Background: Putative host regulators of inflammation, like the recently described primate-restricted POP2, have not been well studied.
Results: POP2 suppresses TNFα and IL-1β responses in macrophages, a function that requires its first 19 residues.
Conclusion: POP2 employs dual regulation of NF-κB and inflammasome functions to restrict inflammatory cytokines.
Significance: Knowledge regarding host molecule limitation of inflammatory signals is fundamental to understanding innate immunity.
Keywords: Cytokine, Inflammation, Innate Immunity, Macrophages, NF-κB, Toll-like Receptors (TLR), Inflammasome, Interleukin-1β, Nod-like Receptors (NLRs), Pyrin Domain
Abstract
Activation of transcription factor NF-κB and inflammasome-directed caspase-1 cleavage of IL-1β are key processes in the inflammatory response to pathogen or host-derived signals. Pyrin-only proteins (POPs) are restricted to Old World monkeys, apes, and humans and have previously been shown to impair inflammasome assembly and/or NF-κB p65 transcriptional activity in transfected epithelial cells. However, the biological role of POP2 and the molecular basis for its observed functions are not well understood. In this report we demonstrate that POP2 regulates TNFα and IL-1β responses in human monocytic THP-1 cells and in stable transfectants of mouse J774A.1 macrophages. Deletion analysis of POP2 revealed that the first α-helix (residues 1–19) is necessary and sufficient for both inflammasome and NF-κB inhibitory functions. Further, key acidic residues Glu6, Asp8, and Glu16, believed critical for Pyrin/Pyrin domain interaction, are important for inflammasome inhibition. Moreover, these mutations did not reduce the effect of POP2 upon NF-κB, indicating that the inflammasome and NF-κB inhibitory properties of POP2 can be uncoupled mechanistically. Collectively, these data demonstrate that POP2 acts as a regulator of inflammatory signals and exerts its two known functions through distinct modalities employed by its first α-helix.
Introduction
Proinflammatory cytokines and chemokines such as TNFα, IL-6, IL-1β, and IL-8 mediate inflammatory and immune responses during microbial infection, cellular stress, and injury (1, 2). Although inflammation constitutes an integral arm of host immunity and cellular homeostasis, exaggerated or chronic inflammation is deleterious, as seen in arthritis, inflammatory bowel disease, diabetes, and cancer. This potential for inflammation-mediated damage to the host signifies the need for cellular mechanisms to maintain a balance between beneficial and harmful responses.
Host immune and inflammatory responses to pathogens are initiated by the recognition of pathogen-associated molecular patterns by pattern recognition receptors (3) such as cell surface or endosomal membrane-localized Toll-like receptors (TLRs)2 (4). One consequence of TLR signaling is the activation of NF-κB, a critical step in the transcriptional induction of inflammatory and immune response genes. The mammalian NF-κB family comprises transcription factors acting either as homo- or heterodimers; these include NF-κB1 (p105, p50), NF-κB2 (p100, p52), RelA (p65), RelB, and c-Rel (5, 6). Canonical NF-κB signaling is mediated by the p50/p65 heterodimer, the most extensively characterized among the NF-κB transcription factors.
Similar to TLRs, the nucleotide-binding leucine-rich receptor family (NLRs; also known as NALPs, NODs, or CATERPILLER) constitute a second line of immune sensors (7–10). NLRs sense cytosolic pathogens, pathogen products, and environmental and host-derived stress signals (11, 12). Most NLRs contain an N-terminal Pyrin domain (PYD) or a caspase activation and recruitment domain (CARD). These domains belong to the death domain superfamily, characterized by a highly similar secondary structure of five or six α-helices (13–15). Some NLRs are crucial players in the production of the biologically active, mature form of the proinflammatory cytokines IL-1β and IL-18. In contrast to TNFα, IL-6, and IL-8, which are regulated predominantly at the transcript level, IL-1β and IL-18 are synthesized pro-forms requiring processing by caspase-1 (16). Caspase-1 activation results from assembly of the inflammasome, a multiprotein scaffold initiated by certain NLRs (17).
Inflammasome assembly by PYD-containing NLRs (NLRPs) requires PYD/PYD-mediated recruitment of the inflammasome adaptor, ASC (apoptotic speck-like protein containing a CARD) followed by caspase-1 recruitment through CARD/CARD interactions between ASC and caspase-1. The most extensively characterized NLRP inflammasome is that initiated by mouse Nlrp3 during bacterial, viral, or fungal infections (18, 19). The Nlrp3 inflammasome is also activated following exposure to toxins such as nigericin and endogenous danger signals such as ATP and gout-associated monosodium urate (MSU) crystals (20, 21).
In humans, single-nucleotide mutations in the NLRP3 gene are associated with hereditary autoinflammatory diseases such as familial cold urticaria and Muckle-Wells syndrome (22–24), resulting from aberrant inflammasome activity. High levels of circulating IL-1β, IL-18, TNFα, and IFNγ are characteristic of these auto-inflammatory diseases. Such observations highlight the crucial role of host mechanisms in regulating cellular inflammatory responses.
Pyrin domain-only proteins (POPs) and CARD domain-only proteins (COPs) have the potential to disrupt PYD/PYD and CARD/CARD interactions, respectively. Examples of COPs include Pseudo-ICE/COP (25, 26), ICEBERG (25, 27), and INCA (28), known to interfere with caspase-1 activation. Pathogen-encoded POPs like myxoma virus M13L subvert host immune responses by inhibiting NF-κB and caspase-1 activation, leading to higher viral burdens and pathogenesis (29). Similarly, in human cells POPs target NF-κB (POP1 and POP2) and caspase-1 activation via disruption of inflammasome assembly (POP2) (30, 31).
POP2 is a 294-nucleotide single-exon gene on chromosome 3q28 encoding an ∼12-kDa protein with largely diffuse or cytosolic localization. Genome-wide analysis indicates that the POP2 gene is restricted to Old World monkeys, apes, and humans and accordingly absent in rats and mice (32). In humans, although POP2 is expressed at low levels in many hematopoeitic cell types including monocytes, POP2 is more highly expressed in lipopolysaccharide- or phorbol ester-treated monocytes (30, 31). In contrast to POP1, which inhibits IκBα kinases (IKK) (33), POP2 inhibits NF-κB signaling at the level of p65 (RelA) downstream of the IKK complex, resulting in less nuclear NF-κB (30). POP2 also blocks the association of several NLRPs with the inflammasome adaptor ASC, thus limiting inflammasome activation (30, 31). However, given its recent discovery, the cellular consequences and molecular basis of POP2-mediated NF-κB and inflammasome regulation have not been well studied.
In the current study we demonstrate that induction of POP2 leads to a reduction in the inflammatory cytokoines TNFα and IL-1β and provide molecular insight into the seemingly disparate functions of POP2. Specifically, the first N-terminal helix of the POP2 six α-helical bundle structure is both necessary and sufficient for NF-κB p65 and inflammasome inhibition. Further, inflammasome inhibition by POP2 relies upon specific acidic residues within the α1 region, which are not required for NF-κB p65 inhibition. Thus, the two functions of POP2, although encoded in the same region, can be uncoupled mechanistically. Using stable expression of wild-type and functionally sufficient (or impaired) POP2 mutant(s) in the J774A.1 macrophage cell line, which natively lacks the POP2 gene, we have confirmed our molecular findings and also shown that POP2 acts as a potent modifier of the TLR/NF-κB pathway and the NLRP3 inflammasome.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Lipopolysaccharide (LPS) from Escherichia coli serotype O26:B6 was from Sigma; recombinant human TNFα from BD Biosciences; and nigericin, ATP, MSU crystals, and Pam3-CSK4 from Invivogen. Antibodies used were mouse anti-Myc IgG1 (clone 4A6, Millipore), mouse anti-Myc IgG2a (clone 9B11, Cell Signaling), rabbit anti-GFP (Santa Cruz Biotechnology), mouse anti-GAPDH (Santa Cruz Biotechnology), HRP-conjugated anti-mouse or anti-rabbit IgG (Sigma), and FITC-labeled goat anti-mouse IgG2a (Invitrogen).
Tissue Culture Cells, Conditions, and Transfection
Human embryonic kidney epithelial cell lines (HEK293T and HEK293) and mouse macrophage cell line J774A.1 cells (American Type Culture Collection) were cultured in Dulbecco's modified Eagle's medium (with 4.5 g/liter glucose) supplemented with 10% FBS, 5 mm l-glutamine, and 0.1% penicillin/streptomycin. All cells were grown at 37 °C with 5% CO2. Cell numbers and viability were determined by trypan blue exclusion. All transfections were performed using FuGENE 6 (2.5 μl:1 μg of DNA; Roche Applied Science) as per the manufacturer's instructions.
Plasmid Constructs and Mutagenesis
Plasmids encoding the N-terminal Myc-tagged wild-type POP2 and GFP-POP2 fusion proteins have been described previously (30). GAL4-p65 TA1 (34) and GAL4-luciferase (35) plasmids have been described previously. QuikChange mutagenesis (Stratagene) was used to generate Myc-POP2 C-terminal deletion mutants (stop codons (TAA or TAG) were inserted after amino acids Ser19, Gly51, Ser64, and Gln80 (Fig. 3A)) and the substitution mutants A2V, E6Q, L7M, D8G, E16A, and ELD to QMG (residues 6–8). All mutagenesis primer sequences used in the study are provided in supplemental Table 1. pCMV-SPORT6 containing NLRP2 was purchased from Origene. The NLRP2-(1–19) mutant was generated using the primer set for POP2 α1, as the corresponding primer binding sites in POP2 and NLRP2 are identical. POP2 Δα1 was generated by inserting BamHI restriction sites after amino acids Met1 and Glu16 followed by restriction digestion and ligation of the purified DNA fragments with T4 DNA ligase (New England Biolabs). The sequences of all DNA amplification-generated constructs were confirmed by fluorescent dye terminator sequencing (GENEWIZ).
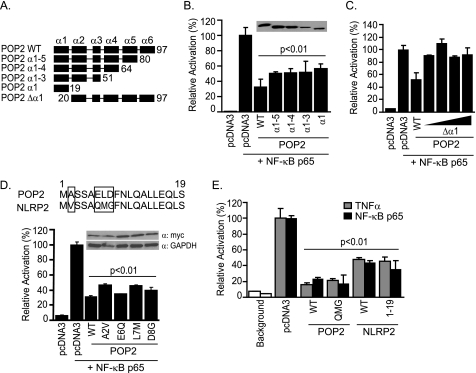
FIGURE 3.
POP2 helix-1 is necessary and sufficient to inhibit NF-κB p65 activity. A, schematic of POP2 deletion mutants showing individual α-helices. B—D, 293T cells were co-transfected with NF-κB luciferase and NF-κB p65 in the presence or absence of WT and POP2 deletion mutants (B), increasing amounts of POP2 Δα1 lacking helix-1 (C), and the indicated POP2 point mutants (D). Alignment of POP2 α1 and the first 19 residues in human NLRP2 with non-conserved residues is shown in box (D). E, NF-κB luciferase assay in 293T cells following TNFα stimulation (open bars) or NF-κB p65 co-transfection (closed bars) in cells expressing WT or mutant versions of POP2 and NLRP2. Results are representative of at least three experiments (error bars, S.D.).
Luciferase Assay
NF-κB luciferase reporter assays were performed as described previously (32). Briefly, 2 × 105 HEK293 cells/well were seeded in 6-well plates. Following overnight incubation, cells were transfected with 100 ng of 3× NF-κB luciferase reporter and 50 ng of NF-κB p65 with 1 μg of wild-type Myc-POP2, mutant POP2, or empty vector. At 18–20 h post-transfection, cells were lysed with 1× reporter lysis buffer (Promega), and luciferase activity was measured using a Victor3V luminometer (PerkinElmer Life Sciences) and normalized to total protein. For TNFα-induced NF-κB activation, cells were stimulated with TNFα (10 ng/ml) for 3 h. The total quantity of DNA in each transfection was kept constant by the addition of empty vector (pcDNA3). GAL4-based luciferase assays were performed similarly by transfection of 100 ng of GAL4-p65 TA1 and 100 ng of GAL4-luciferase in the presence or absence of 1 μg of POP1 or POP2.
Inflammasome Reconstitution Assay
HEK293T cells were used to reconstitute the inflammasome as described previously (32). Cells were seeded at 5 × 104 293T cells/well in 24-well plates. Following overnight culture, the cells were transfected with plasmids encoding pro-caspase-1 (50 ng), pro-IL-1β (200 ng), ASC (10 ng or 400 ng), and NLRP3 (100 ng) in the absence or presence of wild-type or POP2 mutants (500 ng). Approximately 20 h post-transfection, culture supernatants were harvested, centrifuged briefly to remove cellular debris, and used for the measurement of secreted IL-1β by ELISA (Invitrogen).
Francisella novicida Culture and Infection
F. novicida strain U112, obtained from the Microbiology Core Facility at Albany Medical College, was cultured on modified Mueller-Hinton (MH) agar plates or in modified MH broth (Difco Laboratories) as described previously (36). For infection, cells were infected with 100 bacteria/cell (multiplicity of infection = 100) for 24 h in antibiotic-free medium.
Generation and Flow Cytometric Screening of J774A.1 Stable Transfectants
5 × 106 cells were suspended in 500 μl of serum-free DMEM and electroporated with 10 μg of wild-type or POP2 mutant plasmid using a GenePulser (Bio-Rad; 570 V, 25 microfarads, and time constant ∼0.9). Cells were transferred immediately to a 150-mm Petri dish containing complete culture medium. After 2 days, transfectants were selected using 1 mg/ml G418 (Invitrogen) with a complete change of culture medium every 2–3 days for 2 weeks. Stable transfectants were harvested by scraping without trypsin treatment, and ∼1 × 106 cells were fixed with paraformaldehyde (2%) at room temperature for 10 min followed by permeabilization with PBS/Triton X-100 (0.05%) on ice for 30 min. Cells were then stained with anti-Myc IgG2a Ab (Cell Signaling, clone 9B11; 1:500) at room temperature for 1 h in 100 μl of assay buffer (PBS/0.5% bovine serum albumin). Cells were washed twice with assay buffer and incubated with FITC-conjugated secondary Ab (Invitrogen; 1:100) at room temperature for 30 min. After washing twice with assay buffer, cells were suspended in 500 μl of assay buffer and analyzed using a FACSCanto flow cytometer (BD Biosciences). Results were analyzed with Flo-Jo 7.2.2 software (Tree Star Inc., Ashland, OR). To avoid clonal effects, individual clones were collected, pooled, and maintained as a single population in culture medium containing 0.25 mg/ml G418 as the selection antibiotic.
siRNA Knockdown in THP-1 Cells
2.5 × 105 cells/well were seeded in 24-well plates and transfected with either scrambled or POP2-specific siRNA using FuGENE 6 (Roche Applied Science). Briefly, 3 μl of FuGENE 6 was incubated in 50 μl of serum-free RPMI medium for 5 min and then added to the siRNA duplex. The siRNA/FuGENE mix was incubated for 30 min and added to each well. Cells were incubated for 24 h before overnight treatment with 100 nm phorbol 12-myristate 13-acetate (PMA). Cells were washed and stimulated with 100 ng/ml LPS for 6 h, and the culture supernatant was collected for human TNFα and IL-1β quantification using a cytometric bead array (CBA) flex set (BD Biosciences). The siRNA sequences used were: POP2 siRNA (sense, 5′-GGA AGC AAC UGG UAG AAA U-3′; antisense, 5′-AUU UCU ACC AGU UGC UUC C-3′) and scrambled siRNA (sense, 5′-AUU CUC UCA UGA CGU UUC C-3′; antisense, 5′-GGA AAC GUC AUG AGA GAA U-3′).
RNA Isolation and Quantitative Real-time-PCR
Total RNA was isolated using RNeasy RNA purification columns (Qiagen) following the manufacturers protocols and treated with DNase I to remove any residual genomic DNA. Quantitative real-time PCR was performed using the SuperScript III Platinum SyBR Green one-step qRT-PCR kit (Invitrogen) and a CFX96 real-time PCR detection system instrument (Bio-Rad). Intron-spanning primer sequences used in this study were: mouse IL-1β (forward, 5′-CAG GCA GGC AGT ATC ACT CA-3′; reverse, 5′-GAG GAT GGG CTC TTC TTC AA-3′); mouse TNFα (forward, 5′-TCG TAG CAA ACC ACC AAG TG-3′; reverse, 5′-CCT TGT CCC TTG AAG AGA ACC-3′); human IL-8 (forward, 5′-GCT CTG TGT GAA GGT GCA GT-3′; reverse, 5′-CCA GAC AGA GCT CTC TTC CA-3′); and β-actin (forward, 5′-CCC CCA TGC CAT CCT GCG TCT G-3′; reverse, 5′-CTC GGC CGT GGT GGT GAA GC-3′). POP2 real-time primers were: forward, 5′-TTC TGC AGA GCT GGA CTT CA-3′; and reverse, 5′-TGC TTC CCA TTA GCC TTG TC-3′. All reactions were run in triplicate and the specificity of PCR amplification was analyzed by melting curve analysis. Ct values were normalized to β-actin, and the relative copy number was calculated using the standard 2(−ΔΔCt) method. Results are represented as -fold change over untreated control.
Mouse TNFα and IL-1β Measurement
J774A.1 cells expressing pcDNA3 control or different versions of POP2 were seeded at 2.5 × 105 cells/well in 24-well plate. Following overnight incubation, cells were stimulated with 50–100 ng/ml LPS or Pam3-CSK4 for 18–20 h, and the amount of TNFα produced in culture supernatant was measured using a CBA mouse flex set (BD Biosciences). For IL-1β assays, cells were primed with LPS (200 ng/ml) for 3 h followed by treatment with ATP (100 mm) for 30 min, nigericin (10 μm) for 30 min, and MSU (100 μg/ml) for 1 h. Cells were washed twice with PBS and further incubated in fresh culture medium for 6–8 h. The amounts of IL-1β and TNFα were measured using cytokine-specific CBA flex sets as described above. Samples were analyzed using a FACSArray Bioanalyzer and FCAP Array software (BD Biosciences).
FLICA Assay
Caspase-1 activation was detected using carboxyfluorescein-YVAD-fluoromethyl ketone, a caspase-1 FLICATM kit (ImmunoChemistry Technologies, Bloomington, IN), which fluorescently labels activated caspase-1. Briefly, 1 × 106 cells were pulsed with 5 mm ATP for 30 min, washed twice, and labeled with the cell-permeable fluorescent FLICA dye for 1 h as per the manufacturer's instruction. Fluorescence intensity proportional to active capase-1 activity was followed by flow cytometry and analysis using the software described above.
Western Blot
Detection of Myc- or GFP-POP2 by Western blot was performed as described previously (30). Following transfection and lysis in radioimmune precipitation assay buffer (50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1% Triton X-100 (v/v), 2 mm EDTA, and 2 mm DTT supplemented with protease inhibitor (Roche Applied Science)), equal amounts of protein (20–30 μg) were resolved by SDS-PAGE (4–20%; Bio-Rad) and transferred to nitrocellulose membrane (0.2 μm) at 100 V for 1 h. The membrane was probed with anti-Myc (Millipore; 1:500) or anti-GFP (Santa Cruz; 1:1000) and anti-GAPDH antibody (Santa Cruz; 1:500) followed by detection with the HRP-conjugated goat anti-mouse or anti-rabbit secondary antibody (Sigma; 1:5000). Western blots were developed using SuperSignal Pico chemiluminescent substrate (Thermo Scientific).
Statistical Analysis
Experiments were repeated at least three times unless indicated otherwise. Statistical significance between experimental groups was measured by two-tailed Student's t test with p < 0.05 considered significant.
RESULTS
Expression and Function of POP2 in THP-1
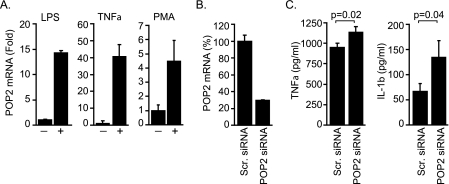
Initial reports identifying the POP2 gene revealed that POP2 interferes with NF-κB signaling and the formation of the inflammasome complex in epithelial cells; thus POP2 is a potential regulator of inflammation (30, 31). However, the impact of POP2 upon proinflammatory cytokine production in monocyte/macrophage cells is not known. Because regulators of inflammation should be expressed during an inflammatory response, we first examined the expression of endogenous POP2 in human monocytic THP-1 cells upon exposure to proinflammatory stimuli. Treatment with E. coli LPS or TNFα led to roughly a 15- or 40-fold increase, respectively, in POP2 mRNA expression as detected by real-time PCR (Fig. 1A). POP2 expression was also induced by ∼5-fold in THP-1 cells following activation with PMA (Fig. 1A), a strong NF-κB inducer (37). Next, we determined the impact of siRNA-mediated POP2 mRNA knockdown upon TNFα and IL-1β production in THP-1 cells. Transfection of POP2-specific siRNA resulted in an ∼70% reduction in POP2 mRNA expression compared with scrambled siRNA-transfected cells (Fig. 1B). In these experiments we could not verify the decrease in POP2 protein level, as a POP2-specific antibody is currently not available. Knockdown of POP2 resulted in a modest, but significant, increase in LPS-induced TNFα and IL-1β production compared with scrambled siRNA-transfected THP-1 cells (Fig. 1C). Although the impact of siRNA knockdown of POP2 upon cytokine response was modest, these results are consistent with a role for POP2 in regulating and/or fine-tuning inflammatory responses. Importantly, these results are also consistent with the previously reported NF-κB and inflammasome inhibitory functions of POP2. Together, these results led us to conclude that POP2 expression is induced in response to a variety of inflammatory signals and that POP2 negatively regulates TNFα and IL-1β production in response to LPS.
FIGURE 1.
Expression and function of POP2 in THP-1 cells. A, quantitative real-time RT-PCR showing POP2 mRNA expression in THP-1 cells stimulated with LPS or TNFα (+) for 1 h and with PMA for 12 h. Results are represented as -fold induction over untreated controls (−). All reactions were run in triplicates, and values were normalized to β-actin as internal control. B, POP2 mRNA knockdown in THP-1 cells following transfection with POP2-specific siRNA or scrambled (Scr.) siRNA as control. C, siRNA-transfected THP-1 cells were treated overnight with PMA, washed, and stimulated with LPS for 6 h. TNFα and IL-1β production was measured in culture supernatant using CBA and is shown as the mean ± S.E. of three experiments.
Stable Expression of POP2 in Mouse J774A.1 Macrophage Regulates Cytokine Response and Caspase-1 Activation
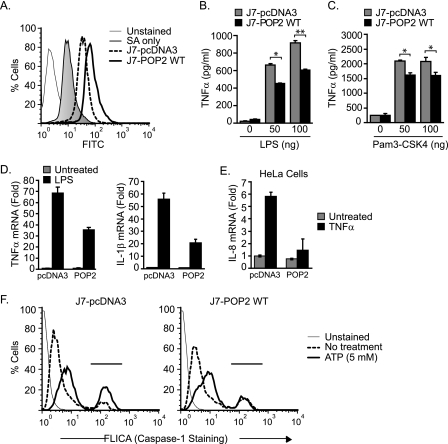
To further investigate a role for POP2 in regulating inflammatory responses, we employed a gain-of-function approach in the J774A.1 mouse macrophage cell line. As the mouse genome lacks a POP2 ortholog (32), J774A.1 cells provide an ideal system for gain-of-function studies in a more biologically relevant context. J774A.1 cells stably expressing Myc-pcDNA3 (control; J7-pcDNA3) or wild-type Myc-POP2 (J7-POP2) were generated, and POP2 expression was confirmed by intracellular staining (Fig. 2A). Using this system, we examined the potential of POP2 to limit production of the TLR-driven, NF-κB-dependent TNFα response. Compared with control cells, J7-POP2 cells produced significantly lower levels of TNFα in response to LPS stimulation, a potent TLR4 agonist (Fig. 2B). J7-POP2 cells also displayed a reduced TNFα response following treatment with Pam3-CSK4, a TLR2 agonist (Fig. 2C). Consistent with the ability of POP2 to inhibit NF-κB, these results demonstrate that POP2 regulates both TLR2- and TLR4-mediated induction of TNFα response. As expected, J7-POP2 cells also had reduced TNFα mRNA induction following LPS stimulation (Fig. 2D). The induction of mRNA for IL-1β, another NF-κB-responsive gene, was similarly down-regulated in J7-POP2 cells compared with control (Fig. 2D). TLR-stimulated IL-1β secretion was not measured because although TLR signals lead to intracellular pro-IL-1β accumulation, they do not cause the inflammasome activation necessary for substantial IL-1β secretion. Additionally, POP2 also inhibits TNFα-induced IL-8 mRNA expression in HeLa cells, further showing the ability of POP2 to regulate the expression of an endogenous NF-κB-responsive gene (Fig. 2E). Beyond regulating NF-κB responses, another function of POP2 is the disruption of the interaction between ASC and the Pyrin domain of NLRP1, NLRP3, or NLRP12 (30). Thus, we also determined the impact of POP2 expression upon inflammasome activation in J774A.1 stable transfectants using the FLICA assay, which labels active caspase-1. Following Nlrp3 inflammasome activation with ATP for 30 min, the percentage of caspase-1 positive cells was found to be lower in J7-POP2 (18.6%) compared with J7-pcDNA3 (30.5%) transfectants (Fig. 2F). In these experiments, we avoided priming the cells with TLR ligands (e.g. LPS) prior to Nlrp3 inflammasome activation with ATP, as POP2 may interfere with the NF-κB-dependent expression of Nlrp3 inflammasome machinery and thus may have confounded the results. Thus, the lower level of caspase-1 activation in J7-POP2 cells was most likely a direct effect upon the inflammasome, consistent with its ability to impair inflammasome assembly in 293T cells (31). Collectively, these results illustrate the utility of J774A.1 cells as a model for POP2 gain-of-function studies and confirm that POP2 acts to regulate the production of proinflammatory cytokines.
FIGURE 2.
Stable expression of POP2 in mouse J774A.1 macrophage regulates cytokine response and caspase-1 activation. A, flow cytometry showing intracellular staining against the myc tag in J774A.1 stable transfectants expressing Myc-pcDNA3 vector control (J7-pcDNA3) or wild-type Myc-POP2 (J7-POP2 WT). SA, secondary antibody only. B and C, J7-pcDNA3 and J7-POP2 WT cells were stimulated with indicated amounts of LPS (B) or Pam3-CSK4 (C) for 18 h. TNFα production was measured in culture supernatants using CBA. Cytokine results are representative of at least three experiments (error bars, S.D.; *, p < 0.05; **, p < 0.001). D, quantitative RT-PCR showing TNFα and IL-1β mRNA induction in J7-pcDNA3 and J7-POP2 WT cells in response to LPS stimulation for 1 h (TNFα) or 6 h (IL-1β). E, IL-8 expression in pcDNA3 or POP2-transfected HeLa cells in response to TNFα stimulation (10 ng/ml, 6 h). All of the mRNA results are represented as -fold induction over untreated controls and are shown as mean ± S.D. of at least three independent experiments. F, capase-1 activation was measured in J7-pcDNA3 and J7-POP2 WT cells using caspase-1 fluorescent FLICA dye and analyzed by flow cytometry. Cells were either left untreated or pulsed with 5 mm ATP for 30 min. The result shown is representative of at least three independent experiments with similar results.
The First α-Helix of POP2 (Residues 1–19) Is Necessary and Sufficient to Inhibit NF-κB p65 Activity
POP2 has previously been shown to mediate two major functions: inhibition of NF-κB signaling by acting at the level (or downstream) of p50/p65 (30) and disruption of ASC/NLR interactions leading to inhibition of inflammasome assembly and subsequent IL-1β processing (30, 31). How POP2 performs these two disparate functions remains unknown. To better understand how POP2 operates at the molecular level, we sought to identify the functional region(s) within POP2 responsible for NF-κB inhibition. The secondary structure of POP2 is predicted to contain six α-helices (31). To maintain the integrity of the individual α-helices, we generated POP2 deletion mutants by inserting a stop codon within the interhelical loops (Fig. 3A). Based on structural studies of the NLRP1 and NLRP7 PYDs (38, 39), POP2 may lack the otherwise short helix 3, thus, helices 2 and 3 were deleted together. The ability of the wild-type and POP2 mutants to inhibit NF-κB was determined by a luciferase assay in 293T cells following expression of NF-κB p65. Surprisingly, the first N-terminal α-helix of POP2 (α1; residues 1–19) significantly inhibited NF-κB p65 activity and was comparable with wild-type POP2 (Fig. 3B), demonstrating that POP2 α1 is sufficient to inhibit NF-κB p65. Deletion of the C-terminal helices did not significantly impact the ability of POP2 to suppress NF-κB p65. To determine whether POP2 α1 was also necessary for p65 inhibition, an N-terminal deletion mutant was generated lacking helix-1 (POP2 Δα1). As expected, POP2 Δα1 failed to inhibit NF-κB p65 activity at all dosages of transfected DNA (Fig. 3C), demonstrating that the first α-helix of POP2 is also necessary for NF-κB p65 inhibition.
POP2 is highly homologous to the Pyrin domain of NLRP2 (30). This is particularly interesting, as NLRP2-PYD also inhibits NF-κB signaling by acting upstream, via interaction with IKKα/β, to block TNFα-induced IκBα phosphorylation and p50/p65 release but does not inhibit transfected NF-κB p65 activity (40). Because the first 19 amino acids of NLRP2-PYD differ at only four residues from the POP2 α1 (Fig. 3D), we reasoned that these differences might account for NF-κB p65 inhibition by POP2. To test this possibility, point mutations were introduced to replace these residues in POP2 with those of NLRP2 (A2V, E6Q, L7M, and D8G). All four POP2 to NLRP2-PYD point mutants equally inhibited the activity of transfected NF-κB p65 (Fig. 3D). This result suggests that either these residues are not involved or that individual mutations alone are not sufficient to disrupt the NF-κB p65 inhibitory surface on POP2. We therefore examined the ability of POP2, carrying the combined E6Q, L7M, and D8G substitutions (POP2 QMG), to mediate NF-κB p65 inhibition. Additionally, we used NLRP2 and a truncated version of NLRP2 containing the N-terminal 19 residues (NLRP2-(1–19)), which essentially resembles POP2 α1, to confirm that NLRP2 indeed lacks the capacity to inhibit NF-κB p65. Expression of wild-type or POP2 QMG significantly inhibited NF-κB p65 activity (Fig. 3E). Similar results were also obtained with TNFα-induced NF-κB activation in these cells. Surprisingly, NLRP2 expression also inhibited NF-κB p65 activity, in contrast with a previous report that NLRP2 fails to inhibit the activity of transfected p65 (40). Nevertheless, from these experiments we concluded that POP2 α1 is both necessary and sufficient to inhibit NF-κB signaling and that Ala2, Glu6, Leu7, and Asp8 side chains are not critical alone or in combination for this function.
Acidic Residues in the First α-Helix of POP2 Mediate Inhibition of the ASC and NLRP3 Inflammasome
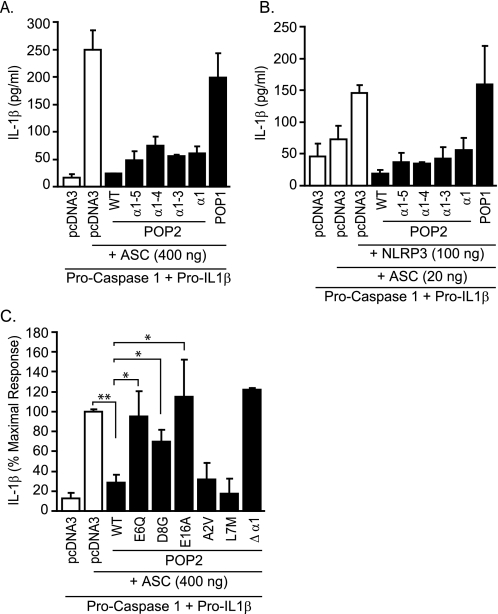
Besides inhibiting NF-κB signaling, POP2 also disrupts ASC/NLR interaction and thus inflammasome-mediated pro-IL-1β processing (30, 31). To determine whether the inflammasome-modifying function of POP2 is mediated by a distinct portion of the protein, we next tested the panel of POP2 truncation mutants for their ability to interfere with pro-IL-1β processing. 293T cells were transfected with plasmids encoding the inflammasome machinery, including pro-caspase-1 (enzyme), pro-IL-1β (substrate), ASC (adaptor), and the NLRP3 (sensor). Because ASC overexpression activates caspase-1 leading to pro-IL-1β processing, most likely via ASC oligomerization or interaction with an endogenous NLR (41), ASC was used in excess (400 ng) without any exogenous NLR. In addition, limiting amounts of ASC (10–20 ng) were transfected with NLRP3 in excess to activate the NLRP3 inflammasome (42). Under both experimental conditions, POP2 α1 expression led to a significant decrease in IL-1β production (Fig. 4, A and B). IL-1β release was similar in cells expressing the α1–3, α1–4, and α1–5 truncations and full-length POP2, suggesting that the presence of helices 2–6 does not enhance the inflammasome-modifying function of POP2 α1. Furthermore, POP2 Δα1 fails to inhibit IL-1β production (Fig. 4C). These results demonstrate that POP2 α1 contains the minimal functional region for inflammasome inhibition. In these experiments, POP1 did not affect either ASC or NLRP3 inflammasome processing of IL-1β (Fig. 4, A and B) even when up to 3 μg of transfected POP1 DNA was used (data not shown). Further, POP1 was functional in these cells, as TNFα-induced NF-κB luciferase activity was inhibited by POP1 (data not shown). Our results are consistent with the report showing that POP1 does not regulate NLPR3 inflammasome activation (43) and suggest that cellular POPs (POP1 and POP2) may exhibit specificity for certain inflammasome complexes.
FIGURE 4.
Acidic residues in POP2 helix-1 mediate inhibition of ASC and the NLRP3 inflammasome in 293T cells. A and B, 293T cells were transfected with plasmids encoding pro-caspase 1 and pro-IL-1β (background control) with high amount (400 ng) of ASC only (A) or low amount of ASC (20 ng) and NLRP3 (B) in the presence or absence of POP1, wild-type POP2, or POP2 deletion mutants. The total amount of transfected DNA per well was kept constant using pcDNA3. The amount of IL-1β produced in culture supernatant was measured using ELISA. Results are representative of at least three experiments (error bars, S.D.). C, 293T cells were transfected as in A in the presence or absence of WT and POP2 mutants, and the amount of IL-1β produced was measured by ELISA. The result is shown as percent maximal IL-1β response by taking the values in (pro-caspase-1 + pro-IL-1β + ASC) transfectant as 100% (error bars, S.E.; n ≥ 3–5; *, p < 0.05; **, p < 0.001).
PYD/PYD interactions are thought to be electrostatic in nature, as suggested by structural studies of the ASC·POP1 and ASC·NLRP3 complexes (43). Thus, we reasoned that charged residues in POP2 α1 could be similarly involved in interaction with ASC resulting in disruption of ASC oligomerization or ASC recruitment to NLRP3. Interestingly, the first α-helix of POP2 contains three acidic residues (Glu6, Asp8, and Glu16), but no basic residues (Fig. 3D). Thus, POP2 E6Q, D8G, and E16A point mutants were tested for their ability to inhibit ASC-mediated inflammasome processing of IL-1β. All of the POP2 point mutants displayed a significant loss of function compared with WT POP2 (Fig. 4C). As controls, POP2 A2V and L7M inhibition of IL-1β generation was comparable with WT POP2. Notably, although POP2 E6Q and D8G mutants are markedly impaired in disrupting inflammasome function, their ability to inhibit NF-κB p65 is comparable with that of WT POP2 (Fig. 3D). The same was observed for POP2 E16A (data not shown). These results demonstrate that the acidic residues in the first α-helix of POP2 are involved in inflammasome inhibition but are not required for NF-κB p65 inhibition. As mutation of these acidic residues does not affect the ability of POP2 to suppress NF-κB p65, our findings demonstrate that the two known functions of POP2, although mediated by the first α-helix, can be uncoupled and are thus likely to be distinct, independent functions.
POP2 Inhibits NF-κB p65 Independently of ASC by Interfering with NF-κB p65 Transactivation
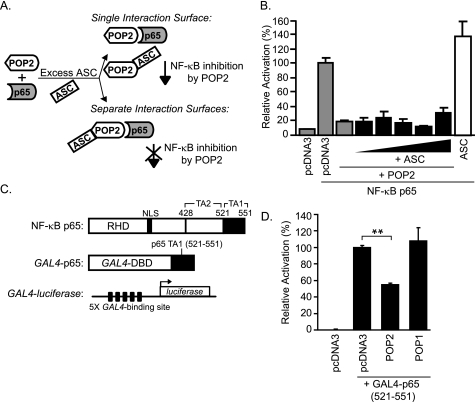
POP2 is known to interact with ASC (30, 31), and our functional mapping studies strongly suggest that the inflammasome inhibitory functions of POP2 are distinct from those affecting NF-κB p65 activity. Further, none of the specific amino acid side chains required for inflammasome inhibition appeared to be important for the impact of POP2 upon NF-κB p65. We therefore reasoned that if the functional surfaces were not truly distinct, the addition of ASC would compete for a single functional surface on POP2 and thus interfere with POP2 inhibition of NF-κB p65 activity (Fig. 5A). To examine this possibility, we tested the ability of POP2 to inhibit NF-κB p65 in 293T cells (which do not express ASC) in the absence or presence of increasing amounts of transfected ASC. POP2 inhibits p65 activity alone and there is no reduction in POP2 function in the presence of concentrations of ASC sufficient to form POP2·ASC specks (30) suggesting that ASC does not interfere with POP2-mediated inhibition of NF-κB p65 (Fig. 5B). These results confirm that ASC does not block or compete with the functional surface of POP2 in mediating NF-κB inhibition and supports our conclusion that POP2 α1 contains two distinct functional surfaces, one that mediates NF-κB inhibition and the other for ASC interaction and inflammasome inhibition.
FIGURE 5.
POP2 inhibits NF-κB p65 independently of ASC by interfering with NF-κB p65 transactivation. A, schematic showing two functional outcomes of ASC overexpression upon POP2-mediated inhibition of NF-κB p65. B, NF-κB luciferase assay in 293T cells, as described in the legend for Fig. 3, in the absence or presence of increasing amounts of transfected ASC (100 ng-2 μg). The result shown is representative of two experiments (error bars, S.D.). C, schematic of full-length NF-κB p65(1–551) with Rel homolgy domain (RHD) and transactivation (TA) domains 1 and 2; GAL4-p65(521–551) fusion construct containing the DNA-binding domain (DBD) of GAL4 and the TA1 of NF-κB p65; and GAL4-luciferase construct containing five tandem repeats of GAL4-binding site upstream of firefly luciferase. D, GAL4-luciferase assay in 293 cells transfected with pcDNA3 control, POP1, or POP2. Results are represented as percentage luciferase activity and shown as mean ± S.D. of one representative experiment of 3–4 independent transfections giving similar results (**, p < 0.001).
Previously, we reported that POP2 blocks the activity of transfected NF-κB p65, demonstrating that POP2 acts at the level of p65, the distal end of the NF-κB signaling pathway (30). This step is downstream of the convergence point of the NF-κB signaling pathway activated in response to TLR or TNF receptor signals. To determine whether POP2 interferes with the transactivation potential of NF-κB p65, we employed a GAL4-based luciferase assay using a GAL4-p65-(521–551) chimeric construct containing the transactivation (TA) domain 1 of NF-κB p65 (Fig. 5C). The TA1 region of p65 is sufficient to activate transcription and is critical for NF-κB p65 transcriptional activity (34, 44). Surprisingly, POP2 expression led to significant inhibition of luciferase activity driven by GAL4-p65 TA1 (Fig. 5D). As a control, POP1 failed to inhibit, consistent with its known action at the level of IKK upstream of NF-κB p65 (33). All of the POP2 deletion mutants including POP2 α1 were similarly effective in suppressing GAL4-p65 TA1-driven luciferase activity (not shown), consistent with the results shown in Fig. 3B. These results suggest that POP2 interferes with the transactivation of NF-κB p65 and provide additional molecular insight into how POP2 acts to regulate NF-κB signaling.
The First α-Helix of POP2 Is Functionally Sufficient to Regulate the TNFα Response to LPS and F. novicida Infection
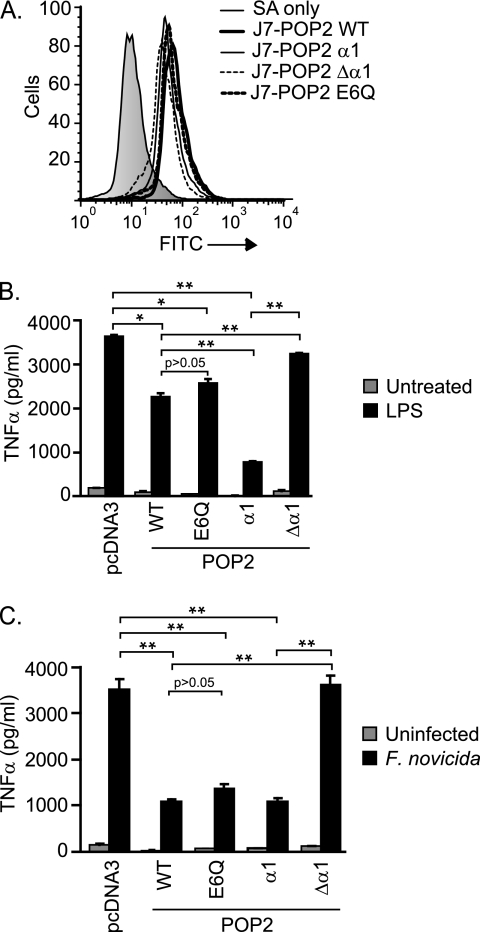
To confirm our molecular findings obtained using 293T cells in a more physiologically relevant setting, we again used J774A.1 cells. The gain-of-function approach provides the advantage of functionally validating POP2 mutants identified in transient transfection studies. Stable transfectants of a functionally sufficient or impaired POP2 mutant(s) were generated. Expression of POP2 α1, Δα1, and E6Q was comparable with wild-type POP2 in J774A.1 stable transfectants (Fig. 6A). Because the first α-helix of POP2 was both necessary and sufficient to inhibit NF-κB p65 activity in 293T cells, we examined the sufficiency of POP2 α1 and the Δα1 mutant in J774A.1 to regulate TNFα production. Because POP2 E6Q retained NF-κB inhibition (Fig. 3D) but was impaired for inflammasome regulation in 293T cells (Fig. 4C), we also considered whether POP2 E6Q would be comparable with WT POP2. Following LPS stimulation, J7-POP2 α1 showed significantly reduced TNFα production compared with J7-pcDNA3 control and more pronounced inhibition than the wild type (Fig. 6B). Secretion of TNFα by J7-POP2 Δα1 cells was similar to J7-pcDNA3 control cells. Moreover, J7-POP2 E6Q and J7-POP2 WT cells were comparable in their ability to impair TNFα production. Similar results were also observed in response to infection with F. novicida, an intracellular bacterium known to activate TLR2 (Fig. 6C). These results confirm that the first α-helix of POP2 contains the functional region responsible for the impact of POP2 upon TLR2 and -4-mediated TNFα production, further establishing the capacity of POP2 to act as a regulator of proinflammatory responses in macrophage.
FIGURE 6.
The first α-helix of POP2 is functionally sufficient to regulate TNFα response to LPS and F. novicida infection. A, flow cytometry analysis showing intracellular staining against the indicated stable transfectants in J774A.1 cells. The plots described in the legend for Fig. 2A and this panel are from the same experiment but are shown separately for clarity. B and C, J774A.1 cells stably expressing pcDNA3, POP2 WT, or the indicated POP2 mutants were treated with LPS for 18 h (B) or infected with F. novicida for 24 h (C) followed by TNFα measurement in culture supernatant. A representative experiment is shown of least three experiments showing similar results (error bars, S.D.). Statistics were performed using two-tailed Student's t test (*, p < 0.05; **, p < 0.001).
POP2 Controls Nlrp3 Inflammasome-mediated IL-1β Response
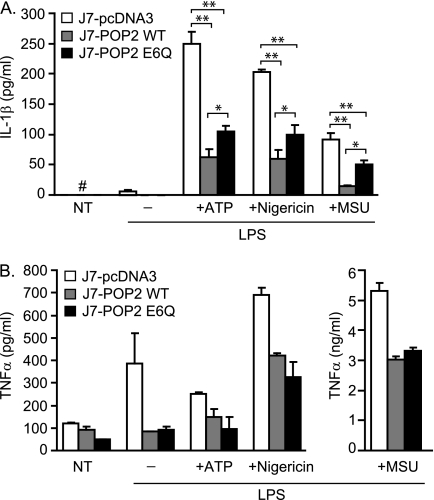
As POP2 exhibits a substantial impact upon inflammasome activation in inflammasome reconstitution experiments, we examined the potential of POP2 to limit IL-1β production in J774A.1 cells stimulated with activators of the Nlrp3 inflammasome. Following Nlrp3 inflammasome activation by ATP, nigericin, or MSU, J7-POP2 WT cells produced significantly less IL-1β compared with the J7-pcDNA3 control (Fig. 7A). As POP2 was observed to limit IL-1β mRNA expression, the impact on IL-1β transcription versus inflammasome activation must be considered. The POP2 E6Q mutant retains its NF-κB inhibitory properties but has a diminished capacity to inhibit the inflammasome. Thus, the E6Q mutant would be predicted to display an increase in IL-1β production only if impairment of inflammasome activation is relieved. As expected, upon stimulation with ATP, nigericin, or MSU, J7-POP2 E6Q cells displayed a modest but significant increase (∼20–30%) in IL-1β release compared with J7-POP2 WT. This finding is consistent with the partial inability of POP2 E6Q to inhibit the NLRP3 inflammasome in 293T cells (Fig. 4C). In the same experiments, J7-POP2 WT and J7-POP2 E6Q cells displayed a similar inhibition of TNFα production (Fig. 7B). These experiments confirm that POP2 regulates the Nlrp3 inflammasome in a manner dependent upon the same acidic residues required in 293T cells. Further, these results strongly implicate POP2 as an endogenous regulator of the IL-1β response.
FIGURE 7.
Mutation of Glu6 compromises ability of POP2 to control Nlrp3 inflammasome. J7-pcDNA3, J7-POP2 WT, and J7-POP2 E6Q cells were primed with LPS for 4 h followed by mock treatment or brief pulse with nigericin (5 μm), ATP (5 mm), and MSU (10 μg/ml) for 30 min. Cells were washed twice and incubated in fresh medium for 6 h. The amounts of IL-1β (A) and TNFα (B) in culture supernatants were measured using CBA. Results are shown for one representative experiment (error bars, S.D.; n ≥ 2–3). NT (#), non-detectable levels; *, p < 0.05; **, p < 0.001.
DISCUSSION
TLRs and NLRs are the critical players mediating inflammation and host immunity (12, 45). Of particular interest are host proteins that regulate NLR-initiated inflammasome activity. However, endogenous host factors controlling this arm of immunity are not widely appreciated and have been poorly studied. Several candidates are proposed that are thought to interfere with PYD/PYD or CARD/CARD interactions, although the details and consequences of their action is less clear. These potential regulators include the small CARD-only proteins such as ICEBERG (27) and Pyrin-only proteins such as POP2 (30, 31), the focus of this study. Here we have addressed the function of POP2, a candidate regulator of Pyrin-containing inflammasomes (e.g. NLRP3), which also limits NF-κB p65 activity, exploring both its mechanism of action and its role in regulating the inflammatory responses.
Our functional mapping studies illustrate that the first α-helix of POP2 is both necessary and sufficient for NF-κB p65 inhibition. As the helical secondary structure of a protein (or a part thereof) is dictated by the properties of the amino acids it comprises, it is very likely that POP2-(1–19) retains the helical structure in isolation. Although we contend that the secondary structure of helix-1 is maintained, this does not mean that the inhibitory events observed are necessarily dependent upon a helical structure. For NF-κB inhibition in particular, it is possible that a short stretch of amino acids in this region is responsible independent of the helix structure. Multiple Pyrin-containing proteins, including NLRP2, -3, -4, -6, and -12, are known to either positively or negatively modulate NF-κB activation, although the mechanistic details are limited (40, 46–50). POP2 is most closely related to the PYD of NLRP2/Pypaf2 (40) with only four amino acid differences within α1. NLRP2/Pypaf2 is reported to block NF-κB activation by blocking IKKα/β activity upstream of p50/p65 release and with no suppressive effect on transfected NF-κB p65 (40). Surprisingly, single and multiple point mutations converting the α1 region of POP2 into that of NLRP2 had no effect upon NF-κB inhibition, and the full-length NLRP2 was also inhibitory in our hands. Although the initial NLRP2/Pypaf2 study used NLRP2-PYD-(1–112), we found that the truncated version of NLRP2 containing only the N-terminal 19 residues was similarly effective at suppressing NF-κB p65 activity, suggesting that NLRP2 is indeed inhibitory (Fig. 3E). Although it is difficult to adequately account for the observed differences, we found that none of the residues that differ between POP2 and NLRP2 account for the impact of POP2 upon NF-κB p65. Moreover, mutation of acidic residues Glu6, Asp8, and Glu16 within α1, important for inflammasome inhibition, also has no apparent impact on NF-κB inhibition by POP2. These studies support the idea that either other residues not yet mutated or a generalized structural feature of POP2 is important. Mechanistically, POP2 appears to actively inhibit NF-κB p65 by interfering with the transactivation potential of p65 TA1; but the precise mechanism and whether POP2 acts directly or indirectly upon p65 is not known. For example, PDLIM2 also suppresses NF-κB but does so by promoting the degradation of promoter-bound p65 in the nucleus (51). Further, it remains unclear whether POP2 is selective for canonical NF-κB signaling. Future studies will be needed to better understand how POP2 inhibits NF-κB.
Most importantly, POP2 modulates the induction of the NF-κB-responsive proinflammatory cytokines TNFα and IL-1β in J774A.1 macrophages. The same trend was also observed following siRNA knockdown in THP-1 cells. Further, inflammatory signals induced POP2 expression with timings reminiscent of inflammatory proteins, as would be expected for regulatory molecules. These findings support the hypothesis that POP2 is an additional regulator of NF-κB signaling in monocytic cells. Several protein, acting either directly or indirectly, modify NF-κB responses in monocyte/macrophages, including Tollip (52), IRAK-M (53, 54), CYLD (55), SOCS-1 (56), Trim30α (57), A20 (58), and more recently TIPE2 (59, 60). Most of these regulators, unlike POP2, act at early steps in the NF-κB signal transduction cascade by targeting either the signaling adaptors or kinases (61, 62). In this regard POP2 appears unique, as it acts distally to IKKα/β activation at the level of p65 (30). Furthermore, POP2 is apparently unique because the POP2 gene is present only in Old World monkeys, apes, and human (i.e. absent in mice), suggesting a relatively recent selective pressure driving its evolution (32). This primate restriction also appears to be true for other solitary POP and COP proteins (32). Exactly why primates need POPs and COPs is not clear, although human autoinflammatory diseases suggest that appropriate physiological regulation of inflammatory mediators is quite essential (23). Beyond reducing TNFα and IL-1β, the ability to regulate NF-κB-dependent processes suggests a number of potential advantages. For example, tightly regulated NF-κB responses in hematopoietic cells could limit aberrant cell proliferation and/or apoptosis and protect against malignant transformation associated with uncontrolled inflammation (63).
As with NF-κB inhibition, POP2 α1 is likewise necessary and sufficient to disrupt inflammasome function (Fig. 4, A and B). Nuclear magnetic resonance studies performed on ASC-PYD (64), NLRP1-PYD (38), and POP1 (65) reveal that these molecules are highly polar and that surface-exposed charged residues mediate the PYD/PYD interaction by forming an electrostatic potential surface patch (EPSP). This interaction is analogous to the CARD/CARD interaction between Apaf-1 and caspase-9 (66, 67). Recently, Srimathi et al. (43) reported an interaction model for ASC-PYD/POP1 in which the negative EPSP formed by helices 1 and 4 on ASC-PYD interact with positive EPSPs on helices 2 and 3 of POP1. Consistent with these structural studies, we found that the acidic residues Glu6, Asp8, and Glu16 within POP2 α1 are clearly important for the regulation of ASC-dependent IL-1β processing. In light of our previous work (30) and that of Dorfleutner et al. (31), POP2 inhibition of the NLRP3 and ASC inflammasomes likely results from competitive inhibition by POP2 for the PYD of NRLP3 and/or ASC. As the ASC-PYD has a bipartite surface (solvent-exposed) charge distribution (negative on α1 and α4 and positive on α2 and α3) (43), we propose that POP2 interacts with the positive EPSP on α2 and α3 of the ASC-PYD. Whether POP2 broadly inhibits NLRP- and other Pyrin-containing inflammasome initiators and whether the pattern of electrostatic interactions reflects a “barcode” determining which inflammasomes are potentially regulated by POP2 remain to be addressed. Furthermore, we find that POP2 and POP1 differ in their ability to target ASC and NLRP3 inflammasome, suggesting some specificity for POP functions. Beyond POPs, host NLR proteins NLRP10/PyNOD (68) and NLRP7/Pypaf3 (69) can interact with ASC and suppress inflammasome, although the biological implications at present are not very clear.
Inflammasome-mediated IL-1β response in monocytes require two signals; signal 1 occurs via cell surface receptors (mostly TLRs) leading to NF-κB and/or MAPK activation driving intracellular pro-IL-1β accumulation, and signal 2 comprises cytosolic events triggering inflammasome assembly and IL-1β processing (70). POP2 appears to regulate both of these steps during Nlrp3 inflammasome activation in J774A.1 (Figs. 2D and 7A). Why are both signals leading to IL-1β production controlled by POP2? By restricting the intracellular pro-IL-1β pool, POP2 can effectively reduce the magnitude and duration of the IL-1β response. Restriction of the inflammasome activation alone will not completely prevent active caspase-1 from enzymatically processing any available pro-IL-1β until the pool is depleted or the enzyme is inactivated. Thus, the duration of IL-1β release will be influenced by the availability of pro-IL-1β. Thus both functions of POP2 may be essential to limit the inflammatory consequences of IL-1β.
On the other hand, it is an open question as to whether POP2-mediated suppression of the NF-κB and inflammasome pathways is beneficial to the host during pathogen infection. Because IL-1β is crucial to eliminate/control the invading microbes, a number of bacteria and viruses have evolved stealth mechanisms, circumventing host immunity by targeting different components of the inflammasome (71). For example, Yersinia enterocolitica YopE and YopT (72), Mycobacterium tuberculosis Zmp1 (73), Pseudomonas aeruginosa ExoU (74), influenza A virus NS1 (75), cowpox virus CrmA (76), and Shope fibroma virus gp013L (77) are all known to inhibit IL-1β and/or caspase-1 activation. Notably, myxoma virus-encoded M13L is a viral POP that, similar to POP2, co-regulates NF-κB and inflammasome by binding to ASC (29, 78). Infection with myxoma virus leads to death in rabbits resulting from insufficient control of viral load; however, the M13L-deficient virus is nonpathogenic (29). Similarly, it is possible that pathogens may induce POP2 and use this host factor to dampen the host immunity, thus favoring infection. But the evolutionary history of the POP2 gene argues against this interpretation (32). This is a difficult question at present and requires future studies to fully understand the role of POP2 in vivo.
It is intriguing that both POP2 functions localize to α1, with no apparent involvement of the remainder of the molecule. However, it is possible that helices 2–6 may play a role in some yet unidentified function of POP2. Nevertheless, our findings suggest the potential for therapeutic peptides based on POP2. A recent study of an 11-amino acid-long peptide derived from the vaccinia virus A46 protein, VIPER (viral inhibitory peptide of TLR4) (79), nicely illustrates this concept. POP2-based peptidomimetics may be expected to be more potent, as they act at the level of p50/p65. Importantly for IL-1β inhibition, POP2-mimetic peptides could exert effects on both NF-κB-dependent transcription and processing of IL-1β. A similar approach could also be used in cancer, where NF-κB activity contributes to tumor cell survival.
Our results demonstrate the importance of the first α-helix of POP2 in the control of NF-κB activation and inflammasome formation in the context of macrophage activation, strongly implicating POP2 as a regulator of inflammation. Deciphering the role of POP2 in human monocytes/macrophages and generating transgenic mouse models to probe the broad implications of POP2 function in vivo are important goals for future studies. In addition, future studies are needed to determine precisely how POP2 directly or indirectly (e.g. sequestration of transcriptional cofactors/accessory proteins) inhibits transcriptional activation by NF-κB p65.
Supplementary Material
Acknowledgments
We acknowledge the contributions of Yili Lin in the CIMD Immunology Core at Albany Medical College for carrying out the CBA analysis and members of the Harton laboratory for suggestions and continued discussion.
This work was supported, in whole or in part, by National Institutes of Health Grant R01AI072259 (to J. A. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
- TLR
- Toll-like receptor
- NLR
- nucleotide-binding leucine-rich receptor
- PYD
- Pyrin domain
- CARD
- caspase activation and recruitment domain
- NLRP
- PYD-containing NLR
- ASC
- apoptotic speck-like protein containing a CARD
- MSU
- monosodium urate
- POP
- Pyrin domain-only protein
- COP
- CARD domain-only protein
- IKK
- IκBα kinase
- PMA
- phorbol 12-myristate 13-acetate
- EPSP
- electrostatic potential surface patch
- CBA
- cytometric bead array.
REFERENCES
- 1. Medzhitov R. (2008) Nature 454, 428–435 [DOI] [PubMed] [Google Scholar]
- 2. Dinarello C. A. (1997) J. Biol. Regul. Homeost. Agents 11, 91–103 [PubMed] [Google Scholar]
- 3. Medzhitov R. (2007) Nature 449, 819–826 [DOI] [PubMed] [Google Scholar]
- 4. Barton G. M., Medzhitov R. (2002) Curr. Top. Microbiol. Immunol. 270, 81–92 [DOI] [PubMed] [Google Scholar]
- 5. Hayden M. S., Ghosh S. (2004) Genes Dev. 18, 2195–2224 [DOI] [PubMed] [Google Scholar]
- 6. Li N., Karin M. (2000) Methods Enzymol. 319, 273–279 [DOI] [PubMed] [Google Scholar]
- 7. Harton J. A., Linhoff M. W., Zhang J., Ting J. P. (2002) J. Immunol. 169, 4088–4093 [DOI] [PubMed] [Google Scholar]
- 8. Ting J. P., Willingham S. B., Bergstralh D. T. (2008) Nat. Rev. Immunol. 8, 372–379 [DOI] [PubMed] [Google Scholar]
- 9. Ting J. P., Lovering R. C., Alnemri E. S., Bertin J., Boss J. M., Davis B. K., Flavell R. A., Girardin S. E., Godzik A., Harton J. A., Hoffman H. M., Hugot J. P., Inohara N., Mackenzie A., Maltais L. J., Nunez G., Ogura Y., Otten L. A., Philpott D., Reed J. C., Reith W., Schreiber S., Steimle V., Ward P. A. (2008) Immunity 28, 285–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tschopp J., Martinon F., Burns K. (2003) Nat. Rev. Mol. Cell Biol. 4, 95–104 [DOI] [PubMed] [Google Scholar]
- 11. Lamkanfi M., Kanneganti T. D., Franchi L., Núñez G. (2007) J. Leukoc. Biol. 82, 220–225 [DOI] [PubMed] [Google Scholar]
- 12. Kanneganti T. D., Lamkanfi M., Núñez G. (2007) Immunity 27, 549–559 [DOI] [PubMed] [Google Scholar]
- 13. Bertin J., DiStefano P. S. (2000) Cell Death Differ. 7, 1273–1274 [DOI] [PubMed] [Google Scholar]
- 14. Weber C. H., Vincenz C. (2001) Trends Biochem. Sci. 26, 475–481 [DOI] [PubMed] [Google Scholar]
- 15. Fairbrother W. J., Gordon N. C., Humke E. W., O'Rourke K. M., Starovasnik M. A., Yin J. P., Dixit V. M. (2001) Protein Sci. 10, 1911–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thornberry N. A., Bull H. G., Calaycay J. R., Chapman K. T., Howard A. D., Kostura M. J., Miller D. K., Molineaux S. M., Weidner J. R., Aunins J., et al. (1992) Nature 356, 768–774 [DOI] [PubMed] [Google Scholar]
- 17. Martinon F., Burns K., Tschopp J. (2002) Mol. Cell 10, 417–426 [DOI] [PubMed] [Google Scholar]
- 18. Sutterwala F. S., Ogura Y., Szczepanik M., Lara-Tejero M., Lichtenberger G. S., Grant E. P., Bertin J., Coyle A. J., Galán J. E., Askenase P. W., Flavell R. A. (2006) Immunity 24, 317–327 [DOI] [PubMed] [Google Scholar]
- 19. Franchi L., Warner N., Viani K., Nuñez G. (2009) Immunol. Rev. 227, 106–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mariathasan S., Weiss D. S., Newton K., McBride J., O'Rourke K., Roose-Girma M., Lee W. P., Weinrauch Y., Monack D. M., Dixit V. M. (2006) Nature 440, 228–232 [DOI] [PubMed] [Google Scholar]
- 21. Martinon F., Pétrilli V., Mayor A., Tardivel A., Tschopp J. (2006) Nature 440, 237–241 [DOI] [PubMed] [Google Scholar]
- 22. Hoffman H. M., Mueller J. L., Broide D. H., Wanderer A. A., Kolodner R. D. (2001) Nat. Genet. 29, 301–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stojanov S., Kastner D. L. (2005) Curr. Opin. Rheumatol. 17, 586–599 [DOI] [PubMed] [Google Scholar]
- 24. Neven B., Callebaut I., Prieur A. M., Feldmann J., Bodemer C., Lepore L., Derfalvi B., Benjaponpitak S., Vesely R., Sauvain M. J., Oertle S., Allen R., Morgan G., Borkhardt A., Hill C., Gardner-Medwin J., Fischer A., de Saint Basile G. (2004) Blood 103, 2809–2815 [DOI] [PubMed] [Google Scholar]
- 25. Druilhe A., Srinivasula S. M., Razmara M., Ahmad M., Alnemri E. S. (2001) Cell Death Differ. 8, 649–657 [DOI] [PubMed] [Google Scholar]
- 26. Lee S. H., Stehlik C., Reed J. C. (2001) J. Biol. Chem. 276, 34495–34500 [DOI] [PubMed] [Google Scholar]
- 27. Humke E. W., Shriver S. K., Starovasnik M. A., Fairbrother W. J., Dixit V. M. (2000) Cell 103, 99–111 [DOI] [PubMed] [Google Scholar]
- 28. Lamkanfi M., Denecker G., Kalai M., D'hondt K., Meeus A., Declercq W., Saelens X., Vandenabeele P. (2004) J. Biol. Chem. 279, 51729–51738 [DOI] [PubMed] [Google Scholar]
- 29. Johnston J. B., Barrett J. W., Nazarian S. H., Goodwin M., Ricciuto D., Wang G., McFadden G. (2005) Immunity 23, 587–598 [DOI] [PubMed] [Google Scholar]
- 30. Bedoya F., Sandler L. L., Harton J. A. (2007) J. Immunol. 178, 3837–3845 [DOI] [PubMed] [Google Scholar]
- 31. Dorfleutner A., Bryan N. B., Talbott S. J., Funya K. N., Rellick S. L., Reed J. C., Shi X., Rojanasakul Y., Flynn D. C., Stehlik C. (2007) Infect. Immun. 75, 1484–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Atianand M. K., Fuchs T., Harton J. A. (2011) BMC Evol. Biol. 11, 56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stehlik C., Krajewska M., Welsh K., Krajewski S., Godzik A., Reed J. C. (2003) Biochem. J. 373, 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schmitz M. L., Stelzer G., Altmann H., Meisterernst M., Baeuerle P. A. (1995) J. Biol. Chem. 270, 7219–7226 [DOI] [PubMed] [Google Scholar]
- 35. Bewry N. N., Bolick S. C., Wright K. L., Harton J. A. (2007) J. Biol. Chem. 282, 26178–26184 [DOI] [PubMed] [Google Scholar]
- 36. Malik M., Bakshi C. S., Sahay B., Shah A., Lotz S. A., Sellati T. J. (2006) Infect. Immun. 74, 3657–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schwende H., Fitzke E., Ambs P., Dieter P. (1996) J. Leukoc. Biol. 59, 555–561 [PubMed] [Google Scholar]
- 38. Hiller S., Kohl A., Fiorito F., Herrmann T., Wider G., Tschopp J., Grütter M. G., Wüthrich K. (2003) Structure 11, 1199–1205 [DOI] [PubMed] [Google Scholar]
- 39. Pinheiro A. S., Proell M., Eibl C., Page R., Schwarzenbacher R., Peti W. (2010) J. Biol. Chem. 285, 27402–27410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bruey J. M., Bruey-Sedano N., Newman R., Chandler S., Stehlik C., Reed J. C. (2004) J. Biol. Chem. 279, 51897–51907 [DOI] [PubMed] [Google Scholar]
- 41. Srinivasula S. M., Poyet J. L., Razmara M., Datta P., Zhang Z., Alnemri E. S. (2002) J. Biol. Chem. 277, 21119–21122 [DOI] [PubMed] [Google Scholar]
- 42. Agostini L., Martinon F., Burns K., McDermott M. F., Hawkins P. N., Tschopp J. (2004) Immunity 20, 319–325 [DOI] [PubMed] [Google Scholar]
- 43. Srimathi T., Robbins S. L., Dubas R. L., Chang H., Cheng H., Roder H., Park Y. C. (2008) J. Biol. Chem. 283, 15390–15398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schmitz M. L., Baeuerle P. A. (1991) EMBO J. 10, 3805–3817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Creagh E. M., O'Neill L. A. (2006) Trends Immunol. 27, 352–357 [DOI] [PubMed] [Google Scholar]
- 46. O'Connor W., Jr., Harton J. A., Zhu X., Linhoff M. W., Ting J. P. (2003) J. Immunol. 171, 6329–6333 [DOI] [PubMed] [Google Scholar]
- 47. Fiorentino L., Stehlik C., Oliveira V., Ariza M. E., Godzik A., Reed J. C. (2002) J. Biol. Chem. 277, 35333–35340 [DOI] [PubMed] [Google Scholar]
- 48. Grenier J. M., Wang L., Manji G. A., Huang W. J., Al-Garawi A., Kelly R., Carlson A., Merriam S., Lora J. M., Briskin M., DiStefano P. S., Bertin J. (2002) FEBS Lett. 530, 73–78 [DOI] [PubMed] [Google Scholar]
- 49. Wang L., Manji G. A., Grenier J. M., Al-Garawi A., Merriam S., Lora J. M., Geddes B. J., Briskin M., DiStefano P. S., Bertin J. (2002) J. Biol. Chem. 277, 29874–29880 [DOI] [PubMed] [Google Scholar]
- 50. Williams K. L., Lich J. D., Duncan J. A., Reed W., Rallabhandi P., Moore C., Kurtz S., Coffield V. M., Accavitti-Loper M. A., Su L., Vogel S. N., Braunstein M., Ting J. P. (2005) J. Biol. Chem. 280, 39914–39924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tanaka T., Grusby M. J., Kaisho T. (2007) Nat. Immunol. 8, 584–591 [DOI] [PubMed] [Google Scholar]
- 52. Burns K., Clatworthy J., Martin L., Martinon F., Plumpton C., Maschera B., Lewis A., Ray K., Tschopp J., Volpe F. (2000) Nat. Cell Biol. 2, 346–351 [DOI] [PubMed] [Google Scholar]
- 53. Kobayashi K., Hernandez L. D., Galán J. E., Janeway C. A., Jr., Medzhitov R., Flavell R. A. (2002) Cell 110, 191–202 [DOI] [PubMed] [Google Scholar]
- 54. Su J., Zhang T., Tyson J., Li L. (2009) J. Innate Immun. 1, 164–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yoshida H., Jono H., Kai H., Li J. D. (2005) J. Biol. Chem. 280, 41111–41121 [DOI] [PubMed] [Google Scholar]
- 56. Mansell A., Smith R., Doyle S. L., Gray P., Fenner J. E., Crack P. J., Nicholson S. E., Hilton D. J., O'Neill L. A., Hertzog P. J. (2006) Nat. Immunol. 7, 148–155 [DOI] [PubMed] [Google Scholar]
- 57. Shi M., Deng W., Bi E., Mao K., Ji Y., Lin G., Wu X., Tao Z., Li Z., Cai X., Sun S., Xiang C., Sun B. (2008) Nat. Immunol. 9, 369–377 [DOI] [PubMed] [Google Scholar]
- 58. Boone D. L., Turer E. E., Lee E. G., Ahmad R. C., Wheeler M. T., Tsui C., Hurley P., Chien M., Chai S., Hitotsumatsu O., McNally E., Pickart C., Ma A. (2004) Nat. Immunol. 5, 1052–1060 [DOI] [PubMed] [Google Scholar]
- 59. Freundt E. C., Bidere N., Lenardo M. J. (2008) Cell 133, 401–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sun H., Gong S., Carmody R. J., Hilliard A., Li L., Sun J., Kong L., Xu L., Hilliard B., Hu S., Shen H., Yang X., Chen Y. H. (2008) Cell 133, 415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang J., Hu Y., Deng W. W., Sun B. (2009) Microbes Infect. 11, 321–327 [DOI] [PubMed] [Google Scholar]
- 62. Liew F. Y., Xu D., Brint E. K., O'Neill L. A. (2005) Nat. Rev. Immunol. 5, 446–458 [DOI] [PubMed] [Google Scholar]
- 63. Dolcet X., Llobet D., Pallares J., Matias-Guiu X. (2005) Virchows Arch. 446, 475–482 [DOI] [PubMed] [Google Scholar]
- 64. Liepinsh E., Barbals R., Dahl E., Sharipo A., Staub E., Otting G. (2003) J. Mol. Biol. 332, 1155–1163 [DOI] [PubMed] [Google Scholar]
- 65. Natarajan A., Ghose R., Hill J. M. (2006) J. Biol. Chem. 281, 31863–31875 [DOI] [PubMed] [Google Scholar]
- 66. Qin H., Srinivasula S. M., Wu G., Fernandes-Alnemri T., Alnemri E. S., Shi Y. (1999) Nature 399, 549–557 [DOI] [PubMed] [Google Scholar]
- 67. Liu T., Rojas A., Ye Y., Godzik A. (2003) Protein Sci. 12, 1872–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang Y., Hasegawa M., Imamura R., Kinoshita T., Kondo C., Konaka K., Suda T. (2004) Int. Immunol. 16, 777–786 [DOI] [PubMed] [Google Scholar]
- 69. Kinoshita T., Wang Y., Hasegawa M., Imamura R., Suda T. (2005) J. Biol. Chem. 280, 21720–21725 [DOI] [PubMed] [Google Scholar]
- 70. Hornung V., Latz E. Eur. J. Immunol. (2010) 40, 620–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Taxman D. J., Huang M. T., Ting J. P. (2010) Cell Host Microbe 8, 7–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Schotte P., Denecker G., Van Den Broeke A., Vandenabeele P., Cornelis G. R., Beyaert R. (2004) J. Biol. Chem. 279, 25134–25142 [DOI] [PubMed] [Google Scholar]
- 73. Master S. S., Rampini S. K., Davis A. S., Keller C., Ehlers S., Springer B., Timmins G. S., Sander P., Deretic V. (2008) Cell Host Microbe 3, 224–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sutterwala F. S., Mijares L. A., Li L., Ogura Y., Kazmierczak B. I., Flavell R. A. (2007) J. Exp. Med. 204, 3235–3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Stasakova J., Ferko B., Kittel C., Sereinig S., Romanova J., Katinger H., Egorov A. (2005) J. Gen. Virol. 86, 185–195 [DOI] [PubMed] [Google Scholar]
- 76. Ray C. A., Black R. A., Kronheim S. R., Greenstreet T. A., Sleath P. R., Salvesen G. S., Pickup D. J. (1992) Cell 69, 597–604 [DOI] [PubMed] [Google Scholar]
- 77. Dorfleutner A., Talbott S. J., Bryan N. B., Funya K. N., Rellick S. L., Reed J. C., Shi X., Rojanasakul Y., Flynn D. C., Stehlik C. (2007) Virus Genes 35, 685–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Rahman M. M., Mohamed M. R., Kim M., Smallwood S., McFadden G. (2009) PLoS Pathog. 5, e1000635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lysakova-Devine T., Keogh B., Harrington B., Nagpal K., Halle A., Golenbock D. T., Monie T., Bowie A. G. (2010) J. Immunol. 185, 4261–4271 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.