Background: Oxidative stress during inflammation contributes to carcinogenesis.
Results: Exposure of epithelial cells to inflammatory macrophages induces the antioxidative transcription factor Nrf2 resulting in proteasome activation and apoptosis protection.
Conclusion: The cytoprotective actions of Nrf2 contribute to inflammation-associated tumorigenesis.
Significance: This study improves the understanding of the complex nature of Nrf2-mediated effects and their role in human disease.
Keywords: Carcinogenesis, Inflammation, Oxidative Stress, Proteasome, Transcription Factors
Abstract
Adaptation of epithelial cells to persistent oxidative stress plays an important role in inflammation-associated carcinogenesis. This adaptation process involves activation of Nrf2 (nuclear factor-E2-related factor-2), which has been recently shown to contribute to carcinogenesis through the induction of proteasomal gene expression and proteasome activity. To verify this possible link between inflammation, oxidative stress, and Nrf2-dependent proteasome activation, we explored the impact of inflammatory (M1) macrophages on the human colon epithelial cell line NCM460. Transwell cocultures with macrophages differentiated from granulocyte monocyte-colony-stimulating factor-treated monocytes led to an increased activity of Nrf2 in NCM460 cells along with an elevated proteasome activity. This higher proteasome activity resulted from Nrf2-dependent induction of proteasomal gene expression, as shown for the 19 and 20 S subunit proteins S5a and α5, respectively. These effects of macrophage coculture were preceded by an increase of reactive oxygen species in cocultured NCM460 cells and could be blocked by catalase or by the reactive oxygen species scavenger Tiron, whereas transient treatment of NCM460 cells with H2O2 similarly led to Nrf2-dependent proteasome activation. Through the Nrf2-dependent increase of proteasomal gene expression and proteasome activity, the sensitivity of NCM460 cells to tumor necrosis factor-related apoptosis-inducing ligand- or irinotecan-induced apoptosis declined. These findings indicate that inflammatory conditions such as the presence of M1 macrophages and the resulting oxidative stress are involved in the Nrf2-dependent gain of proteasome activity in epithelial cells, e.g. colonocytes, giving rise of greater resistance to apoptosis. This mechanism might contribute to inflammation-associated carcinogenesis, e.g. of the colon.
Introduction
As an important mechanism in inflammation-associated carcinogenesis, epithelial cells undergo a permanent adaptation to oxidative stress conditions coming along with the chronic exposure to inflammatory cells and their released mediators. Along with this adaptation, the stressed epithelium acquires a cellular phenotype paving the way for the development of cancer (1–3). One mechanism of stress adaptation is the activation of the oxidative stress-responsive cap'n collar-bZIP transcription factor Nrf2 (nuclear factor-E2-related factor-2). Under conditions of oxidative and electrophilic stress, Nrf2 is activated through its release from the inhibitory protein Keap1 (Kelch ECH-associating protein 1) and subsequent nuclear accumulation (4–8).
Nrf2 has been primarily recognized as modulator of an antioxidative response by inducing transcription of phase II enzymes such as GST, heme oxygenase-1, or NAD(P)H-quinone oxidoreductase 1, which protect the cell from reactive oxygen species (ROS)-induced2 damage (7). This protection includes the potential of Nrf2-induced phase II enzyme expression to prevent genotoxic insults from oxidative stress, thus making Nrf2 activation a promising tool in chemo prevention of cancer (6). Electrophils and anti-oxidants such as tert-butylhydroquinone (tBHQ) and the phytochemical sulforaphane are well known to induce Nrf2 and thereby to stimulate phase II enzyme expression. Thus, Nrf2 inducing agents have been suggested to be of substantial benefit in cancer prevention (5, 6, 9).
However, evidence accumulates that Nrf2 can also promote tumorigenesis (10–13) and resistance of tumors to chemotherapy, e.g. by inducing detoxification of anti-cancer drugs in a phase II-dependent fashion (14–16). Moreover, Nrf2 is involved in the gain of anti-apoptotic protection by tumor cells (11, 17, 18). To some extent, this protective effect against apoptosis could be attributed to the Nrf2-induced expression of proteasomal genes leading to an elevated proteasome activity in tumor cells (11, 19–24). Whereas this proteasome inducing effect of Nrf2 in response to oxidative and electrophilic stress has been widely reported already (11, 25–30), recent data revealed that the closely related transcription factor Nrf1 (TCF11) plays a particular role in the maintenance of constitutive proteasome activity (31). Moreover, Nrf1 mediates the inducing effect of proteasome inhibitors on proteasomal gene expression to a greater extent than Nrf2 (31, 32). Thus, Nrf1 is regarded as modulator of basal proteasomal gene expression and proteasome activity, whereas Nrf2 is believed to mediate the induced proteasomal gene expression and proteasome activity (33), particularly in response to electrophilic and oxidative stress (34).
Through an enhanced proteasome activity in tumor cells, such as colon cancer cells (11), Nrf2 activation confers resistance not only to anti-cancer drugs, here together with the detoxification by phase II enzymes, but also to death ligands such as TRAIL. In the latter case, we have recently shown that Nrf2-dependent proteasome activation promotes NF-κB activation by TRAIL, an effect that mainly relies on the forced turnover of IκBα as a result of the gain of proteasome activity (11). From TRAIL-induced NF-κB activation, increased expression level of certain anti-apoptotic genes (e.g. cIAP1) emerged (11) through which TRAIL-induced apoptosis is inhibited. In this way, Nrf2 activation and subsequent induction of the proteasome might contribute to the acquisition of a tumorigenic phenotype of epithelial cells during inflammation associated carcinogenesis, e.g. of the colon.
The present study therefore aimed at elucidating whether the presence of inflammatory cells, in particular macrophages that are a major constituent of immune cell infiltrates and a source of ROS in chronically inflamed tissues (35–38), alters proteasomal gene expression and proteasome activity in colonic NCM460 cells in a Nrf2-dependent fashion. Moreover, it was investigated whether under these conditions the protection of NCM460 cells against apoptosis is enhanced. Our data demonstrate that the exposure to inflammatory macrophages leads to a Nrf2-dependent increase of proteasomal gene expression and proteasome activity in NCM460 cells and as a consequence leads to apoptosis resistance. It was also found that ROS play a particular role in this response of NCM460 cells exposed to inflammatory macrophages.
EXPERIMENTAL PROCEDURES
Materials
IL-1β and IL-6 were from Calbiochem (Mannheim, Germany). TNFα, Tiron, and catalase were purchased from Sigma. TNFα, IL-6, TGFβ, and IL-10 ELISAs were from EBioscience (Frankfurt, Germany), and the IL-1β ELISA was from R & D Systems (Wiesbaden, Germany), and N-succinyl-l-leucyl-l-leucyl-l-valyl-l-tyrosyl-7-amido-4-methylcumarin (Suc-LLVY-AMC) and Mg132 were from Biomol (Taunusstein, Germany). Killer-TRAIL was from Enzo Life Science/Alexis (Lörrach, Germany), and irinotecan was from Pfizer (Berlin, Germany). 5-Carboxy-2′,7′-dichlorodihydro-fluorescein diacetate (c-H2DCF-DA) and MitoSOXTM Red were purchased from Molecular Probes via Invitrogen.
Cell Culture
Human NCM460 colonocytes (39) were provided by INCELL (San Antonio, TX) and cultured in M3A medium (INCELL) containing 10% FCS. The cells were cultured at 37 °C, 5% CO2, and 85% humidity. For coculture, NCM460 cells were seeded onto 12-well plates at a density of 1 × 105 cells/well, incubated for 24 h, and then inserts with 5 × 105 macrophages were added for 6–96 h. For the time of siRNA treatment of NCM460 cells (see below), the inserts were placed in a separate 12-well plate; afterward, the coculture of siRNA-treated NCM460 cells and macrophages continued. Transient exposure of NCM460 cells to H2O2 (200 μm) was conducted in prewarmed PBS for 30 min at standard culture conditions, and then the culture medium was replaced.
Generation and Culture of Macrophages
Fifteen × 106 monocytes (95% purity) generated from human peripheral blood mononuclear cells by counterflow centrifugation were transferred into Teflon bags (VueLife Teflon bags; Süd-Laborbedarf, Gauting, Germany) and cultured in RPMI 1640 (Biochrom, Berlin, Germany) supplemented with 1% l-glutamine, 10% FCS, and 100 μg/ml penicillin/streptomycin (PAA Laboratories, Cölbe, Germany) in the presence of 50 ng/ml GM-CSF or M-CSF (RELIATech, Wolfenbüttel, Germany) for 7 days as described (40, 41). Macrophages were detached by placing the Teflon bags on ice for 1 h, and then the cells were resuspended in ice-cold PBS. The phenotype of the macrophages was verified (supplementary material) by real time RT-PCR and ELISA for determination of the expression of pro-inflammatory (IL-1β, TNFα, and IL-6) versus anti-inflammatory (IL-10 and TGFβ) mediators as well as by CD11b, CD14, and CD163 immunoflow cytometry.
Fluorometric Proteasome Assay
For the determination of proteasomal activity in living cells, a fluorometric assay using the proteasome substrate Suc-LLVY-AMC was performed as described recently (27). All of the measurements were carried out in duplicate.
Determination of Intracellular ROS
The medium of NCM460 cells was replaced by 1 ml of prewarmed PBS, and the cells were treated with 10 μm of the cellular ROS indicator c-H2DCF-DA dissolved in Me2SO or with the vehicle alone for 20 min at 37 °C. Then the labeled NCM460 cells were incubated in OPTIMEM (PAA Laboratories) for 5 min at 37 °C followed by mono- or coculture in OPTIMEM for 1, 3, and 6 h. Fluorescence in response to oxidation of c-H2DCF-DA was measured with the Techan 200 infinite microplate reader. Fluorescence units were normalized to the protein content determined in parallel. To discriminate endogenous generation of mitochondrial ROS, NCM460 cells were stained with 250 nm MitoSOXTM Red for 30 min following the manufacturer's instructions. Then cells were mono- or cultured as above, and stained cells were analyzed with the Techan 200 infinite, as well.
Western Blotting
Preparation of nuclear and cytoplasmic extracts or total cell lysates was carried out as described before (11, 42). After electrophoresis and semi-dry electroblotting onto PVDF membranes, the following primary antibodies were used for immunodetection at a 1000-fold dilution in 5% (w/v) nonfat milk powder, 0.05% Tween 20 in TBS (50 mm Tris/HCl, pH 7.6, and 150 mm NaCl): Nrf2 (Epitomics via Biomol), Hsp90 and Nrf1 (Santa Cruz, Heidelberg, Germany), tubulin (Sigma), S5a and α5 (both from Affiniti/Biomol, Hamburg, Germany), and cleaved caspase-3 and PARP1 (Cell Signaling Technology, Frankfurt, Germany). After incubation overnight at 4 °C, the blots were exposed to the appropriate horseradish peroxidase-conjugated secondary antibody (Cell Signaling Technology) diluted (1:1000) in blocking buffer and developed using the Dura detection kit (Perbio Sciences, Bonn, Germany). Data acquisition was done with the Chemidoc-XRSTM gel documentation system (Bio-Rad) using the Quantity One® software (Bio-Rad). Hsp90 served as a loading control and was used for band density normalization.
RNA Preparation and Real Time PCR
Isolation of total RNA and reverse transcription into single-stranded cDNA was carried out as described (27). cDNA was subjected to real time PCR (iCycler; Bio-Rad) using the SYBR-Green assay with gene-specific primers at a final concentration of 0.2 μm. The primer sequences and PCR conditions for the detection of S5a and α5, as well as β-actin, have been described recently (27). For amplification of Nrf1 and Nrf2, the forward/reverse primers 5′-cggtatgcaacaggacattg-3′/5′-actggttggggtcttctgtg-3′ and 5′-ttccctgcacaggtgcctagtg3′/5′-cttccatggcctgcatttccatg-3′ were used, respectively, and amplification was carried out at 95 °C for 15 s, 56 °C for 30 s, and 72 °C for 20 s. The data were collected during annealing steps and were further analyzed by using the iCycler iQ optical system software (Bio-Rad). All of the samples were analyzed in duplicate, and the data are expressed as the amounts of mRNA normalized to β-actin mRNA.
siRNA Treatment
For siRNA transfection, NCM460 cells grown in 12-well plates were submitted to lipofection using 6 μl of the HiperFect reagent (Qiagen) and 150 ng/well of either negative control siRNA (Qiagen) or Nrf2 (SI03246614; Qiagen), Nrf1 (SI00657909; Qiagen), S5a (SI03019331; Qiagen), or α5 (SI100043316; Qiagen) siRNA. After 8 h, the medium was changed, and cells were subjected to further mono- or coculture.
Dual Luciferase Assay
NCM460 cells grown in 12-well plates were subjected to lipofection using Effectene (Qiagen) and either 0.2 μg of pARE and ptkRL or the empty vector and ptk-RL (all vectors from Qiagen/SABioscience). After 8 h, the medium was changed followed by mono- or coculture for up to 72 h or by treatments as indicated. Afterward, the cells were washed with PBS and lysed in 150 μl/well passive lysis buffer (Promega, Mannheim, Germany). The lysates were centrifuged for 2 min at 4 °C at 13,000 rpm. 20 μl of the supernatant were used for the dual luciferase assay procedure, using Dual-Glow luciferase assay system from Promega. Firefly luciferase expression was normalized to constitutive Renilla luciferase expression. All of the measurements were carried out in duplicate.
Living Cell Microscopy
NCM460 cells grown onto coverslips were stained with 2 μm c-H2DCF-DA and 100 nm MitoSOXTM Red for 5 min at 37 °C in prewarmed PBS. Then cells were incubated with OPTIMEM for 5 min at 37 °C followed by mono- and coculture as specified. Analysis was done with an AxioScan fluorescence microscope (Zeiss, Jena, Germany) equipped with a dip-in objective (40× magnification).
Measurement of Apoptosis
Apoptosis induced by Killer-TRAIL or irinotecan was determined by measurement of caspase-3/7 activity (Promega) and of the generation of the cytokeratin 18 neoepitope (M30-Apoptosense® ELISA kit; PEVIVA, Alexis, Grünwald, Germany) according to the manufacturer's instructions and as described (42). All of the assays were done in duplicate. Caspase-3/7 activity and M30 immunoreactivity were normalized to the protein content of the analyzed cell lysates and expressed as activity/mg protein.
Statistics
The data represent the means ± standard deviation. Significances were calculated by Student's t test with data from at least four independent experiments, a p value <0.05 was considered as statistically significant.
RESULTS
Nrf2 Is Activated in NCM460 Colonocytes Exposed to Inflammatory Macrophages
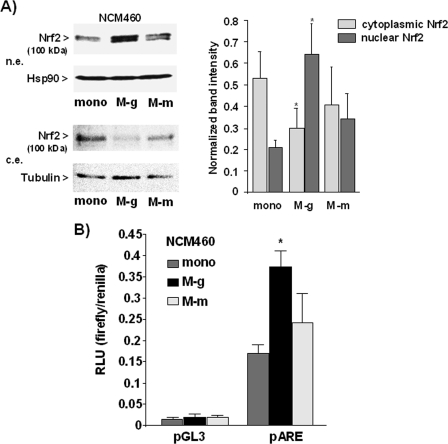
To investigate whether the exposure to macrophages affects the activity of Nrf2 in NCM460 colonocytes, Transwell coculture experiments were conducted using inflammatory macrophages (M1 polarized) derived from elutriated monocytes and differentiated in the presence of GM-CSF for 7 days (43). Verification of the inflammatory phenotype was done by real time PCR analysis and ELISA detecting high level expression of TNFα, IL-1β, and IL-6 and low levels of IL-10 and TGFβ (supplemental Fig. S1). In addition, immunoflow cytometry analysis (supplemental Fig. S2) revealed high CD11b and CD14 expression but the absence of CD163 in these cells indicating M1 polarization (41). After 2 days of coculture with these macrophages (M-g), nuclear protein levels of Nrf2 in NCM460 cells, indicative of its activation, were analyzed by Western blotting. As shown in Fig. 1A, higher Nrf2 protein levels were detectable in nuclear extracts from M-g cocultured NCM460 cells than in nuclear extracts from monocultured NCM460 cells. Conversely, the Nrf2 protein level in cytoplasmic extracts decreased, thus indicating enhanced nuclear accumulation of Nrf2. Next, we looked whether the increased nuclear Nrf2 protein levels correspond to higher Nrf2 transcriptional activity. For this purpose, reporter gene assays using firefly luciferase under the control of the antioxidant response element (ARE) were conducted. As shown in Fig. 1B, M-g cocultured NCM460 cells exhibited a 2–3-fold increase of ARE-driven reporter gene expression in comparison with monocultured NCM460 cells.
FIGURE 1.
Coculture with inflammatory macrophages induces Nrf2 activity in NCM460 cells. Macrophages deriving from monocytes treated with GM-CSF (M-g) or M-CSF (M-m) were used for coculture with NCM460 cells. A, nuclear (n.e.) and cytoplasmic (c.e.) extracts from NCM460 cells cultured for 2 days either in the absence (mono) or presence of these macrophages were submitted to Nrf2 immunoblotting, using Hsp90 and tubulin as loading control. A representative blot (left panel) and the normalized Nrf2 band intensities (right panel) as determined by band densitometry of blots from four independent experiments are shown (means ± S.D.). *, p < 0.05. B, NCM460 cells were transfected with the Nrf2-responsive pARE or the control (pGL3) firefly luciferase reporter vector together with the ptkRL reference vector expressing Renilla luciferase. Afterward, transfectants were mono- or cocultured for 2 days, and NCM460 cell lysates were analyzed for firefly and Renilla luciferase expression. Luminescence induced by firefly luciferase was normalized to the luminescence induced by Renilla luciferase; the data represent the means ± S.D. of four independent experiments performed in duplicate. RLU, relative light units.
To elucidate whether the induction of Nrf2 activation in NCM460 cells depends on the pro-inflammatory phenotype of the macrophages, coculture experiments were also set up using anti-inflammatory macrophages that were obtained from monocytes treated with M-CSF (41). Real time PCR analysis and ELISA (supplemental Fig. S1) revealed lower expression of TNFα, IL-1β, and IL-6 in these M-CSF-derived macrophages (M-m), along with higher expression of IL-10 and TGFβ compared with GM-CSF macrophages. Immunoflow cytometry analysis (supplemental Fig. S2) further detected CD163 positivity of M-CSF macrophages in contrast to GM-CSF macrophages lacking this M2 marker (41). As shown in Fig. 1, after coculture with these macrophages (M-m) both the nuclear and cytoplasmic Nrf2 protein levels as well as Nrf2 transcriptional activity were not significantly altered in NCM460 cells.
Exposure of NCM460 Cells to Inflammatory Macrophages Elevates Proteasome Activity and Proteasomal Gene Expression in a Nrf2-dependent Manner
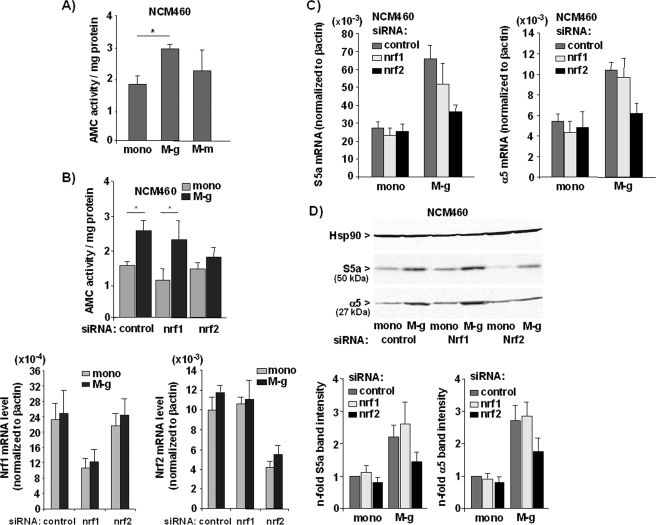
To investigate whether the Nrf2 induction by inflammatory macrophages (M-g) is followed by an altered proteasome activity, NCM460 cells were cocultured for 3 days and then submitted to the AMC-Suc-LLVY proteasome assay. It could be shown that the proteasome activity was significantly increased in M-g cocultured NCM460 cells when compared with monocultured NCM460 cells (Fig. 2A). By contrast, coculture with M-CSF derived macrophages (M-m) did not significantly induce proteasome activity in NCM460 cells (Fig. 2A).
FIGURE 2.
Nrf2-dependent induction of proteasome activity in NCM460 cells by coculture with inflammatory macrophages. A and B, NCM460 cells either monocultured (mono) or cocultured (3 days) with macrophages (M-g and M-m) were submitted to the Suc-LLVY-AMC assay (A) or NCM460 cells treated with control, Nrf1, or Nrf2 siRNA and cultured alone (mono) or with GM-CSF macrophages (M-g) were submitted to the Suc-LLVY-AMC assay (B, upper panel) or real time RT-PCR (B, lower panels). Specific proteasome activity (A and B, upper panel) was determined by the release of fluorogenic AMC normalized to the protein content; the data represent the means ± S.D. of six independent experiments. *, p < 0.05. Real time RT-PCR was conducted using Nrf1, Nrf2, and β-actin primers (B, lower panels), and data from three independent experiments (means ± S.D.) performed in duplicate are shown. C and D, NCM460 cells treated with control, Nrf1, or Nrf2 siRNA and cultured (3 days) alone (mono) or with G-MCSF macrophages (M-g) were submitted to real time PCR using S5a, α5, and β-actin primers (C) or to immunoblotting using S5a, α5, and Hsp90 antibodies (D). Data from three independent experiments (means ± S.D.) performed in duplicate are shown in C, and D shows representative blots and the normalized S5a and α5 band intensities as determined by band densitometry of blots from three independent experiments (means ± S.D.).
The significant induction of proteasome activity after coculture with inflammatory macrophages (M-g), exclusively used in all further coculture experiments, was nearly abrogated in NCM460 cells if Nrf2 expression was specifically knocked down by Nrf2-siRNA treatment (Fig. 2B, upper panel). By contrast, knockdown of Nrf1 only moderately altered proteasome activity in cocultured NCM460 cells indicating that the induced proteasome activity depends on Nrf2, but not on Nrf1. The efficacy of both siRNAs was confirmed by real time RT-PCR (Fig. 2B, lower panel) and Western blot analysis (supplemental Fig. S3). Because the Nrf1 protein is almost undetectable in the absence of proteasome inhibitors (32), lysates for detection of Nrf1 protein were generated from NCM460 cells treated with the proteasome inhibitor Mg132 (supplemental Fig. S3).
Because the Nrf2-dependent increase of proteasome activity relies on the induction of proteasomal gene expression (11, 25, 26, 29), real time RT-PCR analyses of the proteasomal genes S5a/psmd4/Rpn10 and α5/psma5 were performed. As shown in Fig. 2C, S5a and α5 mRNAs were detected at 2–2.8-fold higher level in M-g cocultured NCM460 cells when compared with monocultured NCM460 cells (Fig. 2C). Upon Nrf2 siRNA treatment, the increase of S5a and α5 mRNA level in NCM460 cells cocultured with inflammatory macrophages (M-g) could not be detected anymore, thus confirming the Nrf2 dependence. A similar effect of 0coculture on proteasomal gene expression in NCM460 cells was demonstrated by Western blot analysis detecting higher protein level of S5a and α5 (Fig. 2D). Again, the increasing effect by the coculture was markedly reduced after knockdown of Nrf2 expression in NCM460 cells. Under the same conditions, the knockdown of Nrf1 did not alter the coculture-induced expression of S5a and α5 (Fig. 2, C and D). These findings point to an involvement of Nrf2 but not Nrf1 in the enhanced proteasome activation in colonic cells in the presence of inflammatory macrophages.
The Inducing Effect of Coculture with Inflammatory Macrophages on Proteasome Activity and Proteasomal Gene Expression Depends on ROS
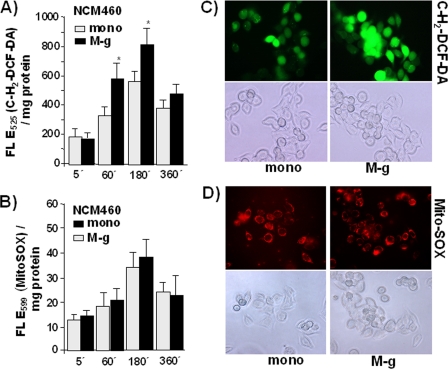
To determine whether coculture with inflammatory macrophages (M-g) increases oxidative stress in NCM460 cells, stainings with the cellular ROS indicator c-H2DCF-DA were performed. Compared with monocultured cells, NCM460 cells exhibited significantly greater fluorescence after M-g coculture for 1 and 3 h (89 and 62% greater fluorescence, respectively), resulting from higher oxidation of the incorporated dye (Fig. 3A). Longer mono- and coculture (6 h) resulted in a decreased fluorescence caused by the decay of the dye, but at this time the differences between mono- and cocultured NCM460 cells turned to be less pronounced, indicating compensation of the oxidative stress in cocultured NCM460 cells. Under the same conditions, MitoSOXTM staining for mitochondrial ROS revealed no significant differences of fluorescence intensities between mono- and M-g cocultured NCM460 cells over the entire period (Fig. 3B). Living cell fluorescence microscopy (Fig. 3C) revealed stronger cellular c-H2DCF-DA staining in cocultured (1 h) than in monocultured NCM460 cells. When staining the cells with MitoSOXTM (Fig. 3D), staining patterns were almost similar, indicating no differences in the mitochondrial ROS activity between cocultured and monocultured NCM460 cells.
FIGURE 3.
The exposure to inflammatory macrophages increases ROS level in NCM460 cells. After labeling with 10 μm cH2DCF-DA (A) or 250 nm MitoSOXTM (B) for 30 min at 37 °C, NCM460 cells were either mono or cocultured (M-g) for the indicated periods. Then fluorescence was determined; the data represent the means ± S.D. of four independent experiments. *, p < 0.05 compared with the corresponding monocultured cells. In parallel, living cell fluorescence microscopy of NCM460 cells stained with 1 μm cH2DCF-DA (C) or 250 nm MitoSOXTM (D) for 10 min at 37 °C followed by mono or coculture for 1 h. Images of 40× magnification and representative results of two independent experiments are shown.
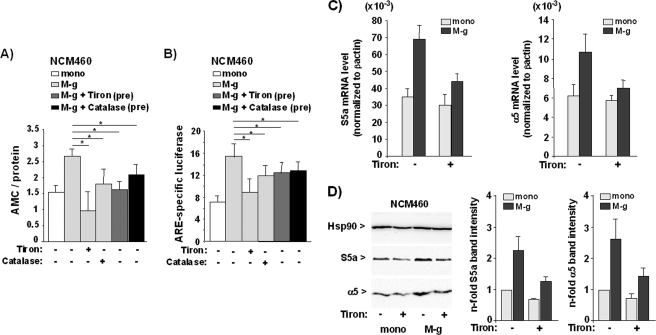
To investigate whether ROS are involved in the induction of Nrf2-dependent proteasome activity in NCM460 cells exposed to inflammatory macrophages, coculture settings were conducted in the absence or presence of the ROS scavenger Tiron (44) or catalase. As shown by AMC assay, the increase of proteasome activity in NCM460 cells cocultured with inflammatory macrophages (M-g) was abolished in the presence of 0.2 mm Tiron (Fig. 4A) and partially reduced by 200 units of catalase. As further shown by ARE luciferase assay, the presence of Tiron suppressed the coculture-induced reporter gene expression (Fig. 4B), and a similar but slightly weaker effect was seen with catalase. When conducting only Tiron or catalase pretreatment of inflammatory macrophages prior to the coculture, the blockade of proteasome and Nrf2 activation in NCM460 cells was still significant (Fig. 4, A and B) as compared with NCM460 cells cocultured with untreated inflammatory macrophages but somewhat less pronounced when compared with the coculture in the presence of Tiron or catalase. Thus, the effect of inflammatory macrophages on Nrf2 and the proteasome in NCM460 cells largely depends on exogenous ROS, but to some extent endogenous ROS may also be involved.
FIGURE 4.
The Nrf2-induced proteasome activity in NCM460 cells by inflammatory macrophages depends on ROS. A, NCM460 cells were either monocultured (mono) or cocultured with G-MCSF macrophages (M-g) for 3 days in the absence or presence of 0.2 mm Tiron or 200 units of catalase, or NCM460 cells were cocultured with G-MCSF macrophages pretreated (pre) with Tiron or catalase for 16 h. Upon either treatment, NCM460 cells were submitted to the Suc-LLVY-AMC assay. The data represent the means ± S.D. of four independent experiments performed in duplicate. *, p < 0.05. B, NCM460 cells either monocultured (mono) or cocultured (M-g) for 3 days in the absence or presence of Tiron or catalase or cocultured with Tiron or catalase pretreated (pre) GM-CSF macrophages (M-g) were transfected with pARE or the control (pGL3) firefly luciferase reporter vector together with ptkRL. After further culture for 24 h, NCM460 cell lysates were analyzed for firefly and Renilla luciferase expression. The data indicate the n-fold ARE-specific firefly luciferase (upon normalization to Renilla luciferase) and represent the means ± S.D. of four independent experiments performed in duplicate. *, p < 0.05. C and D, NCM460 cells cultured either without (mono) or with GM-CSF macrophages (M-g) for 3 days in the absence or presence of Tiron were analyzed for the expression of S5a and α5 by real time PCR (D) and Western blot (E). Data from three independent experiments (means ± S.D.) performed in duplicate are shown in C, and D shows a representative blot and the normalized S5a and α5 band intensities as determined by band densitometry of blots from three independent experiments (means ± S.D.).
Because Tiron was most efficient in the blockade of Nrf2 and proteasome activation, the impact of this ROS scavenger on proteasomal gene expression was analyzed. As shown by real time PCR and Western blotting, the increase of the mRNA and protein level of S5a and α5 in NCM460 cells after coculture was markedly reduced in the presence of the ROS scavenger (Fig. 4, C and D).
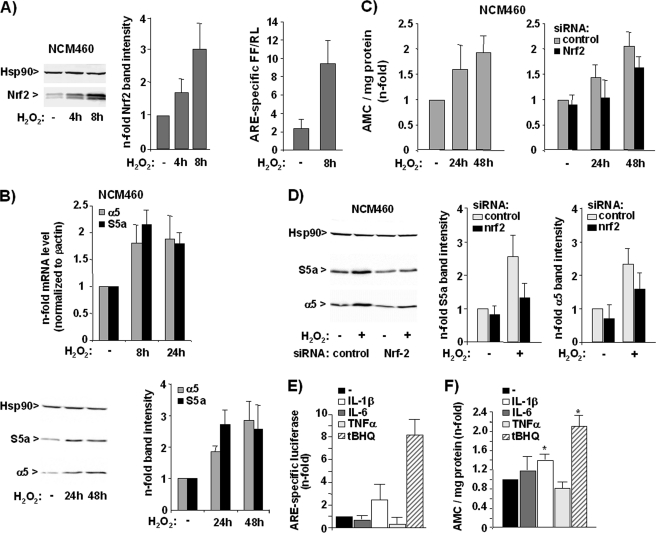
The impact of exogenous ROS on Nrf2-dependent proteasome activation was further verified by the effect of mild oxidative stress imposed on NCM460 cells by transient administration of 200 μm H2O2. As shown in Fig. 5A, the nuclear level of Nrf2 rise in NCM460 cells if treated with H2O2 (left and middle panels) and ARE-dependent luciferase expression markedly increased in NCM460 cells subject of H2O2 treatment (right panel). Real time PCR and Western blot analyses revealed that the expression level of S5a and α5 were elevated in NCM460 cells if treated with H2O2 (Fig. 5B). Moreover, the proteasome activity of NCM460 cells increased 1.5–2-fold in response to transient H2O2 treatment after 24 or 48 h (Fig. 5C, left panel). Knockdown of Nrf2 expression by siRNA transfection diminished the inducing effects of H2O2 on the proteasome activity (Fig. 5C, right panel) and proteasomal gene expression (Fig. 5D).
FIGURE 5.
H2O2 induces Nrf2-dependent proteasome activity in NCM460 cells. A, NCM460 cells were transiently treated with H2O2 at 200 μm, and nuclear extracts were analyzed by immunoblotting using Nrf2 and Hsp90 antibodies (left panel, a representative blot; middle panel, normalized Nrf2 band intensities from three independent experiments; means ± S.D.), or NCM460 cells were transfected with pARE or the control (pGL3) firefly luciferase reporter vector together with ptkRL prior to H2O2 treatment, and then cells were submitted to dual luciferase assay (right panel, data from three independent experiments performed in duplicate are shown). B, NCM460 cells were transiently treated with H2O2 at 200 μm, and reverse transcribed mRNA was submitted to real time PCR using S5a, α5, and β-actin primers (upper panel, mean values from three independent experiments performed in duplicate), or cellular extracts were analyzed by immunoblotting using S5a, α5, and Hsp90 antibodies (lower panel, a representative blot and normalized band intensities from three representative experiments; means ± S.D.). C, untreated NCM460 cells (left panel) or NCM460 cells pretreated with control, Nrf1, and Nrf2 siRNA (right panel) were transiently exposed to H2O2 at 200 μm and then submitted to the Suc-LLVY-AMC assay; data (means ± S.D.) from three independent experiments are shown. D, NCM460 cells treated with control, Nrf1, and Nrf2 siRNA and then subjected to treatment with 200 μm H2O2 for 24 h were analyzed by S5a and α5 Western blotting. A representative blot (left panel) and the normalized S5a and α5 band intensities (right panel) from three independent experiments as determined by band densitometry are shown (means ± S.D.). E and F, NCM460 cells were analyzed by ARE luciferase assay and Suc-LLVY-AMC assay after treatment with the indicated stimuli for 24 h; data (means ± S.D.) from three (E) and five (F) independent experiments performed in duplicate are shown. *, p < 0.05 compared with untreated.
To elucidate whether proteasome activity in NCM460 cells is inducible by other inflammatory mediators, besides exogenous ROS, that are released by inflammatory macrophages (35, 36), ARE luciferase and AMC assays were conducted with NCM460 cells subject of treatment (24 h) with IL-1β, IL-6, and TNFα (20 ng/ml each) as well as with the Nrf2 inducer tBHQ (50 μm) for comparison. As shown in Fig. 5E, the greatest increase of ARE luciferase activity in NCM460 cells could be detected after tBHQ treatment, whereas a lower effect was seen with IL-1β, and there was no effect with IL-6 and TNFα. A greater than 2-fold increase of proteasome activity was observed in NCM460 cells treated with tBHQ (Fig. 5F), and a moderate increase was seen with IL-1β-treated NCM460 cells. By contrast, IL-6 and TNFα did not significantly induce proteasome activity (Fig. 5F) in NCM460 cells.
Increased Resistance to TRAIL- and Irinotecan-induced Apoptosis in NCM460 Cells Exposed to Inflammatory Macrophages
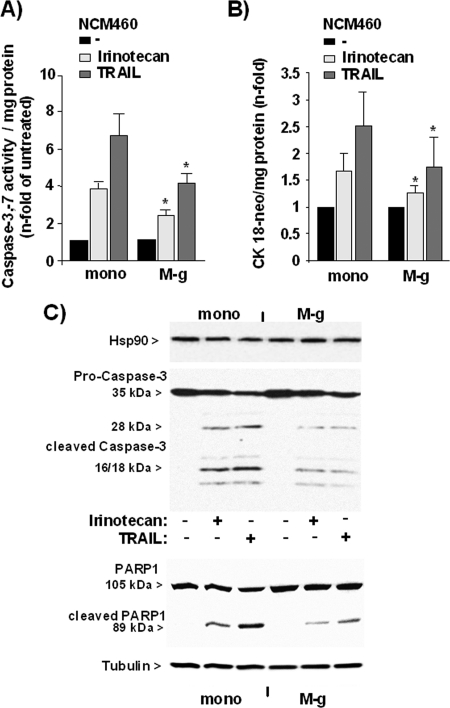
As recently demonstrated, the Nrf2-dependent increase of proteasome activity alters the sensitivity of NCM460 cells to TRAIL-induced apoptosis (11). To elucidate whether exposure of NCM460 cells to inflammatory macrophages (M-g) confers apoptosis protection in such a way, NCM460 cells were cocultured for 3 days followed by treatment with 10 ng/ml TRAIL. It could be shown that the induction of caspase-3/7 activity by TRAIL treatment (Fig. 6A) was significantly lower in M-g cocultured NCM460 cells (4.2 ± 0.6-fold of basal caspase-3/7 activity) than in NCM460 cells cultured without macrophages (6.7 ± 1.1-fold of basal caspase-3/7 activity). Similarly, the caspase-3/7 activation after treatment with the anti-cancer drug irinotecan (Fig. 6A) was lower in NCM460 cells cocultured with inflammatory macrophages (2.4 ± 0.3 versus 3.9 ± 0.4). In line with the caspase assay results, M30 immunoassay detecting an apoptosis-specific cytokeratin-18 neoepitope revealed that the amount of this neoepitope generated from caspase-3- and -7-mediated cleavage after exposition to TRAIL or irinotecan was lower in cocultured than in monocultured NCM460 cells (Fig. 6B). Accordingly, Western blots detected reduced protein levels of cleaved caspase-3 and cleaved PARP1 in TRAIL- or irinotecan-treated NCM460 cells when cocultured with M-g (Fig. 6C).
FIGURE 6.
Coculture with inflammatory macrophages confers apoptosis resistance to NCM460 cells. NCM460 cells either mono- (mono) or cocultured with GM-CSF macrophages (M-g) for 3 days and were then treated with 20 μg/ml irinotecan or 10 ng/ml TRAIL for 24 and 8 h, respectively. A and B, cells submitted to a caspase-3/7 assay (A) or culture supernatants (B) were submitted to the M30 ELISA for the determination of apoptosis. The data represent the means ± S.D. of five independent experiments. *, p < 0.05 compared with the corresponding monocultured cells. C, cell lysates were submitted to Western blots detecting cleaved caspase-3 (upper panel) or detecting PARP1 (lower panel). Hsp90 and tubulin were detected as loading control. Representative results of two independent experiments are shown.
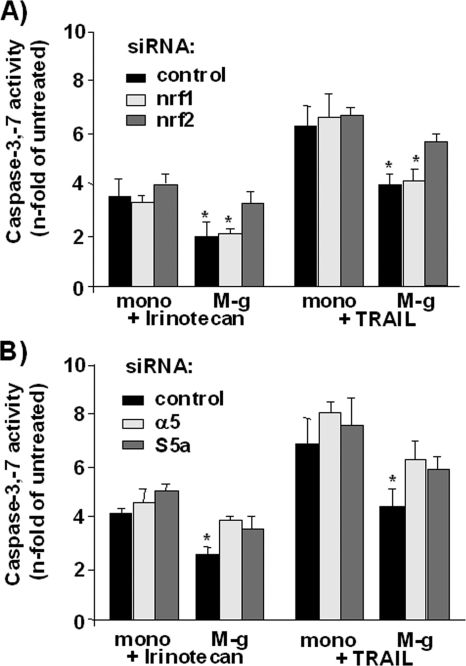
The decrease of TRAIL- or irinotecan-induced apoptosis in NCM460 cells subjected to coculture with inflammatory macrophages was much less pronounced if Nrf2 expression in NCM460 cells was knocked down by siRNA treatment, as determined by caspase-3/7 activity (Fig. 7A). Compared with control siRNA-treated cells exhibiting 4.0 ± 0.4- and 2.0 ± 0.5-fold caspase-3/7 activation by TRAIL and irinotecan, the caspase-3/7 activation in Nrf2 siRNA-treated cells after TRAIL and irinotecan treatment increased to 5.7 ± 0.3- and 3.2 ± 0.4-fold, respectively. No alteration in the resistance inducing effect of the M-g coculture on NCM460 cells against TRAIL- and irinotecan-dependent apoptosis was observed after knockdown of Nrf1 expression (Fig. 7A).
FIGURE 7.
Coculture induced apoptosis resistance in NCM460 cells depends on Nrf2 and proteasomal gene expression. NCM460 cells were transfected with (A) Nrf1 or Nrf2 and (B) α5 or S5a siRNAs and submitted 24 h later to either mono- or coculture (M-g) for 3 days. Then cells were treated with 20 μg/ml irinotecan or 10 ng/ml TRAIL for 24 and 8 h, respectively. The cells were submitted to a caspase-3/7 assay for the determination of apoptosis. All of the data represent the means ± S.D. of five independent experiments. *, p < 0.05 compared with the corresponding monocultured cells.
In addition, NCM460 cells subject to coculture with inflammatory macrophages became also more sensitive to both apoptosis stimuli if they had been treated before with S5a- and α5-specific siRNAs (Fig. 7B). Under these conditions, the irinotecan- or TRAIL-induced caspase-3/7 activity in cocultured NCM460 cells was not significantly different from that in monocultured NCM460 cells.
DISCUSSION
Chronic inflammation plays an important role in the development of cancer, including colorectal cancer (3, 38, 45). Over a long period of time, epithelial cells such as those of the intestinal mucosa are exposed to inflammation-associated stress factors, including ROS released by immune cells infiltrating the inflamed tissue (3, 38). Among these, pro-inflammatory (M1 polarized) macrophages play an important role. These cells are found in high number in the diseased tissue, such as the colonic mucosa, and attract other immune cells such as monocytes, neutrophils, and T-cells to the site of inflammation and activate them (46). In addition to cytokines and chemokines eliciting endogenous ROS production by the epithelial cells, these inflammatory macrophages themselves produce high level of ROS (35, 47, 48) normally contributing to the defense against pathogens. These exogenous ROS also locally affect the surrounding mucosal tissue, including the colonic epithelium, and evoke an adaptation process involving the activation of Nrf2. Accordingly, an increased epithelial Nrf2 activity is widely found along with inflammation (2, 8), and the impairment of Nrf2 activation aggravates inflammatory conditions and manifests in diseases such as inflammatory bowel disease (8, 49, 50).
Our data here demonstrate that colonic NCM460 cells gain an increased Nrf2 activity in the presence of inflammatory (M1) macrophages generated from elutriated monocytes treated with GM-CSF (40, 43) but not in the presence of M-CSF generated macrophages exhibiting a M2 polarized phenotype (41). Depending on the increased Nrf2 activity, M1 macrophage cocultured NCM460 cells express greater level of proteasomal genes and possess greater proteasome activity, giving rise to an enhanced anti-apoptotic protection (Fig. 8). Under coculture conditions, the increase of ROS level in NCM460 cells, mainly resulting from exogenous ROS released by inflammatory macrophages, accounts for the Nrf2-induced proteasome activity in NCM460 cells, as shown by the blocking effects of the ROS scavenger Tiron (44, 51) or catalase. The influence of exogenous ROS is underscored by our observation that the transient exposure of NCM460 cells to moderate concentrations of H2O2 induced Nrf2-dependent proteasome activity and proteasomal gene expression. This finding points to an adaptation of the proteasome function to oxidative stress (52, 53) and is in line with the activation of Nrf2 by H2O2 (44, 54) through Nrf2 translational control (55) and Keap1 oxidation (56) as well as with recent reports demonstrating H2O2-dependent proteasome activation and proteasomal gene expression (57, 58).
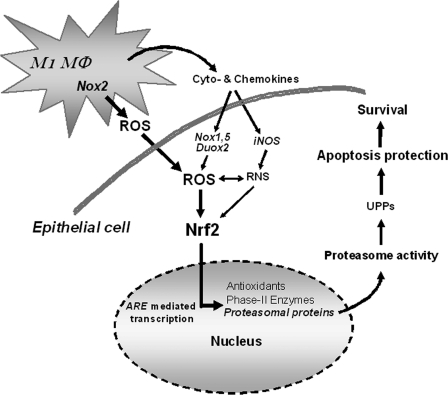
FIGURE 8.
Scheme of the inducing effect of inflammatory macrophages on Nrf2-dependent proteasome activity in epithelial cells. ROS released by inflammatory (M1 polarized) macrophages directly affect neighboring epithelial cells and activate Nrf2. This is followed by activation of ARE-mediated transcription of target genes including proteasomal subunit proteins, which account for an increase of proteasome activity promoting apoptosis protection and thereby cellular survival. In addition, cyto- and chemokines released by the macrophages, as well, may induce endogenous ROS production, e.g. through Nox1 or Duox2 activation (65). Duox, dual oxidase; M1Mφ, M1 macrophage; Nox, NADPH oxidase; RNS, reactive nitrogen species; UPPs, ubiquitin proteasome pathways.
These findings as well as the observation that Nrf1 is not involved in the macrophage-induced proteasome activity in NCM460 cells support the current view that Nrf2 is responsible for the inducible proteasome activity that is part of a cellular stress response, e.g. toward oxidative stress (33) caused by ROS production during inflammation. This property discriminates Nrf2 from Nrf1, which rather constitutively drives proteasomal gene expression and mediates compensation of a dampened proteasome activity under the influence of proteasome inhibitors (31, 32). Because persistent oxidative stress is particularly present during chronic inflammation (2, 3, 38), the inducible proteasome activity under the control of Nrf2 plays a predominant role here and contributes to inflammation-associated carcinogenesis.
Thus, our findings underscore the tumor-promoting potential of Nrf2 that is given by the fact that amplifying mutations in the Keap1/Nrf2 pathway are frequently found in various types of cancer (10) but are in an apparent contrast to the tumor preventing properties that have been attributed to Nrf2, as well. As long as an early Nrf2 activation and subsequent expression of phase II enzymes confer protection from ROS-mediated damage of DNA, proteins, and membrane lipids, epithelial cells exposed to oxidative stress are excluded from molecular alterations, giving rise to a tumorigenic phenotype, e.g. gene mutations. However, an extended induction of Nrf2 leads also to an enhanced proteasome activity that confers several growth advantages to epithelial cells, such as apoptosis protection, and thereby contributes to a tumorigenic phenotype (11, 19–24). Accordingly, Nrf2 when permanently activated, e.g. during chronic inflammation, may turn out to be a proto-oncogene (2, 59), and its targeting by inhibitory compounds may greatly assist in cancer therapy. This includes the resistance of cancer cells against anti-cancer drugs that are often weakened in their efficacy because of detoxification processes mediated in part by phase II enzymes, too (14, 15). Therefore, combined treatment with anti-cancer drugs and Nrf2 blockade may be of great value in cancer therapy (59–61). Moreover, the therapeutic induction of Nrf2 by anti-oxidative phytochemicals, such as sulforaphane, as has been proposed for chemoprevention may turn to be disadvantageous because under these conditions Nrf2-dependent oncogenic mechanisms are triggered, as well (12).
In summary, our findings underscore the potential of Nrf2 to induce proteasomal gene expression and proteasome activity already in nontransformed epithelial cells, such as NCM460 colonocytes. This relates to an anti-oxidative response of epithelial cells against exposure to inflammatory cells, such as inflammatory macrophages, and their released mediators, in particular ROS. Under these conditions, the enhanced proteasome activity in epithelial cells favors cellular survival and thereby a tumorigenic phenotype. In this fashion, Nrf2 might play a crucial role in inflammation-associated carcinogenesis (2, 7), and its therapeutic targeting might be useful for cancer treatment. This is further underlined by a growing number of reports demonstrating an enhanced activity of Nrf2 and its involvement in therapy resistance in various tumor entities, e.g. ovarian, endometrial, pancreas, lung, or colon cancer (60–64). Thus, a better knowledge of the mechanisms leading to an exaggerated Nrf2 activation, either through mutations in the Keap1 pathway or through inflammation-associated oxidative stress, as well as the mechanisms accounting for the oncogenic potential of Nrf2, will significantly add to the understanding of the complex nature of Nrf2-mediated effects and its dual role in human disease.
Supplementary Material
Acknowledgments
We thank Dagmar Leisner for excellent technical assistance and Erika Kaltenhäuser and Christina Trabandt for the preparation of elutriated monocytes.
This work was supported by Deutsche Forschungsgemeinschaft Grant SCHA677/9-1 and the Cluster of Excellence “Inflammation at Interfaces.”

The on-line version of this article (available at http://www.jbc.org) contains supplemental text and Figs. S1–S3.
- ROS
- reactive oxygen species
- c-H2DCF-DA
- 5-carboxy-2′,7′-dichlorodihydro-fluoresceine diacetate
- GM-CSF
- granulocyte monocyte-colony stimulating factor
- Suc-LLVY-AMC
- N-succinyl-l-leucyl-l-leucyl-l-valyl-l-tyrosyl-7-amido-4-methylcumarin
- M-CSF
- monocyte-colony stimulating factor
- M-g
- GM-CSF macrophage
- M-m
- M-CSF macrophage
- tBHQ
- tert-butyl-hydroquinone
- TRAIL
- tumor necrosis factor-related apoptosis-inducing ligand
- ARE
- antioxidant response element.
REFERENCES
- 1. Visconti R., Grieco D. (2009) Curr. Opin. Drug Discov. Devel. 12, 240–245 [PubMed] [Google Scholar]
- 2. Reuter S., Gupta S. C., Chaturvedi M. M., Aggarwal B. B. (2010) Free Radic. Biol. Med. 49, 1603–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ullman T. A., Itzkowitz S. H. (2011) Gastroenterology 140, 1807–1816 [DOI] [PubMed] [Google Scholar]
- 4. Kensler T. W., Wakabayashi N., Biswal S. (2007) Annu. Rev. Pharmacol. Toxicol. 47, 89–116 [DOI] [PubMed] [Google Scholar]
- 5. Osburn W. O., Kensler T. W. (2008) Mutat. Res. 659, 31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hayes J. D., McMahon M., Chowdhry S., Dinkova-Kostova A. T. (2010) Antioxid. Redox Signal. 13, 1713–1748 [DOI] [PubMed] [Google Scholar]
- 7. Singh S., Vrishni S., Singh B. K., Rahman I., Kakkar P. (2010) Free Radic. Res. 44, 1267–1288 [DOI] [PubMed] [Google Scholar]
- 8. Kim J., Cha Y. N., Surh Y. J. (2010) Mutat. Res. 690, 12–23 [DOI] [PubMed] [Google Scholar]
- 9. Zhao C. R., Gao Z. H., Qu X. J. (2010) Cancer Epidemiol. 34, 523–533 [DOI] [PubMed] [Google Scholar]
- 10. Hayes J. D., McMahon M. (2009) Trends Biochem. Sci. 34, 176–188 [DOI] [PubMed] [Google Scholar]
- 11. Arlt A., Bauer I., Schafmayer C., Tepel J., Müerköster S. S., Brosch M., Röder C., Kalthoff H., Hampe J., Moyer M. P., Fölsch U. R., Schäfer H. (2009) Oncogene 28, 3983–3996 [DOI] [PubMed] [Google Scholar]
- 12. Kensler T. W., Wakabayashi N. (2010) Carcinogenesis 31, 90–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lister A., Nedjadi T., Kitteringham N. R., Campbell F., Costello E., Lloyd B., Copple I. M., Williams S., Owen A., Neoptolemos J. P., Goldring C. E., Park B. K. (2011) Mol. Cancer 10, 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lau A., Villeneuve N. F., Sun Z., Wong P. K., Zhang D. D. (2008) Pharmacol. Res. 58, 262–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang X. J., Sun Z., Villeneuve N. F., Zhang S., Zhao F., Li Y., Chen W., Yi X., Zheng W., Wondrak G. T., Wong P. K., Zhang D. D. (2008) Carcinogenesis 29, 1235–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Akhdar H., Loyer P., Rauch C., Corlu A., Guillouzo A., Morel F. (2009) Eur. J. Cancer 45, 2219–2227 [DOI] [PubMed] [Google Scholar]
- 17. Rushworth S. A., Bowles K. M., MacEwan D. J. (2011) Cancer Res. 71, 1999–2009 [DOI] [PubMed] [Google Scholar]
- 18. Du Z. X., Yan Y., Zhang H. Y., Liu B. Q., Gao Y. Y., Niu X. F., Meng X., Wang H. Q. (2011) J. Clin. Endocrinol. Metab. 96, E763–E771 [DOI] [PubMed] [Google Scholar]
- 19. Ren S., Smith M. J., Louro I. D., McKie-Bell P., Bani M. R., Wagner M., Zochodne B., Redden D. T., Grizzle W. E., Wang N., Smith D. I., Herbst R. A., Bardenheuer W., Opalka B., Schütte J., Trent J. M., Ben-David Y., Ruppert J. M. (2000) Oncogene 19, 1419–1427 [DOI] [PubMed] [Google Scholar]
- 20. Chen L., Madura K. (2005) Cancer Res. 65, 5599–5606 [DOI] [PubMed] [Google Scholar]
- 21. Chen W., Hu X. T., Shi Q. L., Zhang F. B., He C. (2009) Oncol. Rep. 21, 531–537 [PubMed] [Google Scholar]
- 22. Rho J. H., Qin S., Wang J. Y., Roehrl M. H. (2008) J. Proteome Res. 7, 2959–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Murata S., Yashiroda H., Tanaka K. (2009) Nat. Rev. Mol. Cell Biol. 10, 104–115 [DOI] [PubMed] [Google Scholar]
- 24. Hellwinkel O. J., Asong L. E., Rogmann J. P., Sültmann H., Wagner C., Schlomm T., Eichelberg C. (2011) Prostate Cancer Prostatic Dis. 14, 38–45 [DOI] [PubMed] [Google Scholar]
- 25. Kwak M. K., Wakabayashi N., Greenlaw J. L., Yamamoto M., Kensler T. W. (2003) Mol. Cell Biol. 23, 8786–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kwak M. K., Kensler T. W. (2006) Biochem. Biophys. Res. Commun. 345, 1350–1357 [DOI] [PubMed] [Google Scholar]
- 27. Arlt A., Minkenberg J., Kruse M. L., Grohmann F., Fölsch U. R., Schäfer H. (2007) Biochem. J. 402, 367–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Malhotra D., Thimmulappa R., Vij N., Navas-Acien A., Sussan T., Merali S., Zhang L., Kelsen S. G., Myers A., Wise R., Tuder R., Biswal S. (2009) Am. J. Respir. Crit. Care Med. 180, 1196–1207 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29. Schaedler S, Krause J., Himmelsbach K., Carvajal-Yepes M., Lieder F., Klingel K., Nassal M., Weiss T. S., Werner S., Hildt E. (2010) J. Biol. Chem. 285, 41074–41086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kapeta S, Chondrogianni N, Gonos E. S. (2010) J. Biol. Chem. 285, 8171–8184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Radhakrishnan S. K., Lee C. S., Young P., Beskow A., Chan J. Y., Deshaies R. J. (2010) Mol. Cell 38, 17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Steffen J., Seeger M., Koch A., Krüger E. (2010) Mol. Cell 40, 147–158 [DOI] [PubMed] [Google Scholar]
- 33. Xie Y. (2010) Cancer Metastasis Rev. 29, 687–693 [DOI] [PubMed] [Google Scholar]
- 34. Nguyen T., Nioi P., Pickett C. B. (2009) J. Biol. Chem. 284, 13291–13295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Laskin D. L. (2009) Chem. Res. Toxicol. 22, 1376–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martinez F. O., Sica A., Mantovani A., Locati M. (2008) Front. Biosci. 13, 453–461 [DOI] [PubMed] [Google Scholar]
- 37. Mosser D. M, Edwards J. P. (2008) Nat. Rev. Immunol. 8, 958–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Roessner A., Kuester D., Malfertheiner P., Schneider-Stock R. (2008) Pathol. Res. Pract. 204, 511–524 [DOI] [PubMed] [Google Scholar]
- 39. Moyer M. P., Manzano L. A., Merriman R. L., Stauffer J. S., Tanzer L. R. (1996) In Vitro Cell Dev. Biol. Anim. 32, 315–317 [DOI] [PubMed] [Google Scholar]
- 40. Krausgruber T., Blazek K., Smallie T., Alzabin S., Lockstone H., Sahgal N., Hussell T., Feldmann M., Udalova I. A. (2011) Nat. Immunol. 12, 231–238 [DOI] [PubMed] [Google Scholar]
- 41. Verreck F. A., de Boer T., Langenberg D. M., Hoeve M. A., Kramer M., Vaisberg E., Kastelein R., Kolk A., de Waal-Malefyt R., Ottenhoff T. H. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4560–4565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Müerköster S., Arlt A., Sipos B., Witt M., Grossmann M., Klöppel G., Kalthoff H., Fölsch U. R., Schäfer H. (2005) Cancer Res. 65, 1316–1324 [DOI] [PubMed] [Google Scholar]
- 43. Fleetwood A. J., Lawrence T., Hamilton J. A., Cook A. D. (2007) J. Immunol. 178, 5245–5252 [DOI] [PubMed] [Google Scholar]
- 44. Pi J., Qu W., Reece J. M., Kumagai Y., Waalkes M. P. (2003) Exp. Cell Res. 290, 234–245 [DOI] [PubMed] [Google Scholar]
- 45. O'Connor P. M., Lapointe T. K., Beck P. L., Buret A. G. (2010) Inflamm. Bowel Dis. 16, 1411–1420 [DOI] [PubMed] [Google Scholar]
- 46. Wendelsdorf K., Bassaganya-Riera J., Hontecillas R., Eubank S. (2010) J. Theor. Biol. 264, 1225–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Keshavarzian A., Banan A., Farhadi A., Komanduri S., Mutlu E., Zhang Y., Fields J. Z. (2003) Gut 52, 720–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Baumgart D. C., Carding S. R. (2007) Lancet 369, 1627–1640 [DOI] [PubMed] [Google Scholar]
- 49. Khor T. O., Huang M. T., Kwon K. H., Chan J. Y., Reddy B. S., Kong A. N. (2006) Cancer Res. 66, 11580–11584 [DOI] [PubMed] [Google Scholar]
- 50. Arisawa T., Tahara T., Shibata T., Nagasaka M., Nakamura M., Kamiya Y., Fujita H., Yoshioka D., Okubo M., Sakata M., Wang F. Y., Hirata I., Nakano H. (2008) Hepatogastroenterology 55, 394–397 [PubMed] [Google Scholar]
- 51. Han Y. H., Park W. H. (2009) Oncol. Rep. 21, 253–261 [PubMed] [Google Scholar]
- 52. Thomas S., Kotamraju S., Zielonka J., Harder D. R., Kalyanaraman B. (2007) Free Radic. Biol. Med. 42, 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Grimm S., Höhn A., Grune T. (2010) Amino Acids, in press [DOI] [PubMed] [Google Scholar]
- 54. Angeloni C., Motori E., Fabbri D., Malaguti M., Leoncini E., Lorenzini A., Hrelia S. (2011) Am. J. Physiol. Heart Circ. Physiol. 300, H2196–2205 [DOI] [PubMed] [Google Scholar]
- 55. Purdom-Dickinson S. E., Sheveleva E. V., Sun H., Chen Q. M. (2007) Mol. Pharmacol. 72, 1074–1081 [DOI] [PubMed] [Google Scholar]
- 56. Fourquet S., Guerois R., Biard D., Toledano M. B. (2010) J. Biol. Chem. 285, 8463–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gomes-Marcondes MC, Tisdale M. J. (2002) Cancer Lett. 180, 69–74 [DOI] [PubMed] [Google Scholar]
- 58. Pickering A. M., Koop A. L., Teoh C. Y., Ermak G., Grune T., Davies K. J. (2010) Biochem. J. 432, 585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Taguchi K., Motohashi H., Yamamoto M. (2011) Genes Cells 16, 123–140 [DOI] [PubMed] [Google Scholar]
- 60. Shim G. S., Manandhar S., Shin D. H., Kim T. H., Kwak M. K. (2009) Free Radic. Biol. Med. 47, 1619–1631 [DOI] [PubMed] [Google Scholar]
- 61. Jiang T., Chen N., Zhao F., Wang X. J., Kong B., Zheng W., Zhang D. D. (2010) Cancer Res. 70, 5486–5496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hong Y. B., Kang H. J., Kwon S. Y., Kim H. J., Kwon K. Y., Cho C. H., Lee J. M., Kallakury BV, Bae I. (2010) Pancreas 39, 463–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Solis L. M., Behrens C., Dong W., Suraokar M., Ozburn N. C., Moran C. A., Corvalan AH, Biswal S., Swisher S. G., Bekele B. N., Minna J. D., Stewart D. J., Wistuba I. I. (2010) Clin. Cancer Res. 16, 3743–3753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ebert B., Kisiela M., Wsól V., Maser E. (2011) Chem. Biol. Interact. 191, 239–249 [DOI] [PubMed] [Google Scholar]
- 65. Bedard K., Krause K. H. (2007) Physiol. Rev. 87, 245–313 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.