Since the role of PKD3 in cardiac hypertrophy reported in this report was performed mostly in neonatal myocytes, whether PKD3 is relevant to adult disease remains an open question until further experiments are performed in adult heart cells.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants (HL55426 and AI058173, to D. L. G.), grants from the Tianjin Science and Technology Support Project (08ZCKFSH04500, to X. D. T.), Tianjin Natural Science Foundation (10JCYBJC14800, to J. L.), NSFC Grants (30871011, to X. D. T.; 81072629 to J. L.; 30870988, to X. W. Z.), The Project “973” grant (2010CB945001, to X. D. T.), and Chinese Ministry of Education (MOE) Doctorial Training Funds (200800550036, to X. D. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S4 and Figs. S1–S5.
- PCH
- pathological cardiac hypertrophy
- HDAC
- histone deacetylase
- MEF2
- myocyte enhancer factor-2
- NFAT
- nuclear factor of activated T-cells
- Nkx2.5
- NK family of transcription factor 2.5
- TAC
- transcription activation complex
- CoRC
- co-repressor complexes
- CaN
- calcineurin
- PKD
- protein kinase D
- ANF
- atrial natriuretic factor
- βMHC
- β-myosin heavy chain
- CsA
- cyclosporine
- NRVM
- neonatal rat ventricular myocytes
- ARVM
- adult rat ventricular myocytes
- siRNA
- small RNA interference
- NC siRNA
- negative control siRNA
- qPCR
- quantitative real-time polymerase chain reaction
- ISO
- isoproterenol.
REFERENCES
- 1. Mathew J., Sleight P., Lonn E., Johnstone D., Pogue J., Yi Q., Bosch J., Sussex B., Probstfield J., Yusuf S. (2001) Circulation 104, 1615–1621 [DOI] [PubMed] [Google Scholar]
- 2. Sambrano G. R., Fraser I., Han H., Ni Y., O'Connell T., Yan Z., Stull J. T. (2002) Nature 420, 712–714 [DOI] [PubMed] [Google Scholar]
- 3. McKinsey T. A. (2007) Cardiovasc Res. 73, 667–677 [DOI] [PubMed] [Google Scholar]
- 4. MacDonald M. R., Petrie M. C., Hawkins N. M., Petrie J. R., Fisher M., McKelvie R., Aguilar D., Krum H., McMurray J. J. (2008) Eur. Heart J. 29, 1224–1240 [DOI] [PubMed] [Google Scholar]
- 5. Putt M. E., Hannenhalli S., Lu Y., Haines P., Chandrupatla H. R., Morrisey E. E., Margulies K. B., Cappola T. P. (2009) Circ. Cardiovasc Genet. 2, 212–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nishida W., Nakamura M., Mori S., Takahashi M., Ohkawa Y., Tadokoro S., Yoshida K., Hiwada K., Hayashi K., Sobue K. (2002) J. Biol. Chem. 277, 7308–7317 [DOI] [PubMed] [Google Scholar]
- 7. Ni Y. G., Berenji K., Wang N., Oh M., Sachan N., Dey A., Cheng J., Lu G., Morris D. J., Castrillon D. H., Gerard R. D., Rothermel B. A., Hill J. A. (2006) Circulation 114, 1159–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ozgen N., Obreztchikova M., Guo J., Elouardighi H., Dorn G. W., 2nd, Wilson B. A., Steinberg S. F. (2008) J. Biol. Chem. 283, 17009–17019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Luedde M., Katus H. A., Frey N. (2006) Recent Pat Cardiovasc Drug Discov 1, 1–20 [DOI] [PubMed] [Google Scholar]
- 10. Heineke J., Molkentin J. D. (2006) Nat. Rev. Mol. Cell Biol. 7, 589–600 [DOI] [PubMed] [Google Scholar]
- 11. Barrick D., Kopan R. (2006) Cell 124, 883–885 [DOI] [PubMed] [Google Scholar]
- 12. Lejon S., Thong S. Y., Murthy A., AlQarni S., Murzina N. V., Blobel G. A., Laue E. D., Mackay J. P. (2011) J. Biol. Chem. 286, 1196–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jepsen K., Rosenfeld M. G. (2002) J. Cell Sci. 115, 689–698 [DOI] [PubMed] [Google Scholar]
- 14. Morin S., Charron F., Robitaille L., Nemer M. (2000) EMBO J. 19, 2046–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang C. L., McKinsey T. A., Chang S., Antos C. L., Hill J. A., Olson E. N. (2002) Cell 110, 479–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Song K., Backs J., McAnally J., Qi X., Gerard R. D., Richardson J. A., Hill J. A., Bassel-Duby R., Olson E. N. (2006) Cell 125, 453–466 [DOI] [PubMed] [Google Scholar]
- 17. Olson E. N. (2006) Science 313, 1922–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang T., Kohlhaas M., Backs J., Mishra S., Phillips W., Dybkova N., Chang S., Ling H., Bers D. M., Maier L. S., Olson E. N., Brown J. H. (2007) J. Biol. Chem. 282, 35078–35087 [DOI] [PubMed] [Google Scholar]
- 19. Harrison B. C., Kim M. S., van Rooij E., Plato C. F., Papst P. J., Vega R. B., McAnally J. A., Richardson J. A., Bassel-Duby R., Olson E. N., McKinsey T. A. (2006) Mol. Cell Biol. 26, 3875–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Molkentin J. D., Lu J. R., Antos C. L., Markham B., Richardson J., Robbins J., Grant S. R., Olson E. N. (1998) Cell 93, 215–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dai Y. S., Xu J., Molkentin J. D. (2005) Mol. Cell Biol. 25, 9936–9948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ellwanger K., Pfizenmaier K., Lutz S., Hausser A. (2008) BMC Dev. Biol. 8, 47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cohn J. N., Levine T. B., Olivari M. T., Garberg V., Lura D., Francis G. S., Simon A. B., Rector T. (1984) N. Engl. J. Med. 311, 819–823 [DOI] [PubMed] [Google Scholar]
- 24. Morisco C., Zebrowski D. C., Vatner D. E., Vatner S. F., Sadoshima J. (2001) J. Mol. Cell Cardiol 33, 561–573 [DOI] [PubMed] [Google Scholar]
- 25. Heather L. C., Catchpole A. F., Stuckey D. J., Cole M. A., Carr C. A., Clarke K. (2009) J. Physiol. Pharmacol. 60, 31–39 [PubMed] [Google Scholar]
- 26. Li C., Cai X., Sun H., Bai T., Zheng X., Zhou X. W., Chen X., Gill D. L., Li J., Tang X. D. (2011) Biochem. Biophys. Res. Commun. 409, 125–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Avkiran M., Rowland A. J., Cuello F., Haworth R. S. (2008) Circ. Res. 102, 157–163 [DOI] [PubMed] [Google Scholar]
- 28. Goodall M. H., Wardlow R. D., 2nd, Goldblum R. R., Ziman A., Lederer W. J., Randall W., Rogers T. B. (2010) J. Biol. Chem. 285, 41686–41700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bossuyt J., Helmstadter K., Wu X., Clements-Jewery H., Haworth R. S., Avkiran M., Martin J. L., Pogwizd S. M., Bers D. M. (2008) Circ. Res. 102, 695–702 [DOI] [PubMed] [Google Scholar]
- 30. Fielitz J., Kim M. S., Shelton J. M., Qi X., Hill J. A., Richardson J. A., Bassel-Duby R., Olson E. N. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3059–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meredith E. L., Beattie K., Burgis R., Capparelli M., Chapo J., Dipietro L., Gamber G., Enyedy I., Hood D. B., Hosagrahara V., Jewell C., Koch K. A., Lee W., Lemon D. D., McKinsey T. A., Miranda K., Pagratis N., Phan D., Plato C., Rao C., Rozhitskaya O., Soldermann N., Springer C., van Eis M., Vega R. B., Yan W., Zhu Q., Monovich L. G. (2010) J. Med. Chem. 53, 5422–5438 [DOI] [PubMed] [Google Scholar]
- 32. Matthews S. A., Liu P., Spitaler M., Olson E. N., McKinsey T. A., Cantrell D. A., Scharenberg A. M. (2006) Mol. Cell Biol. 26, 1569–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang Y., Deng X., Mancarella S., Hendron E., Eguchi S., Soboloff J., Tang X. D., Gill D. L. (2010) Science 330, 105–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tang X. D., Xu R., Reynolds M. F., Garcia M. L., Heinemann S. H., Hoshi T. (2003) Nature 425, 531–535 [DOI] [PubMed] [Google Scholar]
- 35. Gschwendt M., Dieterich S., Rennecke J., Kittstein W., Mueller H. J., Johannes F. J. (1996) FEBS Lett. 392, 77–80 [DOI] [PubMed] [Google Scholar]
- 36. Ago T., Yang Y., Zhai P., Sadoshima J. (2010) J. Cardiovasc Transl Res. 3, 304–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dahlöf B., Sever P. S., Poulter N. R., Wedel H., Beevers D. G., Caulfield M., Collins R., Kjeldsen S. E., Kristinsson A., McInnes G. T., Mehlsen J., Nieminen M., O'Brien E., Ostergren J. (2005) Lancet 366, 895–906 [DOI] [PubMed] [Google Scholar]
- 38. Schug J. (2008) in Current Protocols in Bioinformatics (Baxevanis A. D. ed) Vol. 21, pp. 2.6.1–2.6.15, J. Wiley and Sons [Google Scholar]
- 39. Bourajjaj M., Armand A. S., da Costa Martins P. A., Weijts B., van der Nagel R., Heeneman S., Wehrens X. H., De Windt L. J. (2008) J. Biol. Chem. 283, 22295–22303 [DOI] [PubMed] [Google Scholar]
- 40. Molkentin J. D., Black B. L., Martin J. F., Olson E. N. (1995) Cell 83, 1125–1136 [DOI] [PubMed] [Google Scholar]
- 41. Higazi D. R., Fearnley C. J., Drawnel F. M., Talasila A., Corps E. M., Ritter O., McDonald F., Mikoshiba K., Bootman M. D., Roderick H. L. (2009) Mol. Cell 33, 472–482 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.